GC-MS Based Metabolite Profiling, and Anti-Inflammatory Activity of Aqueous Extract of Myrica esculenta through In Vitro and In Silico Approach †
Abstract
:1. Introduction
2. Material and Methods
2.1. Plant Collection and Authentication
2.2. Plant Extraction
2.3. Total Phenolic Content (TPC)
2.4. Total Flavonoid Content (TFC)
2.5. Gas Chromatography and Mass Spectroscopy
2.6. In Vitro Anti-Inflammatory Activity
2.6.1. Lipoxygenase (LOX) Inhibition Assay
2.6.2. 15-LOX Inhibitory Assay
2.6.3. Hylurenadease (HYA) Inhibition Assay
2.7. In Silico Computational Study
2.7.1. Preparation of Target Protein/Macromolecules and Ligands
2.7.2. Quantitative Structure-Activity Relationship (QSAR) Analysis
2.7.3. Molecular Docking Analysis
2.7.3.1. AutoDock 4.2.6
2.7.3.2. AutoDock Vina
2.7.3.3. iGMDOCK
2.7.4. Molecular Dynamic Simulation
2.7.4.1. LARMD Online Server
2.7.4.2. MD Simulation Methodology Using Schrodinger Software
2.7.4.3. Statistical Analysis
3. Results
3.1. Extractive Value of Bark and Root Extract of Myrica esculenta Plant
3.2. Determination of Total Phenolic and Total Flavonoid Content
3.3. Identification of Phytoconstituents by GC-MS Analysis
3.4. In Vitro Anti-Inflammatory Activity
3.5. In Silico Computational Analysis
3.5.1. Quantitative Structure–Activity Relationship (QSAR) Studies
3.5.2. Molecular Docking
3.5.3. Molecular Dynamic (MD) Simulation
3.5.4. Schrodinger Molecular Dynamic Simulation
3.5.4.1. Stability Analysis of Complex by RMSD
3.5.4.2. Stability Analysis by RMSF
3.5.4.3. Protein-Ligand Contact Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Min, Z.; Navam, S.H.; Ronny, H.; Arvind, K.; Apputhury, P.M.; Arumugam, M.; Rasekhara, R.M. Phytochemicals, antioxidant and antimicrobial activity of Hibiscus sabdariffa, Centella asiatica, Moringa oleifera and Murraya koenigii leaves. J. Med. Plants Res. 2011, 5, 6672–6680. [Google Scholar]
- Utami, W.; Aziz, H.; Fitriani, I.; Zikri, A.; Mayasri, A.; Nasrudin, D. In silico anti-inflammatory activity evaluation of some bioactive compound from ficus religiosa through molecular docking approach. J. Phys. Conf. Ser. 2020, 1563, 012024. [Google Scholar] [CrossRef]
- Ul Hassan, S.S.; Zhang, W.-D.; Jin, H.-Z.; Basha, S.H.; Priya, S.S. In silico anti-inflammatory potential of guaiane dimers from Xylopia vielana targeting COX-2. J. Biomol. Struct. Dyn. 2022, 40, 484–498. [Google Scholar] [CrossRef] [PubMed]
- Dinarello, C.A. Anti-inflammatory agents: Present and future. Cell 2010, 140, 935–950. [Google Scholar] [CrossRef] [PubMed]
- Oboh, G. Effect of blanching on the antioxidant properties of some tropical green leafy vegetables. LWT Food Sci. Technol. 2005, 38, 513–517. [Google Scholar] [CrossRef]
- Refaey, M.S.; Abouelela, M.E.; El-Shoura, E.A.M.; Alkhalidi, H.M.; Fadil, S.A.; Elhady, S.S.; Abdelhameed, R.F.A. In Vitro Anti-Inflammatory Activity of Cotula anthemoides Essential Oil and In silico Molecular Docking of Its Bioactives. Molecules 2022, 27, 1994. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, M.; Dwivedi, U.N.; Kakkar, P. Suppression of oxidative stress and pro-inflammatory mediators by Cymbopogon citratus D. Stapf extract in lipopolysaccharide stimulated murine alveolar macrophages. Food Chem. Toxicol. 2010, 48, 2913–2919. [Google Scholar] [CrossRef]
- K Rana, R.; K Patel, R. Pharmacological evaluation of antiasthmatic activity of Myrica nagi bark extracts. Anti-Inflamm. Anti-Allergy Agents Med. Chem. 2016, 15, 145–152. [Google Scholar] [CrossRef]
- Kabra, A.; Martins, N.; Sharma, R.; Kabra, R.; Baghel, U.S. Myrica esculenta Buch.-Ham. ex D. Don: A Natural Source for Health Promotion and Disease Prevention. Plants 2019, 8, 149. [Google Scholar] [CrossRef]
- Kabra, A.; Sharma, R.; Hano, C.; Kabra, R.; Martins, N.; Baghel, U.S. Phytochemical Composition, Antioxidant, and Antimicrobial Attributes of Different Solvent Extracts from Myrica esculenta Buch.-Ham. ex. D. Don Leaves. Biomolecules 2019, 9, 357. [Google Scholar] [CrossRef]
- Sood, P.; Shri, R. A review on ethnomedicinal, phytochemical and pharmacological aspects of Myrica esculenta. Indian J. Pharm. Sci. 2018, 80, 2–13. [Google Scholar] [CrossRef]
- Fang, Z.; Zhang, Y.; Lü, Y.; Ma, G.; Chen, J.; Liu, D.; Ye, X. Phenolic compounds and antioxidant capacities of bayberry juices. Food Chem. 2009, 113, 884–888. [Google Scholar] [CrossRef]
- Srivastava, B.; Sharma, V.C.; Pant, P.; Pandey, N.; Jadhav, A. Evaluation for substitution of stem bark with small branches of Myrica esculenta for medicinal use–A comparative phytochemical study. J. Ayurveda Integr. Med. 2016, 7, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Mahato, R.; Chaudhary, R. Ethnomedicinal study and antibacterial activities of selected plants of Palpa district, Nepal. Sci. World 2005, 3, 26–31. [Google Scholar]
- Rauf, A.; Abu-Izneid, T.; Rashid, U.; Alhumaydhi, F.A.; Bawazeer, S.; Khalil, A.A.; Aljohani, A.S.M.; Abdallah, E.M.; Al-Tawaha, A.R.; Mabkhot, Y.N.; et al. Anti-inflammatory, Antibacterial, Toxicological Profile, and In silico Studies of Dimeric Naphthoquinones from Diospyros lotus. Biomed Res. Int. 2020, 2020, 7942549. [Google Scholar] [CrossRef]
- Singh, J.; Lan, V.K.; Trivedi, V.P. Pharmacognostic Evaluation of Katphala (The bark of Myrica esculenta Buch—Ham). Anc. Sci. Life 1986, 6, 85–87. [Google Scholar]
- Middha, S.K.; Usha, T.; Babu, D.; Misra, A.K.; Lokesh, P.; Goyal, A.K. Evaluation of antioxidative, analgesic and anti-inflammatory activities of methanolic extract of Myrica nagi leaves-an animal model approach. Symbiosis 2016, 70, 179–184. [Google Scholar] [CrossRef]
- Yadav, D.K.; Mudgal, V.; Agrawal, J.; Maurya, A.K.; Bawankule, D.U.; Chanotiya, C.S.; Khan, F.; Thul, S.T. Molecular docking and ADME studies of natural compounds of Agarwood oil for topical anti-inflammatory activity. Curr. Comput. Aided Drug. Des. 2013, 9, 360–370. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, B.; Peng, Y.; Xiong, X.; Jing, W.; Wang, J.; Gao, H. In silico exploration of the molecular mechanism of cassane diterpenoids on anti-inflammatory and immunomodulatory activity. J. Chem. Inf. Model. 2019, 59, 2309–2323. [Google Scholar] [CrossRef]
- Iqbal, D.; Khan, M.S.; Waiz, M.; Rehman, M.T.; Alaidarous, M.; Jamal, A.; Alothaim, A.S.; AlAjmi, M.F.; Alshehri, B.M.; Banawas, S. Exploring the Binding Pattern of Geraniol with Acetylcholinesterase through In silico Docking, Molecular Dynamics Simulation, and In Vitro Enzyme Inhibition Kinetics Studies. Cells 2021, 10, 3533. [Google Scholar] [CrossRef]
- Kabra, A.; Sharma, R.; Singla, S.; Kabra, R.; Baghel, U.S. Pharmacognostic characterization of Myrica esculenta leaves. J. Ayurveda Integr. Med. 2019, 10, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Chludil, H.D.; Corbino, G.B.; Leicach, S.R. Soil quality effects on Chenopodium album flavonoid content and antioxidant potential. J. Agric. Food Chem. 2008, 56, 5050–5056. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, A.K.; Thapa, S.; Shrestha, L.; Mehta, R.K.; Gupta, A.; Koirala, N. Phytochemical screening and the effect of Trichosanthes dioica in high-fat diet induced atherosclerosis in Wistar rats. Food Front. 2021, 2, 527–536. [Google Scholar] [CrossRef]
- Olivia, N.U.; Goodness, U.C.; Obinna, O.M. Phytochemical profiling and GC-MS analysis of aqueous methanol fraction of Hibiscus asper leaves. Future J. Pharm. Sci. 2021, 7, 59. [Google Scholar] [CrossRef]
- Truong, D.H.; Ta, N.T.A.; Pham, T.V.; Huynh, T.D.; Do, Q.T.G.; Dinh, N.C.G.; Dang, C.D.; Nguyen, T.K.C.; Bui, A.V. Effects of solvent—Solvent fractionation on the total terpenoid content and in vitro anti-inflammatory activity of Serevenia buxifolia bark extract. Food Sci. Nutr. 2021, 9, 1720–1735. [Google Scholar] [CrossRef]
- Paun, G.; Neagu, E.; Albu, C.; Savin, S.; Radu, G.L. In vitro evaluation of antidiabetic and anti-inflammatory activities of polyphenolic-rich extracts from anchusa officinalis and melilotus officinalis. ACS Omega 2020, 5, 13014–13022. [Google Scholar] [CrossRef]
- Ahmad, R. Steroidal glycoalkaloids from Solanum nigrum target cytoskeletal proteins: An In silico analysis. Peerj 2019, 7, 6012. [Google Scholar] [CrossRef]
- Contreras-Puentes, N.; Alviz-Amador, A. Virtual screening of natural metabolites and antiviral drugs with potential inhibitory activity against 3CL-PRO and PL-PRO. Biomed. Pharmacol. J. 2020, 13, 933–941. [Google Scholar] [CrossRef]
- Khan, T.; Azad, I.; Ahmad, R.; Raza, S.; Dixit, S.; Joshi, S.; Khan, A.R. Synthesis, characterization, computational studies and biological activity evaluation of Cu, Fe, Co and Zn complexes with 2-butanone thiosemicarbazone and 1, 10-phenanthroline ligands as anticancer and antibacterial agents. Excli J. 2018, 17, 331. [Google Scholar]
- Okoli, B.J.; Eltayb, W.A.; Gyebi, G.A.; Ghanam, A.R.; Ladan, Z.; Oguegbulu, J.C.; Abdalla, M. In silico Study and Excito-Repellent Activity of Vitex negundo L. Essential Oil against Anopheles gambiae. Appl. Sci. 2022, 12, 7500. [Google Scholar] [CrossRef]
- Mohapatra, R.K.; Dhama, K.; El–Arabey, A.A.; Sarangi, A.K.; Tiwari, R.; Emran, T.B.; Azam, M.; Al-Resayes, S.I.; Raval, M.K.; Seidel, V. Repurposing benzimidazole and benzothiazole derivatives as potential inhibitors of SARS-CoV-2: DFT, QSAR, molecular docking, molecular dynamics simulation, and In silico pharmacokinetic and toxicity studies. J. King Saud. Univ. Sci. 2021, 33, 101637. [Google Scholar] [CrossRef]
- Alameen, A.A.; Abdalla, M.; Alshibl, H.M.; AlOthman, M.R.; Alkhulaifi, M.M.; Mirgany, T.O.; Elsayim, R. In silico studies of glutathione peroxidase4 activators as candidate for multiple sclerosis management. J. Saudi Chem. Soc. 2022, 26, 101554. [Google Scholar] [CrossRef]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.M.; Chen, C.C. GEMDOCK: A generic evolutionary method for molecular docking. Proteins 2004, 55, 288–304. [Google Scholar] [CrossRef] [PubMed]
- Dolinsky, T.J.; Czodrowski, P.; Li, H.; Nielsen, J.E.; Jensen, J.H.; Klebe, G.; Baker, N.A. PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007, 35, W522–W525. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-F.; Wang, F.; Chen, Y.-Z.; Hao, G.-F.; Yang, G.-F. LARMD: Integration of bioinformatic resources to profile ligand-driven protein dynamics with a case on the activation of estrogen receptor. Brief. Bioinform. 2020, 21, 2206–2218. [Google Scholar] [CrossRef]
- Kausar, M.A.; Anwar, S.; Eltayb, W.A.; Kuddus, M.; Khatoon, F.; El-Arabey, A.A.; Khalifa, A.M.; Rizvi, M.R.; Najm, M.Z.; Thakur, L.; et al. MD Simulation Studies for Selective Phytochemicals as Potential Inhibitors against Major Biological Targets of Diabetic Nephropathy. Molecules 2022, 27, 4980. [Google Scholar] [CrossRef]
- Baby, K.; Maity, S.; Mehta, C.H.; Suresh, A.; Nayak, U.Y.; Nayak, Y. Targeting SARS-CoV-2 RNA-dependent RNA polymerase: An In silico drug repurposing for COVID-19. F1000Research 2020, 9, 1166. [Google Scholar] [CrossRef]
- Trivedi, A.; Ahmad, R.; Siddiqui, S.; Misra, A.; Khan, M.A.; Srivastava, A.; Ahamad, T.; Khan, M.F.; Siddiqi, Z.; Afrin, G. Prophylactic and therapeutic potential of selected immunomodulatory agents from Ayurveda against coronaviruses amidst the current formidable scenario: An In silico analysis. J. Biomol. Struct. Dyn. 2022, 40, 9648–9700. [Google Scholar] [CrossRef]
- Patel, K.; Rao, N.; Gajera, V.; Bhatt, P.; Patel, K.; Gandhi, T. Anti-allergic activity of stem bark of Myrica esculenta Buch.-Ham (Myricaceae). J. Young Pharm. 2010, 2, 74–78. [Google Scholar] [CrossRef]
- Patel, T.; Dudhpejiya, A.; Sheath, N. Anti inflammatory activity of Myrica nagi Linn. Bark. Anc. Sci. Life 2011, 30, 100. [Google Scholar] [PubMed]
- Patel, M. Analgesic and anti-inflammatory activity of ethanolic extract of stem bark of Myrica nagi (T.). World J. Pharm. Res. 2017, 6, 844–857. [Google Scholar] [CrossRef]
- Patel, T.; Rajshekar, C.; Parmar, R. Mast cell stabilizing activity of bark of Myrica nagi. J. Pharm. Stud. Res. 2011, 2, 1–6. [Google Scholar]
- Shresta, S.; Bhattarai, B.R.; Adhikari, B.; Rayamajhee, B.; Poudel, P.; Khanal, S.; Marasini, B.P.; Aryal, B.; Bhandari, S.; Parajuli, N. Evaluation of phytochemical, antioxidant and antibacterial activities of selected medicinal plants. Nepal J. Biotechnol. 2021, 9, 50–62. [Google Scholar] [CrossRef]
- Anjum, N.; Tripathi, Y. evaluation of total polyphenols, flavonoids and antioxidant activity of Myrica esculenta buch-ham. ex d. don fruits. World J. Pharm. Med. Res. 2021, 7, 186–192. [Google Scholar]
- Rawat, S.; Jugran, A.; Giri, L.; Bhatt, I.D.; Rawal, R.S. Assessment of antioxidant properties in fruits of Myrica esculenta: A popular wild edible species in Indian Himalayan region. Evid. Based Complement. Altern. Med. 2011, 2011, 512787. [Google Scholar] [CrossRef] [PubMed]
- Langhansova, L.; Landa, P.; Kutil, Z.; Tauchen, J.; Marsik, P.; Rezek, J.; Lou, J.D.; Yun, Z.L.; Vanek, T. Myrica rubra leaves as a potential source of a dual 5-LOX/COX inhibitor. Food Agric. Immunol. 2017, 28, 343–353. [Google Scholar] [CrossRef]
- Kumar, H.P.; Panda, P.; Karunakar, P.; Shiksha, K.; Singh, L.; Ramesh, N.; Usha, T.; Middha, S.K. Potential Cyclooxygenase (COX-2) enzyme inhibitors from Myrica nagi-from In silico to in vitro investigation. Pharmacogn. Mag. 2019, 15, 280. [Google Scholar]







| Conc. (mg/mL) | Mean Absorbance of TPC at 765 nm and TFC at 510 nm | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bark Extract | Root Extract | |||||||||||
| Abs. of Extract | Conc. of Gallic Acid (mg/mL) | TPC (mg GAE/g) | Abs. of Extract | Conc. of Quercetin (mg/mL) | TFC (mg of Quercetin/g) | Abs. of Extract | Conc. of Gallic Acid (mg/mL) | TPC (mg GAE/g) | Abs. of Extract | Conc. of Quercetin (mg/mL) | TFC (mg of Quercetin/g) | |
| 0.1 | 0.332 | 28.63 ± 1.14 | 286.34 ± 11.40 | 0.238 | 15.84 ± 0.09 | 158.40 ± 0.98 | 0.245 | 21.06 ± 0.05 | 210.69 ± 0.50 | 0.168 | 11.11 ± 0.12 | 111.10 ± 1.25 |
| 0.2 | 0.459 | 39.70 ± 2.06 | 397.07 ± 20.61 | 0.353 | 23.94 ± 0.26 | 239.48 ± 2.65 | 0.317 | 27.38 ± 1.10 | 273.88 ± 11.06 | 0.289 | 19.28 ± 0.30 | 191.28 ± 3.03 |
| 0.4 | 0.560 | 51.67 ± 2.26 | 484.89 ± 22.63 | 0.452 | 29.10 ± 0.05 | 291.05 ± 5.85 | 0.462 | 39.96 ± 0.77 | 399.68 ± 7.78 | 0.379 | 25.34 ± 0.28 | 253.44 ± 2.81 |
| 0.8 | 0.688 | 56.54 ± 2.01 | 596.20 ± 20.15 | 0.571 | 38.31 ± 0.64 | 387.455 ± 6.41 | 0.533 | 46.14 ± 0.28 | 461.42 ± 2.85 | 0.477 | 32.01 ± 0.21 | 320.11 ± 2.14 |
| 1.0 | 0.841 | 71.15 ± 1.47 | 729.24 ± 14.74 | 0.682 | 45.97 ± 2.90 | 459.75 ± 29.07 | 0.627 | 54.31 ± 0.27 | 543.15 ± 2.76 | 0.529 | 35.52 ± 0.45 | 355.06 ± 4.52 |
| 2.0 | 0.953 | 82.69 ± 2.07 | 826.92 ± 20.76 | 0.714 | 47.97 ± 0.33 | 480.02 ± 3.31 | 0.736 | 63.82 ± 0.71 | 638.23 ± 7.10 | 0.645 | 43.31 ± 0.07 | 433.17 ± 0.78 |
| Mean ± SEM | 553.44 ± 18.38 | 336.02 ± 8.04 | 421.17 ± 5.34 | 277.65 ± 2.42 | ||||||||
| S.N. | RT (min) | Peak Width 50% (min) | Compound Name | Molecular Formula | Molecular Weight | Structure |
|---|---|---|---|---|---|---|
| 1 | 4.094 | 0.485 | Methyl salicylate | C8H8O3 | 152.047 |  |
| 2 | 4.86 | 0.485 | O-Amino benzohydroxamic acid | C7H8N2O2 | 152.059 |  |
| 3 | 6.672 | 0.324 | Pyridine, 4-(1,1-dimethylethyl)- | C11H17NS | 135.105 |  |
| 4 | 7.147 | 0.432 | Benzoic acid, 4-amino-, hydrazide | C7H9N3O | 151.075 |  |
| 5 | 7.6 | 0.486 | 1,2,4-Triazolo(4,3-a) pyrimidine | C5H4N4 | 120.044 |  |
| 6 | 12.12 | 0.432 | 2-Chlorobenzimidazole | C5H5ClN2 | 152.014 |  |
| 7 | 13.501 | 0.486 | 5-Chlorobenzimidazole | C7H5ClN2 | 152.014 |  |
| 8 | 14.343 | 0.593 | Benzenemethanamine, N-methyl- | C8H11N | 121.089 |  |
| 9 | 15.184 | 0.593 | 2-Isocyanatopyridine | C6H4N2O | 120.032 |  |
| 10 | 17.072 | 0.486 | N,N,N′,N′-Tetraethyl-1,2-di-furan-2-yl-ethane-1,2-diamine | C4H12N2 | 304.215 |  |
| 11 | 19.953 | 0.863 | Eicosane | C20H42 | 282.329 |  |
| 12 | 21.916 | 0.378 | Octadecane | C18H38 | 254.297 |  |
| 13 | 22.251 | 0.378 | Tetratetracontane | C44H90 | 618.704 |  |
| 14 | 23.179 | 0.378 | Hexatriacontane | C36H74 | 506.579 |  |
| 15 | 24.171 | 0.378 | Docosane, 7-hexyl- | C28H58 | 394.454 |  |
| 16 | 28.541 | 0.378 | Tetrapentacontane | C54H110 | 758.861 |  |
| 17 | 30.558 | 0.432 | Triacontane, 1-bromo- | C30H60Br | 500.396 |  |
| 18 | 31.141 | 0.378 | Hentriacontane | C31H64 | 436.501 |  |
| 29 | 38.056 | 0.539 | Tetracosane | C24H50 | 338.391 |  |
| 20 | 41.314 | 0.539 | Octadecane, 3-ethyl-5-(2-ethylbutyl)- | C26H54 | 366.423 |  |
| 21 | 46.558 | 1.025 | Dotriacontane | C32H66 | 450.516 |  |
| 5-LOX (IC50) | 15-LOX (IC50) | HYA (IC50) | |
|---|---|---|---|
| Bark extract | 11.26 ± 3.93 | 25.57 ± 8.94 | 21.61 ± 8.27 |
| Root extract | 23.024 ± 8.04 | 16.95 ± 5.92 | 40.24 ± 15.41 |
| Indomethacin | 9.87 ± 3.78 | 12.19 ± 4.67 | 7.82 ± 2.99 |
| Celecoxib | 14.07 ± 5.38 | 8.62 ± 3.30 | 17.96 ± 6.87 |
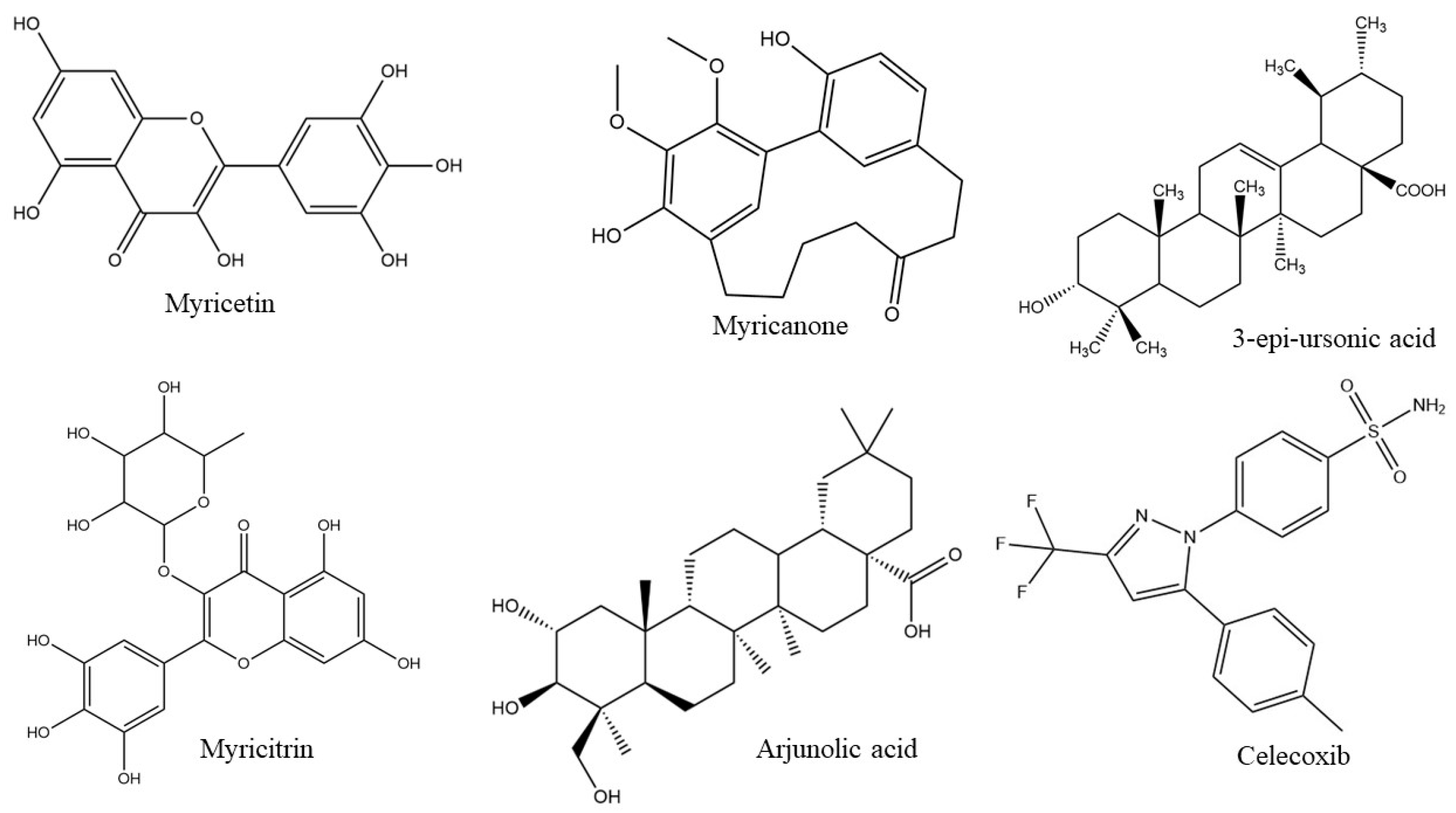
| Function | 3-Epi-Ursolic Acid | Arjunolic Acid | Celecoxib | Myrecitin | Myricanone | Myricitrin |
|---|---|---|---|---|---|---|
| Surface area (Approx) (Å2) | 539.38 | 592.70 | 529.12 | 395.28 | 450.85 | 523.82 |
| Surface area (Grid) (Å2) | 649.10 | 680.61 | 595.51 | 485.46 | 550.54 | 644.51 |
| Volume (Å3) | 1239.43 | 1323.95 | 987.13 | 802.37 | 993.22 | 1138.09 |
| Hydration energy (Kcal/mole) | −4.67 | −11.58 | −10.90 | −40.64 | −12.50 | −45.17 |
| Log P | 9.37 | 7.84 | 7.86 | 4.05 | 5.51 | 3.27 |
| Refractivity (Å3) | 122.50 | 132.91 | 37.41 | 20.81 | 52.62 | 51.96 |
| Polarizability (Å3) | 53.12 | 54.59 | 32.80 | 29.18 | 38.47 | 41.96 |
| Mass (amu) | 456.71 | 490.72 | 381.37 | 318.24 | 356.42 | 464.38 |
| Total energy (kcal/mol) | 74.3512 | 109.971 | 45.3604 | 7.90082 | 22.7156 | 22.8694 |
| Dipole moment (Debye) | 1.766 | 0 | 2.792 | 3.297 | 1.536 | 0 |
| RMS gradient (kcal/Å mol) | 0.09703 | 0.09558 | 0.09688 | 0.0953 | 0.09526 | 0.09747 |
| S.N. | Ligands | COX-1 (PDB ID: 4O1Z) | COX-2 (PDB ID: 4M11) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| B.E. (kcal/mol) | Diss. Constant (Kd) | Interacting Amino Acid | H-Atom | B.E. (kcal/mol) | Diss. Constant (Kd) | Interacting Amino Acid | H-Atom | ||
| 1. | Myricetin | −9.95 | 50.71 nM | Ser143, Arg374, Asn375, Gly533, Gly533, Asn537, Asn537, Asn53, Val228, Val228, Val228, Gly227 | 6 | −6.97 | 7.79 μM | Phe361,Phe36,Lys360, Trp545,Arg61 | 4 |
| 2. | Myricanone | −7.65 | 2.46 μM | Ser143, Trp139, Arg376, Arg37 | 1 | −6.51 | 16.78 μM | Trp545,Asp362, Asn560 | 2 |
| 3. | 3-epi-ursonic acid | −6.88 | 9.08 μM | Arg376, Asn375, Gly225 | 2 | −6.72 | 11.89 μM | Asp239,His242, Lys253 | 3 |
| 4. | Myricitrin | −7.64 | 2.51 μM | Asp229, Trp139, Ser143,Arg376, Phe142, Arg374, Val145, Asn375 | 6 | −4.37 | 624.03 μM | Glu346,Arg109,Lys342, Glu553,Trp545,Asp362, Lys360 | - |
| 5. | Arjunolic acid | −9.25 | 165.34 nM | Arg374, Asn375, Asn537,Val228, His226, Val145, Phe142 | 1 | −6.92 | 8.5 μM | Arg61, Asn560 | - |
| 6. | Celecoxib | −7.9 | 1.61 μM | Asn375, Asn37, Asn375,Trp139, Ser143, Arg374, Gly225 | 1 | −5.72 | 64.32 μM | Lys342,Lys360,Lys557,Glu553, Glu553 | 1 |
| S.N. | Ligands | Tumor Necrosis Factor (TNF)- α (PDB ID: 2AZ5) | Interleukin (IL)-10 (PDB ID: 2H24) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| B.E. (kcal/mol) | Diss. Constant (Kd) | Interacting Amino Acid | H-Atom | B.E. (kcal/mol) | Diss. Constant (Kd) | Interacting Amino Acid | H-Atom | ||
| 1. | Myricetin | −7.3 | 4.42 μM | Lys11,Lys11 | 2 | −5.78 | 57.5 μM | Arg110,Phe111, Phe56 | 1 |
| 2. | Myricanone | −7.78 | 2.0 μM | Gly121 | 2 | −7.09 | 6.32 μM | Phe56, Phe111 | 2 |
| 3. | 3-epi-ursonic acid | −6.97 | 7.84 μM | Leu120,Ser60 | - | −4.51 | 491.31 μM | Arg102,Arg102, Arg106,Gln70 | 1 |
| 4. | Myricitrin | −5.55 | 85.03 μM | Ser60,Gln61, Leu120 | - | −5.09 | 184.36 μM | Glu74,Arg102, Glu115,Gln63 | - |
| 5. | Arjunolic acid | −7.34 | 4.2 μM | Leu120, Leu57 | - | −6.76 | 11.11 μM | Gly61, Gly58, Cys62 | - |
| 6. | Celecoxib | −6.52 | 16.55 μM | Tyr59,Tyr59,Tyr151, Tyr151,Tyr151, Gln61 | - | −5.44 | 102.12 μM | Glu115,Asn116, Arg102,Arg102, Gln70, Glu74 | 1 |
| S.N. | Ligands | COX-1 (PDB ID: 4O1Z) | COX-2 (PDB ID: 4M11) | ||||
|---|---|---|---|---|---|---|---|
| B.E. (kcal/mol) | Diss. Constant (Kd) | Interacting Amino Acid | B.E. (kcal/mol) | Diss. Constant (Kd) | Interacting Amino Acid | ||
| 1. | Myricetin | −9.7 | 79.61 nM | Cys47,His43,Gln44,Gln461 | −9.9 | 53.8 nM | Cys41, Arg44, Gln461 |
| 2. | Myricanone | −8.4 | 290.78 nM | Gln372,Glu543 | −8.9 | 314.99 nM | Gln543,Arg44 |
| 3. | 3-epi-ursonic acid | −9.3 | 150.85 nM | Asp135,Gln327, Arg157 | −9.2 | 181.8 nM | Gln372,Gln370, Gln543, Arg44 |
| 4. | Myricitrin | −10.1 | 38.59 nM | Gln327, Asn34, Asp135 | −10.5 | 13.51 nM | Glu322, Val132 |
| 5. | Arjunolic acid | −9.0 | 229.41 nM | Asn34, Arg157 | −9.5 | 109.5 nM | Gly225, Tyr373 |
| 6. | Celecoxib | −9.5 | 102.91 nM | Lys532,Gln372,Pro542,Arg61, Lys546 | −9.3 | 150.85 nM | Tyr130, Cys41 |
| S.N. | Ligands | Tumor Necrosis Factor (TNF)- α (PDB ID: 2AZ5) | Interleukin (IL)-10 (PDB ID: 2H24) | ||||
|---|---|---|---|---|---|---|---|
| B.E. (kcal/mol) | Diss. Constant (Kd) | Interacting Amino Acid | B.E. (kcal/mol) | Diss. Constant (Kd) | Interacting Amino Acid | ||
| 1. | Myricetin | −7.8 | 1.91 µM | Ser95,Arg82 | −6.3 | 24.29 µM | Arg110, Cys62 |
| 2. | Myricanone | −8.2 | 945.3 nM | Tyr151, Tyr59 | −8.0 | 1.62 µM | Phe56, Phe111 |
| 3. | 3-epi-ursonic acid | −9.2 | 181.81 nM | Leu157 | −7.4 | 3.95 µM | Arg106, Arg102 |
| 4. | Myricitrin | −8.0 | 9.95 µM | Gly121,Tyr151 | −6.8 | 11.55 µM | Ala139 |
| 5. | Arjunolic acid | −9.4 | 120.71 nM | Tyr151,Gly121, Leu120 | −8.3 | 848.93 nM | Gln63, Glu67 |
| 6. | Celecoxib | −8.2 | 945.3 nM | Gln125, Gly121 | −8.1 | 1.1 µM | Phe30 |
| S.N. | Ligands | COX-1 (PDB ID: 4O1Z) | COX-2 (PDB ID: 4M11) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| T.E. (kcal/mol) | vDW | HB | E.I. | T.E. (kcal/mol) | vDW | HB | E.I. | ||
| 1. | Myricetin | −132.492 | −105.267 | −27.2253 | 0 | −125.784 | −91.263 | −34.5206 | 0 |
| 2. | Myricanone | −87.3133 | −75.4082 | −11.9051 | 0 | −107.541 | −97.6307 | −9.90984 | 0 |
| 3. | 3-epi-ursonic acid | −103.976 | −87.1912 | −16.4963 | −0.288703 | −86.4932 | −69.4132 | −17.08 | 0 |
| 4. | Myricitrin | −125.517 | −93.0059 | −32.5107 | 0 | −114.496 | 79.4543 | −35.0418 | 0 |
| 5. | Arjunolic acid | −83.2344 | −67.5726 | −15.6618 | 0 | −87.3774 | −71.8736 | −15.5038 | 0 |
| 6. | Celecoxib | −108.282 | −100.838 | −7.44436 | 0 | −90.8942 | −83.167 | −7.72725 | 0 |
| S.N. | Ligands | Tumor Necrosis Factor (TNF)- α (PDB ID: 2AZ5) | Interleukin (IL)-10 (PDB ID: 2H24) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| T.E. (kcal/mol) | vDW | HB | E.I. | T.E. (kcal/mol) | vDW | HB | E.I. | ||
| 1. | Myricetin | −91.4497 | −65.7845 | −25.6652 | 0 | −75.9232 | −60.0728 | −15.8504 | 0 |
| 2. | Myricanone | −87.7068 | −73.3761 | −14.3307 | 0 | −77.8226 | −68.6794 | −9.14318 | 0 |
| 3. | 3-epi-ursonic acid | −85.9916 | −78.5542 | −7.43742 | 0 | −82.3913 | −76.6764 | −5.71494 | 0 |
| 4. | Myricitrin | −108.992 | −79.9469 | −29.0448 | 0 | −87.1763 | −73.3057 | −13.8706 | 0 |
| 5. | Arjunolic acid | −95.3887 | −81.6511 | −13.425 | −0.312637 | −86.1421 | −78.8447 | −5.99124 | −1.3061 |
| 6. | Celecoxib | −86.9344 | −80.4249 | −6.50946 | 0 | −77.6205 | −76.2528 | −1.36773 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shrivastava, A.K.; Chaudhary, D.; Shrestha, L.; Awadalla, M.E.; Al-Shouli, S.T.; Palikhey, A.; Eltayb, W.A.; Gupta, A.; Gupta, P.P.; Parab, M.; et al. GC-MS Based Metabolite Profiling, and Anti-Inflammatory Activity of Aqueous Extract of Myrica esculenta through In Vitro and In Silico Approach. Med. Sci. Forum 2023, 21, 52. https://doi.org/10.3390/ECB2023-14079
Shrivastava AK, Chaudhary D, Shrestha L, Awadalla ME, Al-Shouli ST, Palikhey A, Eltayb WA, Gupta A, Gupta PP, Parab M, et al. GC-MS Based Metabolite Profiling, and Anti-Inflammatory Activity of Aqueous Extract of Myrica esculenta through In Vitro and In Silico Approach. Medical Sciences Forum. 2023; 21(1):52. https://doi.org/10.3390/ECB2023-14079
Chicago/Turabian StyleShrivastava, Amit Kumar, Dipendra Chaudhary, Laxmi Shrestha, Maaweya E. Awadalla, Samia T. Al-Shouli, Anjan Palikhey, Wafa Ali Eltayb, Anamika Gupta, Pramodkumar P. Gupta, Mala Parab, and et al. 2023. "GC-MS Based Metabolite Profiling, and Anti-Inflammatory Activity of Aqueous Extract of Myrica esculenta through In Vitro and In Silico Approach" Medical Sciences Forum 21, no. 1: 52. https://doi.org/10.3390/ECB2023-14079
APA StyleShrivastava, A. K., Chaudhary, D., Shrestha, L., Awadalla, M. E., Al-Shouli, S. T., Palikhey, A., Eltayb, W. A., Gupta, A., Gupta, P. P., Parab, M., Trivedi, A., Srivastava, A., & Abdalla, M. (2023). GC-MS Based Metabolite Profiling, and Anti-Inflammatory Activity of Aqueous Extract of Myrica esculenta through In Vitro and In Silico Approach. Medical Sciences Forum, 21(1), 52. https://doi.org/10.3390/ECB2023-14079










