Abstract
Spiramycin is a 16-membered macrolide used in human medicine as an antibacterial and antiparasitic agent. Spiramycin is effective against bacterial pathogens including Gram-positive and Gram-negative bacteria. In contrast, Pseudomonas aeruginosa is intrinsically resistant to macrolides including azithromycin and spiramycin. Despite the results of in vitro susceptibility tests, interest in macrolides in the treatment of some pseudomonal infections arose from both clinical and preclinical studies. For this reason, macrolides have drawn attention as adjunct therapy against chronic and/or biofilm-mediated P. aeruginosa infections. While most of the studies on the antivirulence activity of macrolides focus on erythromycin and its derivative azithromycin, there is no information on spiramycin. We used in vitro methods to test the ability of spiramycin to inhibit certain phenotypic features or traits associated with virulence in P. aeruginosa. Moreover, we tested spiramycin using Galleria mellonella as an in vivo model.
1. Introduction
Macrolides are a class of antibiotics produced by bacteria, such as those of the genus Streptomyces. These natural compounds consist of a large macrocyclic lactone ring bound to deoxy sugars. Spiramycin is a 16-membered macrolide used in human medicine as an antibacterial and antiparasitic agent [1,2,3]. The antibacterial activity of this antibiotic is enacted via its ability to bind the 50S ribosomal subunit and block the path through which nascent peptides exit the ribosome [4]. Spiramycin is effective against Gram-positive bacterial pathogens and is also effective against bacteria belonging to the genera Neisseria, Legionella, Mycoplasma, Chlamydia, and against Toxoplasma spp. [5]. In contrast, Pseudomonas aeruginosa is considered intrinsically resistant to macrolides including spiramycin [6].
Macrolides were used in the treatment of infections caused by P. aeruginosa in both clinical trials and a mouse model of bacteremia [7,8]. Erythromycin and its derivative azithromycin can inhibit the activity of many virulence factors of P. aeruginosa including exotoxin A, protease, elastase, lipase, phospholipase C, lecithinase, gelatinase, DNase, and pyocyanin [8,9,10,11,12,13,14,15,16]. These findings led to the hypothesis that macrolides may act by downmodulating either the inflammatory response or P. aeruginosa virulence [9].
We used several in vitro methods to test the effect of spiramycin on P. aeruginosa. Phenotypic traits under study included pigment synthesis, rhamnolipid synthesis, bacterial motility, and biofilm formation. We also used an in vivo infection model, Galleria mellonella larvae, to test the effect of spiramycin on P. aeruginosa in vivo.
2. Materials and Methods
2.1. Strain and Culture Conditions
We isolated Pseudomonas aeruginosa GG-7 in a previous study [17]. The medium used for the growth of the bacterium was LB (composition per liter: NaCl 10 g, Tryptone 10 g, Yeast Extract 5 g). This medium was used both as a liquid and solid by adding 15 g per liter of agar. We cultured the bacterium at 37 °C with shaking if necessary (180 rpm).
2.2. Pyocyanin, Pioverdine, Rhamnolipids and Motility Measurement
Pyocyanin production was determined as described previously [18,19] with minor modification. Moreover, the fluorescence intensity was measured to estimate the amount of pyoverdine (Ex: 405 nm, Em: 450 nm) [20]. We used the colorimetric analysis of the orcinol reaction to measure the rhamnolipids content [19]. Finally, we performed the motility assay as previously described [19].
2.3. Biofilm Formation and Biomass Estimation
We used hydroxyapatite discs to form the biofilm. We initially grew P. aeruginosa in LB liquid overnight (37 °C, 180 rpm). The next morning, we diluted the bacterial suspension 1 in 100. We filled a 24-multiwell plate (1 mL per well) with the new medium. In each well, we added a hydroxyapatite disk. We incubated the multiwell plate at 37 °C and 120 rpm for 72 h. After this time, we sampled the hydroxyapatite discs, washed the discs in sterile physiological water three times, and used two biofilm measurement methods. The first measurement we conducted was biomass estimation by counting the CFU/mL with the dilution method. The second measurement we performed was the biomass estimation using crystal violet, as previously reported [21].
2.4. In Vivo Expreriment
We used Galleria mellonella larvae as an in vivo infection model [22]. We used the procedures employed previously to carry out the infection of the larvae [23]. We prepared three different samples: (1) control; (2) infected larvae; (3) infected larvae, to which we injected a suspension of both P. aeruginosa and spiramycin (60 μg/mL). After 24 h (at 37 °C) we counted the dead and diseased larvae.
2.5. In Silico Expreriment
We performed docking simulation using SwissDock (http://www.swissdock.ch/, accessed on 8 September 2022). The simulations were performed using spiramycin A as the ligand. We used three protein structures as receptors. These proteins were regulators involved in Quorum Sensing: LasR (4ng2), PqsR (4jvd), and RhlR (AF-P54292-F1).
3. Results
3.1. Influence of Spiramycin on Pigment Production
Initially, we performed MIC (minimal inhibitory concentration) experiments to test the resistance of P. aeruginosa to four antibiotics (ampicillin, streptomycin, rifampicin and spiramycin). We inoculated the bacterium in 1 mL of LB and then administered 10 different concentrations of antibiotics: 500 μg/mL; 250 μg/mL; 125 μg/mL; 62.5 μg/mL; 31.25 μg/mL; 15.62 μg/mL; 7. 8 μg/mL; 3.9 μg/mL; 1.95 μg/mL and 0.98 μg/mL. After 24 h, we detected the optical density (O.D.) at 600 nm to estimate the biomass. We noted that spiramycin inhibits the production of pyocyanin, so we measured the absorbance at 520 nm to quantify these green pigments, see Figure 1. Spiramycin has a small effect on biomass (Figure 1A). There is a marked reduction in pyocyanin (Figure 1B) produced by P. aeruginosa, and 30 μg/mL of spiramycin already leads to a marked decrease in the pigments produced.
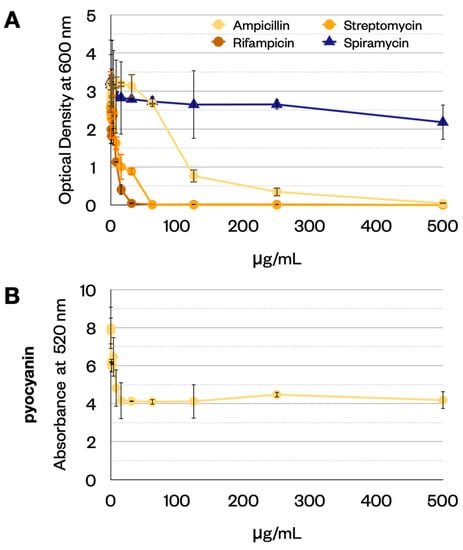
Figure 1.
(A) Measure of biomass (OD at 600 nm) in the MIC experiment. (B) Measure of pyocyanin (absorbance at 520 nm). Error bars show the standard deviation (three replicates).
3.2. Influence of Spiramycin on Biofilm Formation
Some studies have reported that macrolides inhibited the ability of P. aeruginosa to form biofilms. We performed biofilm growth experiments to confirm that spiramycin had the same effect. The administration of spiramycin (60 μg/mL) causes a marked reduction in the synthesis of pyocyanin (Table 1) and pyoverdine (Figure 2). Furthermore, the administration of spiramycin (60 μg/mL) inhibits biofilm formation (Table 1) and increases the number of planktonic cells (Table 1).

Table 1.
Measurements performed during biofilm growth experiments.
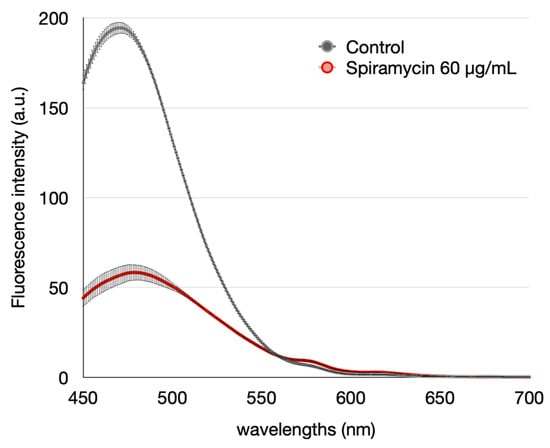
Figure 2.
Effect of spiramycin on pyoverdine produced by P. aeruginosa in the experiment involving the biofilm formation (Fluorescence intensity, Excitation 405 nm). Error bars show the standard deviation (three replicates).
3.3. Effect of Spiramycin on Motility and Rhamnolipids Production by P. aeruginosa
Some authors had previously shown that azithromycin reduced the motility of P. aeruginosa grown on a solid medium [19]. We performed the same experiment using spiramycin to confirm this effect. In the presence of spiramycin (60 μg/mL), the motility of the bacterium is considerably reduced (Figure 3A,B).
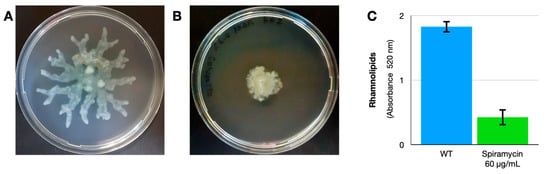
Figure 3.
Effect of spiramycin on motility and rhamnolipids production of P. aeruginosa. (A,B) P. aeruginosa on the medium described by Gödeke et al., 2013 [19] without (A) or with (B) spiramycin (60 μg/mL). Error bars show the standard deviation (three replicates). (C) Spiramycin (60 μg/mL) also reduces the production of rhamnolipids in liquid broth.
Rhamnolipids influence swarming motility in P. aeruginosa [24], so we measured the rhamnolipids in P. aeruginosa cultures. The results confirm that spiramycin (60 μg/mL) also reduces the production of rhamnolipids (Figure 3C).
3.4. Results of Injection of P. aeruginosa in Insect Larvae: Effect of Spiramycin
We observed G. mellonella larvae 24 h after infection with P. aeruginosa. After 24 h of incubation, we counted the dead or healthy larvae in the three groups: the control group, the group infected with P. aeruginosa, and the group infected with P. aeruginosa but treated with spiramycin. The results show that spiramycin reduces mortality by about 60%.
3.5. The In Silico Results Suggest That PqsR Is a New Probable Target for Spiramycin
We hypothesized that possible targets of spiramycin could be the proteins involved in Quorum Sensing. Therefore, we performed some docking simulations to estimate the binding energy between spiramycin and three probable receptors PqsR, RhlR and LasR. SwissDock provides the results as a “cluster”, a set of positions that the ligand can cover in a specific position on the protein. We analyzed the clusters one by one by selecting only those whose location was compatible with the presence of the binding site. We used all the binding energies calculated for each molecular configuration in the cluster to calculate the mean and standard deviation. The results are reported in Figure 4. As can be seen, the lowest bond energy (and therefore the best bond) is between spiramycin and PqsR.
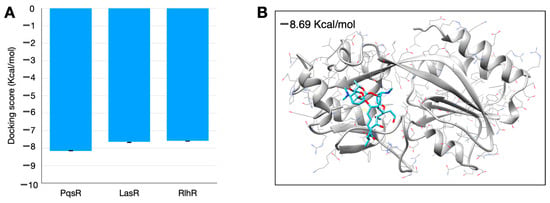
Figure 4.
Results of in silico simulation performed with SwissDock. (A) Binding energy estimation. (B) Structure of spiramycin (blue) docked with PqsR (gray) showing the docking structure with lower binding energy. Error bars show the standard deviation of the results of the better clusters (lower energy score).
4. Discussion and Conclusions
Our results show that spiramycin inhibits the production of pyocyanin, pyoverdine and rhamnolipids in P. aeruginosa. Moreover, the treatment of the bacterium with this antibiotic inhibits biofilm formation in an artificial biomimetic system and blocks the motility on the agar surface. Finally, to test the effect of spiramycin against P. aeruginosa in an in vivo system, we used the insect Galleria mellonella and the preliminary data obtained by this colonization model show a marked reduction in mortality. We explored the probable mechanisms of action of spiramycin against P. aeruginosa using computational modeling and docking simulation. Based on the docking data, PqsR could be a target of spiramycin. However, the effects of this molecule on ribosome activity could affect the functionality of the translation causing the observed phenomena. In conclusion, further data are needed to elucidate the functional mechanisms of spiramycin and other macrolides on P. aeruginosa.
Author Contributions
Conceptualization, P.A.; methodology, M.C.; investigation, M.C.; writing—original draft preparation, M.C.; writing—review and editing, P.A.; project administration, P.A.; funding acquisition, P.A. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partially supported by the Italian Ministry for Education, Universities and Research [grant no. PRIN 2017SFBFER to Pietro Alifano].
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pinnert-Sindico, S.; Ninet, L.; Preud’Homme, J.; Cosar, C. A new antibiotic-spiramycin. In Antibiotics Annual 1954–1955; Welch, H., Marti-Ibanez, F., Eds.; Medical Encyclopedia: New York, NY, USA, 1955; pp. 827–830. [Google Scholar]
- Poulet, P.P.; Duffaut, D.; Barthet, P.; Brumpt, I. Concentrations and in vivo antibacterial activity of spiramycin and metronidazole in patients with periodontitis treated with high-dose metronidazole and the spiramycin/metronidazole combination. J. Antimicrob. Chemother. 2005, 55, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Chew, W.K.; Segarra, I.; Ambu, S.; Mak, J.W. Significant reduction of brain cysts caused by Toxoplasma gondii after treatment with spiramycin coadministered with metronidazole in a mouse model of chronic toxoplasmosis. Antimicrob. Agents Chemother. 2012, 56, 1762–1768. [Google Scholar] [CrossRef] [PubMed]
- Tenson, T.; Lovmar, M.; Ehrenberg, M. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J. Mol. Biol. 2003, 330, 1005–1014. [Google Scholar] [CrossRef]
- Chabbert, Y.A. Early studies on in-vitro and experimental activity of spiramycin: A review. J. Antimicrob. Chemother. 1988, 22 (Suppl. B), 1–11. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, J.D. Macrolide resistance in Pseudomonas aeruginosa: Implications for practice. Eur. Respir. J. 2017, 49, 1700689. [Google Scholar] [CrossRef]
- Kobayashi, H. Biofilm disease: Its clinical manifestation and therapeutic possibilities of macrolides. Am. J. Med. 1995, 99, 26S–30S. [Google Scholar] [CrossRef]
- Hirakata, Y.; Kaku, M.; Tomono, K.; Tateda, K.; Furuya, N.; Matsumoto, T.; Araki, R.; Yamaguchi, K. Efficacy of erythromycin lactobionate for treating Pseudomonas aeruginosa bacteremia in mice. Antimicrob. Agents Chemother. 1992, 36, 1198–1203. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Howe, R.A.; Spencer, R.C. Macrolides for the treatment of Pseudomonas aeruginosa infections? J. Antimicrob. Chemother. 1997, 40, 153–155. [Google Scholar] [CrossRef][Green Version]
- Kita, E.; Sawaki, M.; Oku, D.; Hamuro, A.; Mikasa, K.; Konishi, M.; Emoto, M.; Takeuchi, S.; Narita, N.; Kashiba, S. Suppression of virulence factors of Pseudomonas aeruginosa by erythromycin. J. Antimicrob. Chemother. 1991, 27, 273–284. [Google Scholar] [CrossRef]
- Mizukane, R.; Hirakata, Y.; Kaku, M.; Ishii, Y.; Furuya, N.; Ishida, K.; Koga, H.; Kohno, S.; Yamaguchi, K. Comparative in vitro exoenzyme-suppressing activities of azithromycin and other macrolide antibiotics against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1994, 38, 528–533. [Google Scholar] [CrossRef] [PubMed]
- Molinari, G.; Guzman, C.A.; Pesce, A.; Schito, G.C. Inhibition of Pseudomonas aeruginosa virulence factors by sub- inhibitory concentrations of azithromycin and other macrolide antibiotics. J. Antimicrob. Chemother. 1993, 31, 681–688. [Google Scholar] [CrossRef] [PubMed]
- Schneirson, S.S.; Amsterdam, D.; Perlman, E. Inhibition of Pseudomonas aeruginosa pigment formation by chloramphenicol and erythromycin. Antibiot. Chemother. 1960, 10, 30–33. [Google Scholar]
- Tanaka, E.; Kanthakumar, K.; Cundell, D.R.; Tsang, K.W.; Taylor, G.W.; Kuze, F.; Cole, P.J.; Wilson, R. The effect of erythromycin on Pseudomonas aeruginosa and neutrophil mediated epithelial damage. J. Antimicrob. Chemother. 1994, 33, 765–775. [Google Scholar] [CrossRef]
- Imperi, F.; Leoni, L.; Visca, P. Antivirulence activity of azithromycin in Pseudomonas aeruginosa. Front. Microbiol. 2014, 5, 178. [Google Scholar] [CrossRef]
- Leroy, A.G.; Caillon, J.; Caroff, N.; Broquet, A.; Corvec, S.; Asehnoune, K.; Roquilly, A.; Crémet, L. Could azithromycin be part of Pseudomonas aeruginosa acute pneumonia treatment? Front. Microbiol. 2021, 12, 594. [Google Scholar] [CrossRef]
- Tredici, S.M.; Buccolieri, A.; Tanini, L.; Calcagnile, M.; Manno, D.; Alifano, P. Calcite-forming Bacillus licheniformis thriving on underwater speleothems of a hydrothermal cave. Geomicrobiol. J. 2018, 35, 804–817. [Google Scholar] [CrossRef]
- Essar, D.W.; Eberly, L.E.E.; Hadero, A.; Crawford, I.P. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: Interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 1990, 172, 884–900. [Google Scholar] [CrossRef] [PubMed]
- Gödeke, J.; Pustelny, C.; Häussler, S. Recycling of peptidyl-tRNAs by peptidyl-tRNA hydrolase counteracts azithromycin-mediated effects on Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2013, 57, 1617–1624. [Google Scholar] [CrossRef] [PubMed]
- Dao, K.H.T.; Hamer, K.E.; Clark, C.L.; Harshman, L.G. Pyoverdine production by Pseudomonas aeruginosa exposed to metals or an oxidative stress agent. Ecol. Appl. 1999, 9, 441–448. [Google Scholar] [CrossRef]
- Cheng, X.; Liu, J.; Li, J.; Zhou, X.; Wang, L.; Liu, J.; Xu, X. Comparative effect of a stannous fluoride toothpaste and a sodium fluoride toothpaste on a multispecies biofilm. Arch. Oral Biol. 2017, 74, 5–11. [Google Scholar] [CrossRef]
- Kavanagh, K.; Fallon, J.P. Galleria mellonella larvae as models for studying fungal virulence. Fungal Biol. Rev. 2010, 24, 79–83. [Google Scholar] [CrossRef]
- Jander, G.; Rahme, L.G.; Ausubel, F.M. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J. Bacteriol. 2000, 182, 3843–3845. [Google Scholar] [CrossRef] [PubMed]
- Caiazza, N.C.; Shanks, R.M.Q.; O’Toole, G.A. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J. Bacteriol. 2005, 187, 7351–7361. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).