Working Primers and qPCR Protocols for Rapid eDNA Identification of Four Aquatic Invasive Species Found in the Lower Great Lakes with High Potential for Ballast Transport to Lake Superior
Abstract
1. Introduction
2. Materials and Methods
2.1. Organisms, Loci and Sample Collection
2.2. Primer Development
2.3. Water Filtration Protocol
2.4. DNA Extraction Protocol
2.5. qPCR Detection and Primer Testing Protocol (eDNA and gDNA)
2.6. In Silico and In Vitro Testing
3. Results
3.1. Limit of Detection, Cq Value Data and Microcosm Signal Duration
3.2. In Silico and In Vitro Results
4. Discussion
4.1. Discussion of LOD Data Relative to eDNA Testing in Similar Experiments
4.2. Conservation Relevance of the Target Research
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| NAGL | North American Great Lakes |
| AIS | Aquatic Invasive Species |
References
- Rup, M.P.; Bailey, S.A.; Wiley, C.J.; Minton, M.S.; Miller, A.W.; Ruiz, G.M.; MacIsaac, H.J. Domestic ballast operations on the Great Lakes: Potential importance of Lakers as a vector for introduction and spread of nonindigenous species. Can. J. Fish. Aquat. Sci. 2010, 67, 256–268. [Google Scholar] [CrossRef]
- Mays, N.; Knight, I.; Gruwell, M.; Beesley, K.M.; Balcer, M.; Gebhard, S.; Fanberg, L.; Anders, O.; Cangelosi, A.; TenEyck, M.; et al. Great Waters Research Collaborative: Great Lakes Ship Ballast Monitoring Project; Lake Superior Research Institute: Superior, WI, USA, 2018; Available online: http://digital.library.wisc.edu/1793/78497 (accessed on 15 January 2023).
- Sheehan, R.; Knight, I.T.; Cangelosi, A.; Melendez, A.; Phillips, H.; Welsbacher, A.; Gruwell, M.E. Quantitative assessment of planktonic AIS eDNA signal duration in Great Lakes harbor water microcosms. Manag. Biol. Invasions 2024, 15, 567–580. [Google Scholar] [CrossRef]
- US Coast Guard. Ballast Water Treatment, U.S. Great Lakes Bulk Carrier Engineering and Cost Study—Volume I: Present Conditions. 2013, CG-D-12-13. Available online: https://apps.dtic.mil/sti/tr/pdf/ADA589870.pdf (accessed on 1 June 2023).
- First, M.R.; Drake, L.A. The practicability of ships arriving to the Great Lakes to conduct ballast water exchange plus treatment—Analysis of shipping patterns. J. Great Lakes Res. 2017, 43, 755–761. [Google Scholar] [CrossRef]
- Black, L. Are the great lakes connected? The great lakes watershed. Great Lakes Guide 2019, 1–3. Available online: https://greatlakes.guide/ideas/are-the-great-lakes-connected (accessed on 1 December 2023).
- Vander Zanden, M.J.; Hansen, G.J.; Higgins, S.N.; Kornis, M.S. A pound of prevention, plus a pound of cure: Early detection and eradication of invasive species in the Laurentian Great Lakes. J. Great Lakes Res. 2010, 36, 199–205. [Google Scholar] [CrossRef]
- Havel, J.E.; Kovalenko, K.E.; Thomaz, S.M.; Amalfitano, S.; Kats, L.B. Aquatic invasive species: Challenges for the future. Hydrobiologia 2015, 750, 147–170. [Google Scholar] [CrossRef] [PubMed]
- Lovell, S.J.; Stone, S.F.; Fernandez, L. The economic impacts of aquatic invasive species: A review of the literature. Agric. Resour. Econ. Rev. 2006, 35, 195–208. [Google Scholar] [CrossRef]
- Rosaen, A.L.; Grover, E.A.; Spencer, C.W.; Anderson, P.L. The Costs of Aquatic Invasive Species to Great Lakes States; Anderson Economical Group: East Lansing, MI, USA, 2012. [Google Scholar]
- Cuthbert, R.N.; Pattison, Z.; Taylor, N.G.; Verbrugge, L.; Diagne, C.; Ahmed, D.A.; Leroy, B.; Angulo, E.; Briski, E.; Capinha, C.; et al. Global economic costs of aquatic invasive alien species. Sci. Total Environ. 2021, 775, 145238. [Google Scholar] [CrossRef]
- Klymus, K.E.; Marshall, N.T.; Stepien, C.A. Environmental DNA (eDNA) metabarcoding assays to detect invasive invertebrate species in the Great Lakes. PLoS ONE 2017, 12, e0177643. [Google Scholar] [CrossRef]
- Larson, E.R.; Renshaw, M.A.; Gantz, C.A.; Umek, J.; Chandra, S.; Lodge, D.M.; Egan, S.P. Environmental DNA (eDNA) detects the invasive crayfishes Orconectes rusticus and Pacifastacus leniusculus in large lakes of North America. Hydrobiologia 2017, 800, 173–185. [Google Scholar] [CrossRef]
- Cangelosi, A.; Balcer, M.; Prihoda, K.; Gruwell, M.; TenEyck, M.; Aicher, R.; Lopez-Camacho, Y.; Knight, I.T.; Grey, E.K. Evaluation of eDNA qPCR monitoring as an early detection tool for a non-native mysid in Great Lakes Waters. J. Great Lakes Res. 2024, 50, 102377. [Google Scholar] [CrossRef]
- Fonseca, V.G.; Davison, P.I.; Creach, V.; Stone, D.; Bass, D.; Tidbury, H.J. The application of eDNA for monitoring aquatic non-indigenous species: Practical and policy considerations. Diversity 2023, 15, 631. [Google Scholar] [CrossRef]
- Fediajevaite, J.; Priestley, V.; Arnold, R.; Savolainen, V. Meta-analysis shows that environmental DNA outperforms traditional surveys, but warrants better reporting standards. Ecol. Evol. 2021, 11, 4803–4815. [Google Scholar] [CrossRef] [PubMed]
- GLANSIS. Great Lakes Aquatic Nonindigenous Species Information System. Available online: https://www.glerl.noaa.gov/glansis/index.html (accessed on 1 January 2022).
- Schubert, H.; Blindow, I.; Nat, E.; Korsch, H.; Gregor, T.; Denys, L.; Stewart, N.; van de Weyer, K.; Romanov, R.; Casanova, M.T. (Eds.) Charophytes of Europe; Springer: Cham, Switzerland, 2024. [Google Scholar] [CrossRef]
- Copilaș-Ciocianu, D.; Palatov, D.; Rewicz, T.; Sands, A.F.; Arbačiauskas, K.; Van Haaren, T.; Hebert, P.D.; Grabowski, M.; Marin, I. A widespread Ponto-Caspian invader with a mistaken identity: Integrative taxonomy elucidates the confusing taxonomy of Trichogammarus trichiatus (=Echinogammarus)(Crustacea: Amphipoda). Zool. J. Linn. Soc. 2023, 198, 821–846. [Google Scholar] [CrossRef]
- Dermott, R.; Witt, J.; Um, Y.M.; González, M. Distribution of the Ponto-Caspian amphipod Echinogammarus ischnus in the Great Lakes and replacement of native Gammarus fasciatus. J. Great Lakes Res. 1998, 24, 442–452. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Ostell, J.; Pruitt, K.D.; Sayers, E.W. GenBank. Nucleic Acids Res. 2018, 46, D41–D47. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D. BOLD: The Barcode of Life Data System. Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Ricciardi, A.; MacIsaac, H.J. Recent mass invasion of the north American Great Lakes by Ponto–Caspian species. Trends Ecol. Evol. 2000, 15, 62–65. [Google Scholar] [CrossRef]
- Charlebois, P.M.; Raffenberg, M.J.; Dettmers, J.M. First occurrence of Cercopagis pengoi in Lake Michigan. J. Great Lakes Res. 2001, 27, 258–261. [Google Scholar] [CrossRef]
- Kane, D.D.; Haas, E.M.; Culver, D.A. The Characteristics and Potential Ecological Effects of the Exotic Crustacean Zooplankter Cercopagis pengoi (Cladocera: Cercopagidae), a Recent Invader of Lake Erie. Ohio J. Sci. 2003, 103, 79–83. [Google Scholar]
- Muzinic, C.J. First record of Daphnia lumholtzi Sars in the Great Lakes. J. Great Lakes Res. 2000, 26, 352–354. [Google Scholar] [CrossRef]
- Sanders, E.R.; Karol, K.G.; McCourt, R.M. Occurrence of matK in a trnK group II intron in charophyte green algae and phylogeny of the Characeae. Am. J. Bot. 2003, 90, 628–633. [Google Scholar] [CrossRef]
- Wattoo, J.I.; Saleem, M.Z.; Shahzad, M.S.; Arif, A.; Hameed, A.; Saleem, M.A. DNA Barcoding: Amplification and sequence analysis of rbcl and matK genome regions in three divergent plant species. Adv. Life Sci. 2016, 4, 03–07. [Google Scholar]
- Bucklin, A.; Hopcroft, R.R.; Kosobokova, K.N.; Nigro, L.M.; Ortman, B.D.; Jennings, R.M.; Sweetman, C.J. DNA barcoding of Arctic Ocean holozooplankton for species identification and recognition. Deep Sea Res. Part II Top. Stud. Oceanogr. 2010, 57, 40–48. [Google Scholar] [CrossRef]
- Frisch, D.; Weider, L.J. Seasonal shifts in genotype frequencies in the invasive cladoceran Daphnia lumholtzi in Lake Texoma, USA. Freshw. Biol. 2010, 55, 1327–1336. [Google Scholar] [CrossRef]
- Frisch, D.; Havel, J.E.; Weider, L.J. The invasion history of the exotic freshwater zooplankter Daphnia lumholtzi (Cladocera, Crustacea) in North America: A genetic analysis. Biol. Invasions 2013, 15, 817–828. [Google Scholar] [CrossRef]
- Watkins, J.M.; Collingsworth, P.D.; Saavedra, N.E.; O’Malley, B.P.; Rudstam, L.G. Fine-scale zooplankton diel vertical migration revealed by traditional net sampling and a Laser Optical Plankton Counter (LOPC) in Lake Ontario. J. Great Lakes Res. 2017, 43, 804–812. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal omega. Curr. Protoc. Bioinform. 2014, 48, 3.13.1–3.13.16. [Google Scholar] [CrossRef] [PubMed]
- Cornetti, L.; Fields, P.D.; Van Damme, K.; Ebert, D. A fossil-calibrated phylogenomic analysis of Daphnia and the Daphniidae. Mol. Phylogenetics Evol. 2019, 137, 250–262. [Google Scholar] [CrossRef] [PubMed]
- Karol, K.G.; Skawinski, P.M.; McCourt, R.M.; Nault, M.E.; Evans, R.; Barton, M.E.; Berg, M.S.; Perleberg, D.J.; Hall, J.D. First discovery of the charophycean green alga Lychnothamnus barbatus (Charophyceae) extant in the New World. Am. J. Bot. 2017, 104, 1108–1116. [Google Scholar] [CrossRef]
- McCourt, R.M.; Casanova, M.T.; Karol, K.G.; Feist, M. Monophyly of genera and species of Characeae based on rbcL sequences, with special reference to Australian and European Lychnothamnus barbatus (Characeae: Charophyceae). Aust. J. Bot. 1999, 47, 361–369. [Google Scholar] [CrossRef]
- Pełechaty, M.; Brzozowski, M.; Pietruczuk, K. Overwintering and gyrogonite formation by the rare and endangered indicative macroalga Lychnothamnus barbatus (Meyen) Leonh. in eutrophic conditions. Aquat. Bot. 2017, 139, 19–24. [Google Scholar] [CrossRef]
- Brzozowski, M.; Pełechaty, M.; Pietruczuk, K. Co-occurrence of the charophyte Lychnothamnus barbatus with higher trophy submerged macrophyte indicators. Aquat. Bot. 2018, 151, 51–55. [Google Scholar] [CrossRef]
- Witt, A.M.; Cáceres, C.E. Potential predator-prey relationships between Bythotrephes longimanus and Cercopagis pengoi in southwestern Lake Michigan. J. Great Lakes Res. 2004, 30, 519–527. [Google Scholar] [CrossRef]
- Ptáčníková, R.; Vanderploeg, H.A.; Cavaletto, J.F. Big versus small: Does Bythotrephes longimanus predation regulate spatial distribution of another invasive predatory cladoceran, Cercopagis pengoi? J. Great Lakes Res. 2015, 41, 143–149. [Google Scholar] [CrossRef]
- Richter, S.; Braband, A.; Aladin, N.; Scholtz, G. The phylogenetic relationships of “predatory water-fleas” (Cladocera: Onychopoda, Haplopoda) inferred from 12S rDNA. Mol. Phylogenetics Evol. 2001, 19, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Therriault, T.W.; Grigorovich, I.A.; Cristescu, M.E.; Ketelaars, H.A.; Viljanen, M.; Heath, D.D.; Macisaac, H.J. Taxonomic resolution of the genus Bythotrephes Leydig using molecular markers and re-evaluation of its global distribution. Divers. Distrib. 2002, 8, 67–84. [Google Scholar] [CrossRef]
- Cristescu, M.E.A.; Witt, J.D.S.; Grigorovich, I.A.; Hebert, P.D.N.; MacIsaac, H.J. Dispersal of the Ponto-Caspian amphipod Echinogammarus ischnus: Invasion waves from the Pleistocene to the present. Heredity 2004, 92, 197–203. [Google Scholar] [CrossRef]
- Radulovici, A.E.; Sainte-marie, B.E.R.N.A.R.D.; Dufresne, F. DNA barcoding of marine crustaceans from the Estuary and Gulf of St Lawrence: A regional-scale approach. Mol. Ecol. Resour. 2009, 9, 181–187. [Google Scholar] [CrossRef]
- Lipinskaya, T.; Radulovici, A.; Makaranka, A. First DNA barcoding based record of Echinogammarus trichiatus (Martynov, 1932)(Crustacea, Gammaridae) in Belarus. BioInvasions Rec. 2018, 7, 55–60. [Google Scholar] [CrossRef]
- PrimerDigital In silico PCR Tool. Available online: https://primerdigital.com/tools/pcr.html (accessed on 15 May 2025).
- National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 1 January 2022).
- NOAA Great Lakes Environmental Research Laboratory: Species List Generator. Available online: https://www.glerl.noaa.gov/glansis/nisListGen.php (accessed on 15 May 2025).
- Blackman, R.C.; Ling, K.K.S.; Harper, L.R.; Shum, P.; Hänfling, B.; Lawson-Handley, L. Targeted and passive environmental DNA approaches outperform established methods for detection of quagga mussels, Dreissena rostriformis bugensis in flowing water. Ecol. Evol. 2020, 10, 13248–13259. [Google Scholar] [CrossRef]
- Davison, P.I.; Falcou-Préfol, M.; Copp, G.H.; Davies, G.D.; Vilizzi, L.; Créach, V. Is it absent or is it present? Detection of a non-native fish to inform management decisions using a new highly-sensitive eDNA protocol. Biol. Invasions 2019, 21, 2549–2560. [Google Scholar] [CrossRef]
- Wozney, K.M.; Wilson, C.C. Quantitative PCR multiplexes for simultaneous multispecies detection of Asian carp eDNA. J. Great Lakes Res. 2017, 43, 771–776. [Google Scholar] [CrossRef]
- Hernandez, C.; Bougas, B.; Perreault-Payette, A.; Simard, A.; Côté, G.; Bernatchez, L. 60 specific eDNA qPCR assays to detect invasive, threatened, and exploited freshwater vertebrates and invertebrates in Eastern Canada. Environ. DNA 2020, 2, 373–386. [Google Scholar] [CrossRef]
- Mauvisseau, Q.; Burian, A.; Gibson, C.; Brys, R.; Ramsey, A.; Sweet, M. Influence of accuracy, repeatability and detection probability in the reliability of species-specific eDNA based approaches. Sci. Rep. 2019, 9, 580. [Google Scholar] [CrossRef]
- Carlsson, J.E.; Egan, D.; Collins, P.C.; Farrell, E.D.; Igoe, F.; Carlsson, J. A qPCR MGB probe based eDNA assay for European freshwater pearl mussel (Margaritifera margaritifera L.). Aquat. Conserv. Mar. Freshw. Ecosyst. 2017, 27, 1341–1344. [Google Scholar] [CrossRef]
- Tréguier, A.; Paillisson, J.M.; Dejean, T.; Valentini, A.; Schlaepfer, M.A.; Roussel, J.M. Environmental DNA surveillance for invertebrate species: Advantages and technical limitations to detect invasive crayfish Procambarus clarkii in fresh water ponds. J. Appl. Ecol. 2014, 51, 871–879. [Google Scholar] [CrossRef]
- Buxton, A.S.; Groombridge, J.J.; Zakaria, N.B.; Griffiths, R.A. Seasonal variation in environmental DNA in relation to population size and environmental factors. Sci. Rep. 2017, 7, 46294. [Google Scholar] [CrossRef]
- Great Lakes Commission. Blue Accounting. 2022. Available online: https://www.blueaccounting.org/metric/monitoring (accessed on 1 January 2024).
- U.S. Fish and Wildlife Service. Aquatic Nuisance Species Task Force Strategic Plan for 2020–2025. 2019. Available online: https://www.fws.gov/sites/default/files/documents/ANSTF-Strategic-Plan-2020-2025.pdf (accessed on 1 June 2025).
- Darling, J.A.; Frederick, R.M. Nucleic acids-based tools for ballast water surveillance, monitoring, and research. J. Sea Res. 2018, 133, 43–52. [Google Scholar] [CrossRef]
- Feist, S.M.; Lance, R.F. Advanced molecular-based surveillance of quagga and zebra mussels: A review of environmental DNA/RNA (eDNA/eRNA) studies and considerations for future directions. NeoBiota 2021, 66, 117–159. [Google Scholar] [CrossRef]
- Kirtane, A.; Wieczorek, D.; Noji, T.; Baskin, L.; Ober, C.; Plosica, R.; Chenoweth, A.; Lynch, K.; Sassoubre, L. Quantification of environmental DNA (eDNA) shedding and decay rates for three commercially harvested fish species and comparison between eDNA detection and trawl catches. Environ. DNA 2021, 3, 1142–1155. [Google Scholar] [CrossRef]
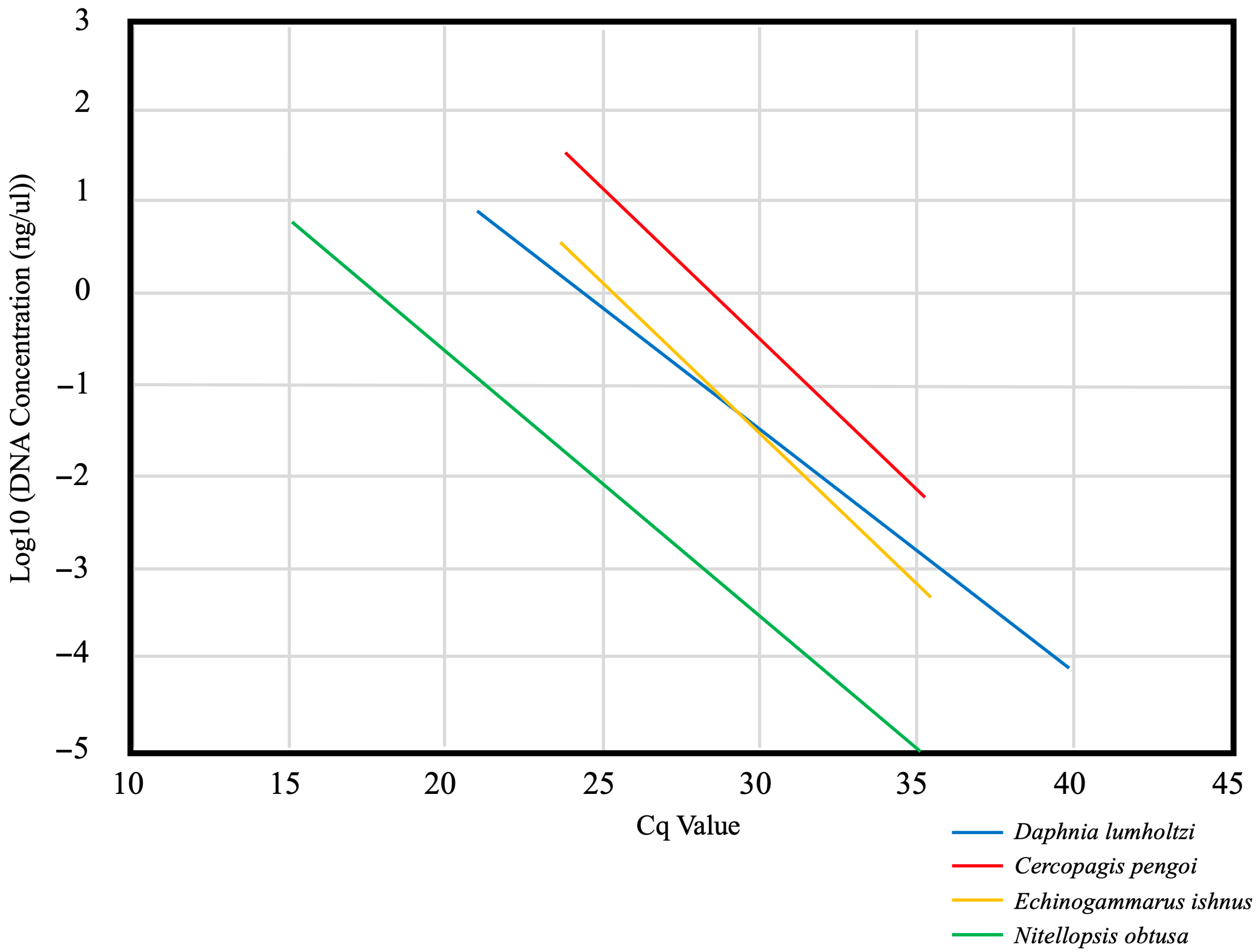
| Species | Reported NAGL | Source of Specimens Used | Primers | Annealing Temp (°C) | Melt Temp (°C) for qPCR | Limit of Detection | Cq Value for 1 ng/μL | Cq Value for 0.01 ng/μL | Cq Value for 0.001 ng/μL |
|---|---|---|---|---|---|---|---|---|---|
| N. obtusa | Ontario, Erie, Huron, Michigan | Lake Erie 42.153629, −80.114770 | Forward: Nobt_458F CTCCTTTAATTCACCAGTTC Reverse: Nobt_695R TGAATTCACCAAATACACTA | 50.0 | 72.0 | 5.60 × 10−6 ng/μL | 17.71 | 24.66 | 28.14 |
| C. pengoi | Ontario, Erie, Huron, Michigan | Lake Ontario planktonic Provided by Dr. James Watkins | Forward: Cpen_115F CAATGTAGTAGTAACAGCCCAC Reverse: Cpen_302R ACCTCCAACTAGAAGTAGAGTTAAA | 53.0 | 79.0 | 1.82 × 10−3 ng/μL | 26.73 | 33.27 | 36.55 |
| D. lumholtzi | Erie, Superior | Provided by Dr. Lawrence Weider’s Lab Culture | Forward: Dlum_163F GGGTTTTGGAAATTGATTAGTT Reverse: Dlum_397R TCCCAGCCAAATGCAAAGA | 51.5 | 78.5 and 83.0 | 7.60 × 10−5 ng/μL | 24.29 | 31.83 | 35.60 |
| E. ishnus | Ontario, Erie, Huron, Michigan, Superior | Lake Erie 42.156317, −80.071064 | Forward: Cisc_357F GCCTCTCTCTAACTCTATAGGC Reverse: Cisc_547R TGGTAAGGACAGGAGAAGCAA | 65.0 | 79.0 | 6.50 × 10−4 ng/μL | 25.40 | 31.53 | 34.62 |
| Target AIS | Test Species | NCBI # | F Primer Match | R Primer Match | Amplification |
|---|---|---|---|---|---|
| Nitellopsis obtusa | |||||
| N. obtusa | AY170447 | 20 of 20 bp—100% | 20 of 20 bp—100% | Success | |
| Primers tested | L. barbatus | AY170448 | 14 of 20 bp—70% | 14 of 20 bp—70% | Fail |
| Nobt_485F—20 bp | C. folia | MZ682285 | 14 of 20 bp—65% | 16 of 20 bp—85% | Fail |
| Nobt_695R—20 bp | |||||
| Cercopagis pengoi | |||||
| C. pengoi | OP830033 | 23 of 23 bp—100% | 25 of 25 bp—100% | Success | |
| Primers tested | B. longimanus | MH321333 | 14 of 23 bp—61% | 16 of 25 bp—64% | Fail |
| Cpen_115F—23 bp | Moina sp. | LC503929 | 14 of 23 bp—61% | 17 of 25 bp—68% | Fail |
| Cpen_302R—25 bp | |||||
| Daphnia lumholtzi | |||||
| D.lumholtzi | AY921417 | 22 of 22 bp—100% | 19 of 19 bp—100% | Success | |
| Primers tested | D. ambigua | MG448989 | 19 of 22 bp—86% | 17 of 19 bp—89% | Fail |
| Dlum_163F—20 | D. catawba | AY380454 | 16 of 22 bp—73% | 15 of 19 bp—79% | Fail |
| Dlum_397R—19 | D. dentifera | MG448806 | 19 of 22 bp—86% | 11 of 19 bp—58% | Fail |
| D. dubia | AY921411 | 16 of 22 bp—73% | 11 of 19 bp—58% | Fail | |
| D. galeata | MH746186 | 17 of 22 bp—77% | 13 of 19 bp—68% | Fail | |
| D. laevis | MG449455 | 17 of 22 bp—77% | 14 of 19 bp—74% | Fail | |
| D. longiremis | AY921413 | 19 of 22 bp—86% | 15 of 19 bp—79% | Fail | |
| D. middenforffiaena | KC502301 | 18 of 22 bp—82% | 14 of 19 bp—74% | Fail | |
| D. parvula | MG936477 | 16 of 22 bp—73% | 15 of 19 bp—79% | Fail | |
| D. pulex | MG315907 | 17 of 22 bp—77% | 14 of 19 bp—74% | Fail | |
| D. pulicara | MG448615 | 17 of 22 bp—77% | 14 of 19 bp—74% | Fail | |
| D. retrocurva | OP830209 | 14 of 22 bp—64% | 15 of 19 bp—79% | Fail | |
| D. sinesis | LS991517 | 18 of 22 bp—82% | 14 of 19 bp—74% | Fail | |
| D. similis | MF346400 | 15 of 22 bp—68% | 15 of 19 bp—79% | Fail | |
| D. magna | MG317471 | 16 of 22 bp—73% | 14 of 19 bp—74% | Fail | |
| Echinogammarus ischnus | |||||
| E. ischnus | FJ581620 | 22 of 22 bp—100% | 21 of 21 bp—100% | Success | |
| Primers tested | G. faciatus | MG734968 | 8 of 22 bp—36% | 13 of 21—62% | Fail |
| Cisc_357F—22 bp | G. lacustris | MG318006 | 10 of 22 bp—45.5% | 17 of 21—81% | Fail |
| Cisc_547R—21 bp | G. tigrinis | FJ581684 | 9 of 22 bp—41% | 14 of 21—66.6% | Fail |
| G. pseudolimnaeus | EU574907 | 10 of 22 bp—45.5% | 13 of 21—62% | Fail |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gruwell, M.E.; Welsbacher, A.; Moore, N.; Cangelosi, A.; Melendez, A.; Sheehan, R.; Knight, I. Working Primers and qPCR Protocols for Rapid eDNA Identification of Four Aquatic Invasive Species Found in the Lower Great Lakes with High Potential for Ballast Transport to Lake Superior. Hydrobiology 2025, 4, 22. https://doi.org/10.3390/hydrobiology4030022
Gruwell ME, Welsbacher A, Moore N, Cangelosi A, Melendez A, Sheehan R, Knight I. Working Primers and qPCR Protocols for Rapid eDNA Identification of Four Aquatic Invasive Species Found in the Lower Great Lakes with High Potential for Ballast Transport to Lake Superior. Hydrobiology. 2025; 4(3):22. https://doi.org/10.3390/hydrobiology4030022
Chicago/Turabian StyleGruwell, Matthew E., Amanda Welsbacher, Noel Moore, Allegra Cangelosi, Abigail Melendez, Ryan Sheehan, and Ivor Knight. 2025. "Working Primers and qPCR Protocols for Rapid eDNA Identification of Four Aquatic Invasive Species Found in the Lower Great Lakes with High Potential for Ballast Transport to Lake Superior" Hydrobiology 4, no. 3: 22. https://doi.org/10.3390/hydrobiology4030022
APA StyleGruwell, M. E., Welsbacher, A., Moore, N., Cangelosi, A., Melendez, A., Sheehan, R., & Knight, I. (2025). Working Primers and qPCR Protocols for Rapid eDNA Identification of Four Aquatic Invasive Species Found in the Lower Great Lakes with High Potential for Ballast Transport to Lake Superior. Hydrobiology, 4(3), 22. https://doi.org/10.3390/hydrobiology4030022






