A Clustered Link-Prediction SEIRS Model with Temporal Node Activation for Modeling Computer Virus Propagation in Urban Communication Systems
Abstract
1. Introduction
2. Model Formulation
2.1. Model Setup and State Definitions
- Susceptible: Node i is healthy and vulnerable to infection.
- Exposed: Node i has been infected but is not yet infectious (latent period).
- Infectious: Node i is actively infected and can transmit the virus to susceptible neighbors.
- Recovered: Node i has recovered and is temporarily immune (will later lose immunity and return to susceptible).
2.2. Contact Network and Link-Prediction Mechanism
- : infection rate for an infectious contact within the same community (intra-community transmission rate),
- : infection rate for an infectious contact across different communities (inter-community transmission rate).
- A direct link uses rate (either or depending on community relation).
- An indirect connection through common neighbors ( and ) uses rate .
2.3. Periodic Activation and Temporal Contact Schedules
2.4. Transition Dynamics and Mean-Field Approximation
- If i and j are directly connected (): the transmission occurs at rate . This equals for intra-community pairs, or for inter-community pairs.
- If i and j are not directly connected () but have one or more mutual neighbors (): they can meet through those neighbors. Each common neighbor provides a potential contact at rate . If there are such mutual connections, the total indirect infection rate is .
2.5. Mean-Field Equations of Motion (Continuous-Time)
- Susceptible : decreases when node i becomes infected (moves to ), and increases when node i loses immunity (moves from back to ). The loss of susceptibility for node i occurs at a rate equal to the sum of infection rates from all infectious contacts (direct and indirect). Given the rules above, node i (if susceptible) experiences infection from an infectious neighbor j at rate and from an infectious non-neighbor j (sharing common neighbors) at rate . Summing over all infectious nodes j, the total infection rate for node i is:Meanwhile, the gain of susceptibility for node i occurs at rate (its own recovery immunity waning). Therefore, the differential equation for is:The second term represents the loss of susceptibles due to infections, where the factor ensures that both i and j are online and in the correct states for transmission. This double sum effectively covers all pairs and includes:
- For each direct neighbor (): a term .
- For each pair with no direct link () but common neighbors (): a term .
If node i or node j is offline ( or ), the term vanishes, reflecting no contact. - Exposed : increases when a susceptible node becomes infected (enters latent state), and decreases when an exposed node progresses to infectious. Any new infection of node i causes an immediate transition from to . Hence, the gain term for is exactly the loss term for derived above (since every susceptible leaving enters ). The loss term for is the progression to at rate . Thus:The first term is the infection force (identical to the term that appears with a minus sign in Equation (9)), and the second term removes individuals from E as they become infectious.
- Infectious : increases when node i finishes the latent period (), and decreases when node i recovers (). Thus:Here is the inflow from exposed to infectious, and is the outflow due to recovery.
- Recovered : increases when node i recovers from infection, and decreases when immunity wanes (node i returns to susceptible). So:The term is the inflow from recovery, and is the outflow to susceptibility.
2.6. Discrete-Time Simulation Equations
- increases by (from recovered) and decreases by the number of new exposures in that interval.
- increases by the number of new exposures and decreases by .
- increases by and decreases by .
- increases by and decreases by .
3. Results and Discussion
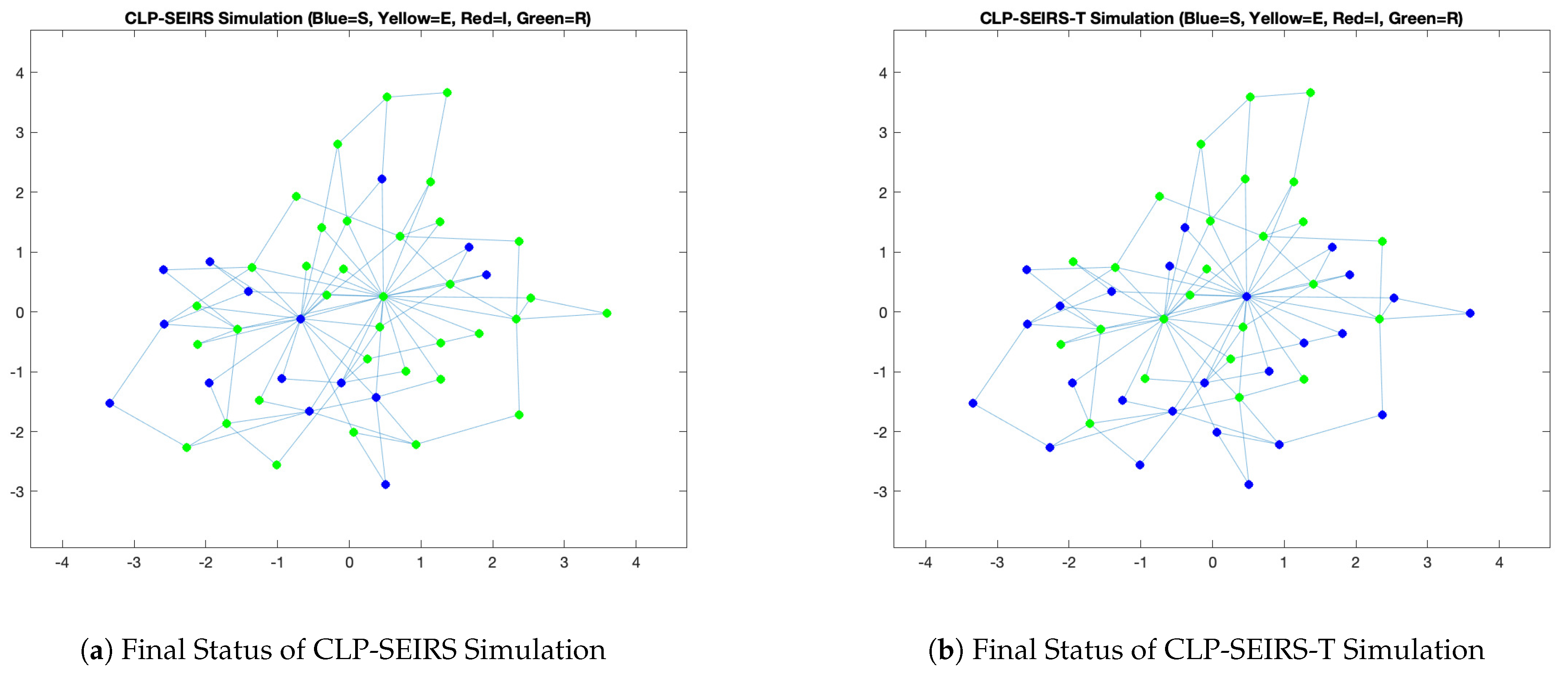
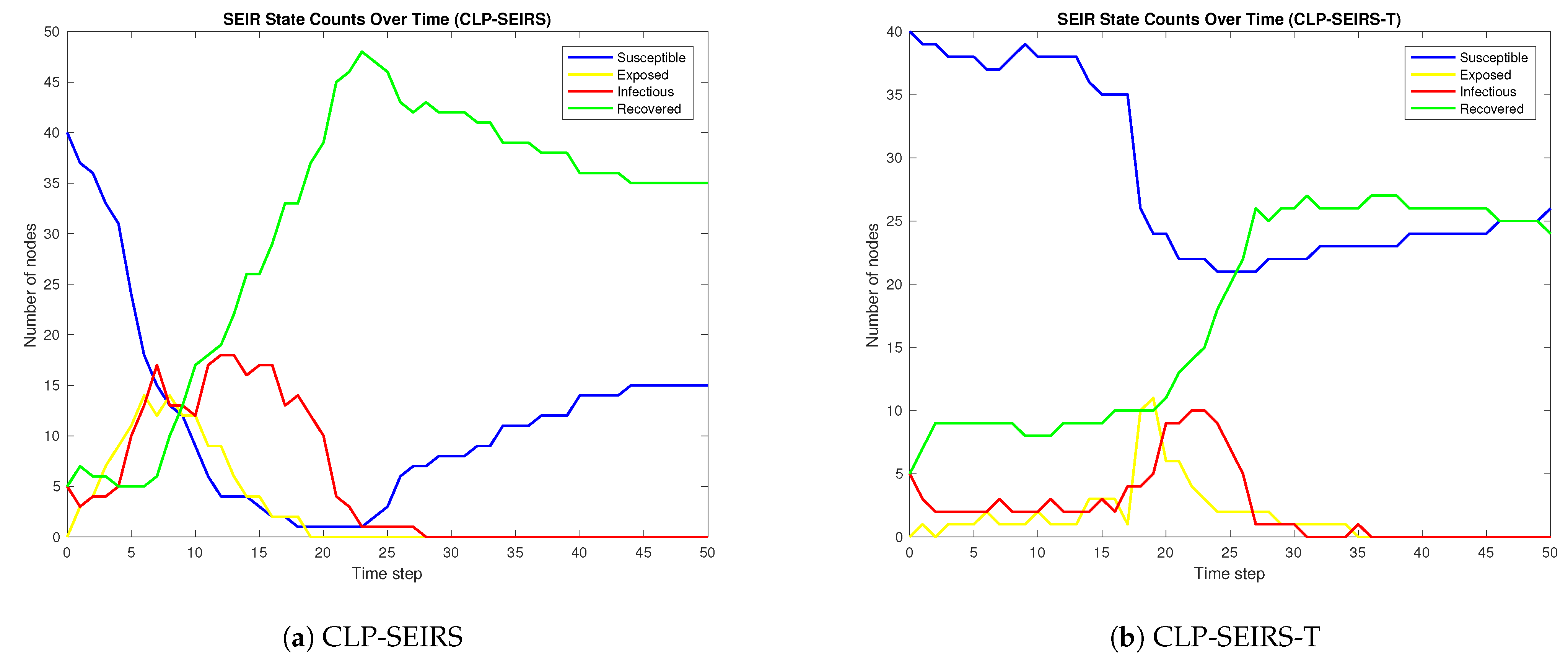
3.1. All-Time Online vs. Time-Varying Availability
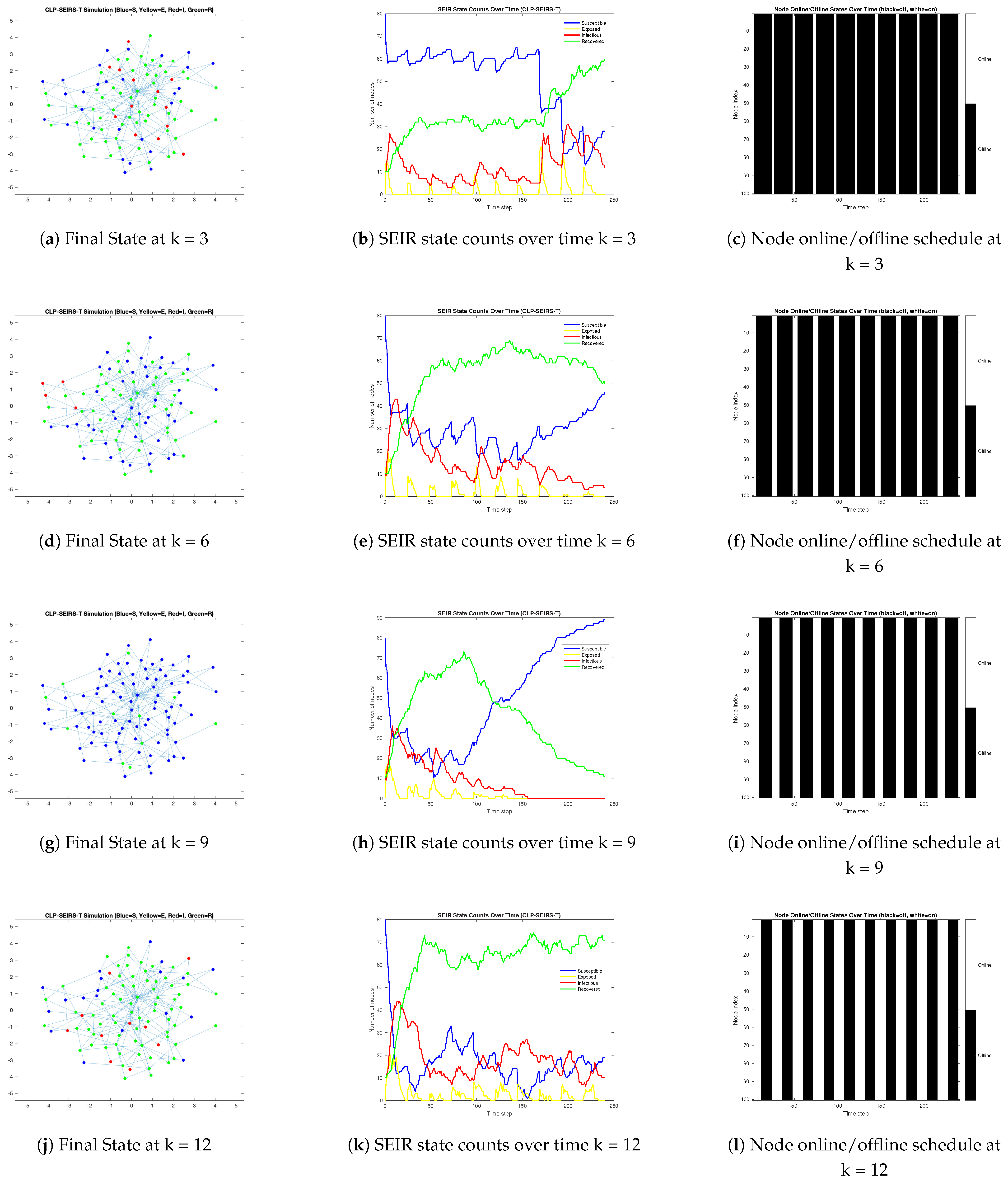
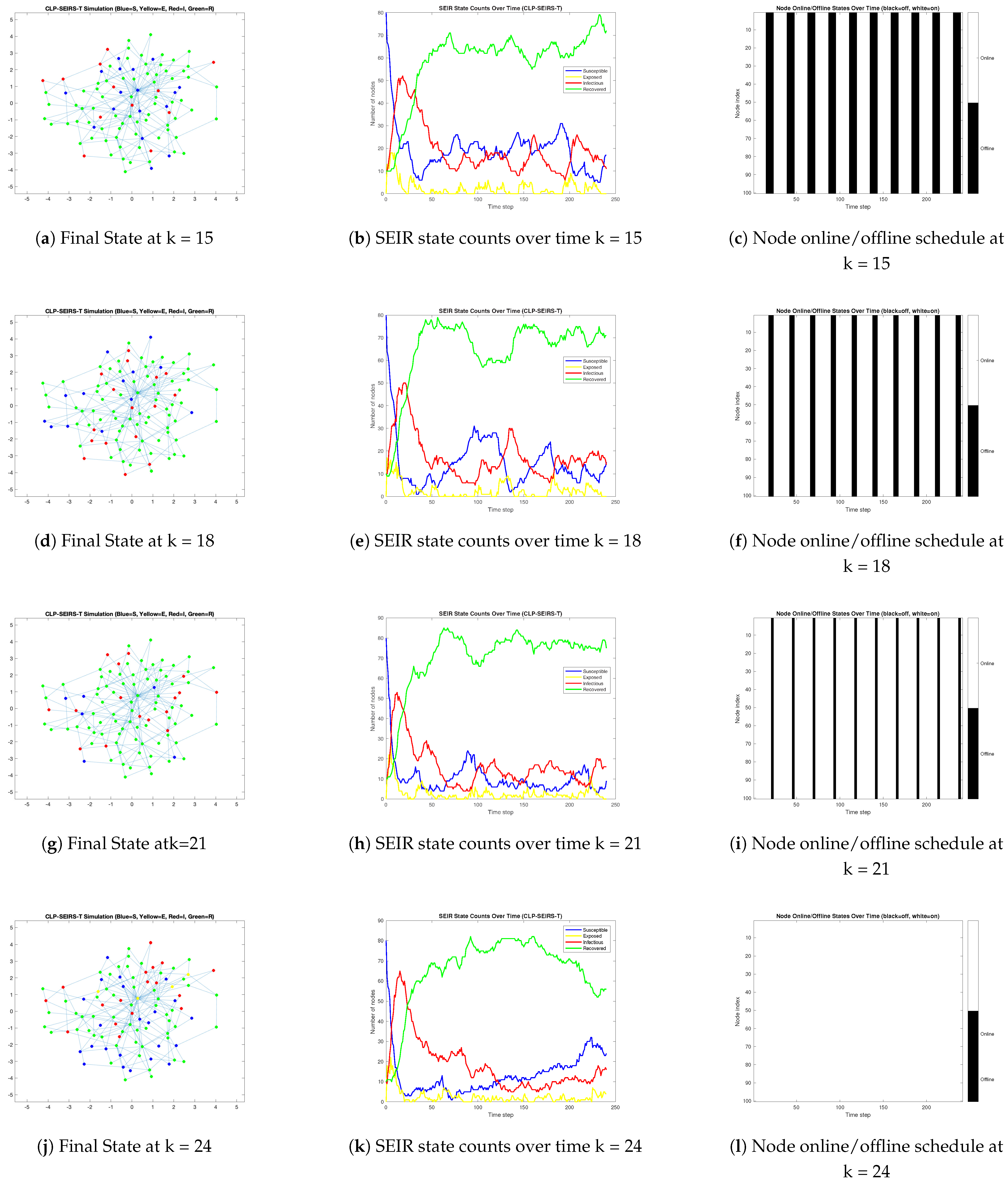
3.2. Propagation Dynamics Under Varying Online Availability Less than 50%
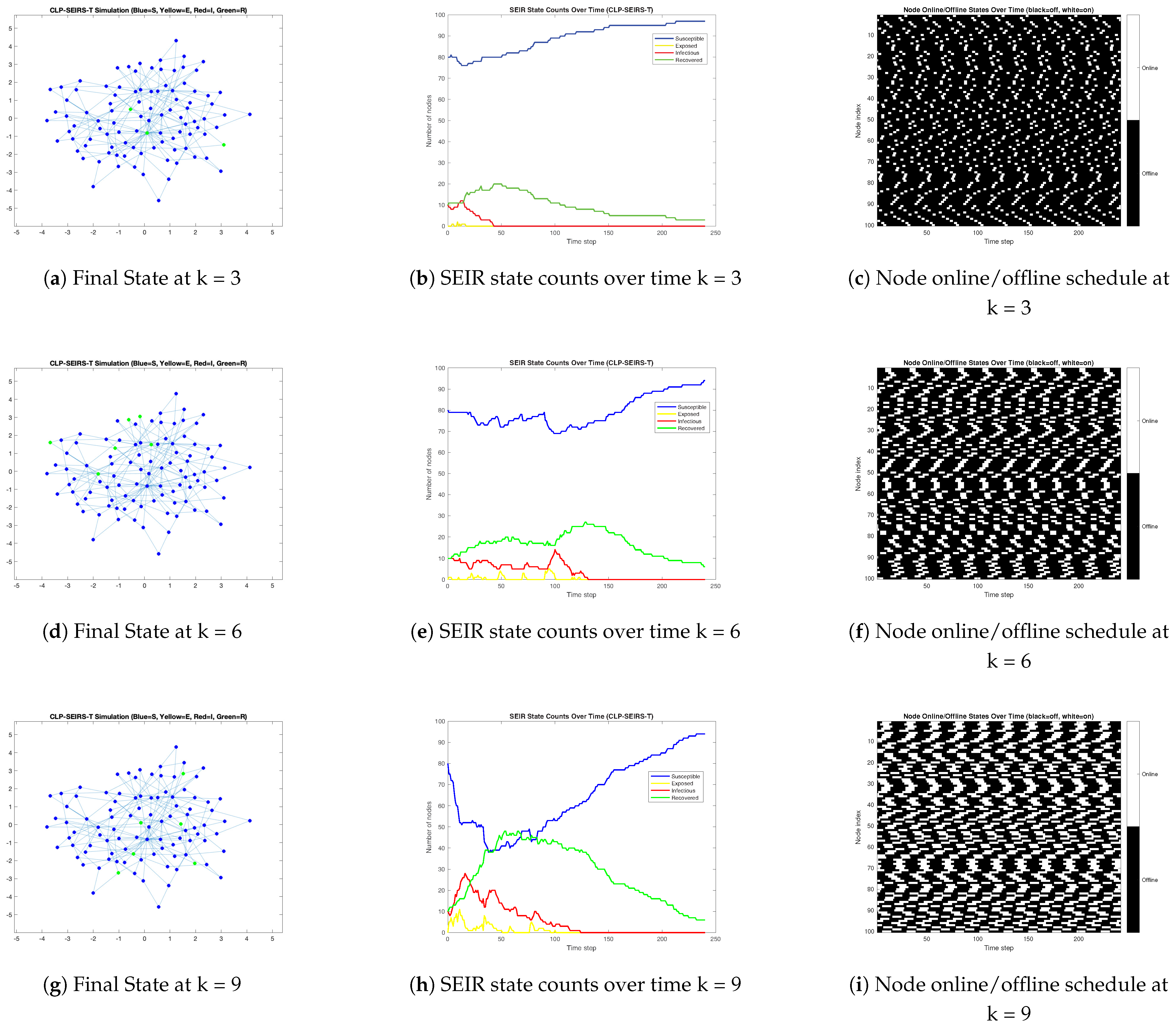
3.3. Propagation Dynamics Under Varying Online Availability Great than 50%
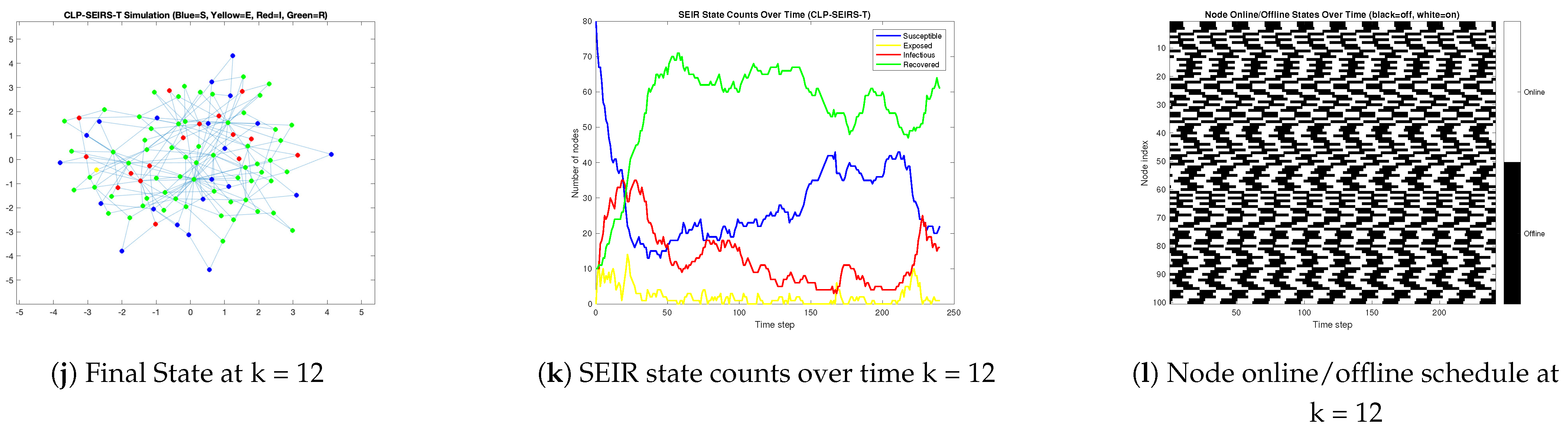
3.4. Impact of Randomized Phase Online Patterns on CLP-SEIRS-T Dynamics
4. Conclusions and Future Work
4.1. Conclusions
- Strong community structure can localize infections and delay inter-community spread, especially when combined with limited or intermittent cross-community contacts. We observed that when nodes follow periodic duty cycles, malware spread is substantially slowed and fragmented. Lower online availability effectively “flattens” the infection curve: outbreaks unfold over longer periods and peak at lower infection levels, often leaving a considerable portion of nodes uninfected. In particular, imposing asynchronous or staggered online schedules across different communities further confined the virus, as misaligned active periods meant that an outbreak in one cluster often failed to synchronize with activity in others.
- When connectivity is near-continuous or highly synchronous, we found that contagion dynamics resemble those in classic epidemic models on homogeneous networks. High overall contact rates (e.g., nodes online nearly 100% of the time) led to fast, explosive outbreaks that infected almost the entire network in a single wave. Community boundaries offer little resistance in such scenarios, as even a small number of inter-community links allow the virus to traverse between clusters when nodes are continuously active. Our numerical simulatoin results revealed a clear threshold effect in node availability: beyond a certain fraction of time online, the malware spread becomes rampant and difficult to contain, whereas below that threshold the outbreak remains partial or dies out after limited waves. Moreover, synchronized activation across nodes produced one dominant outbreak peak followed by die-out, whereas staggered (asynchronous) activation allowed the infection to persist via multiple smaller waves. In the asynchronous cases, the malware could re-emerge when previously offline devices came online, resulting in protracted, oscillatory epidemic behavior. This result underlines the importance of incorporating realistic, non-synchronized usage patterns into virus spread models, as they can sustain contagion even when instantaneous infection rates are low.
- By incorporating a common-neighbor link-prediction mechanism, the CLP-SEIRS-T model captures indirect infection pathways that would be missed in static-network models. Even if two devices are not directly connected, our model allows malware transmission if they share enough neighbors. We found that these predicted links can significantly increase the reach of an outbreak: infections were able to jump across network gaps and infect otherwise isolated communities via “future” connections. Including this effect effectively lowers the epidemic threshold and raises the final fraction of infected nodes, indicating that traditional models which ignore potential link formation may underestimate the true risk and extent of malware spread. From a defensive standpoint, this finding suggests that network administrators should not only secure existing links but also be mindful of likely future connections.
4.2. Future Work and Extensions
- Heterogeneous Duty Cycles: In this study we assumed all nodes share the same duty cycle . However, real networks often contain devices with widely varying activity levels. The model can readily accommodate different values for each node, which would lead to interesting changes in propagation dynamics. For example, a device that is almost always online could act as a super-spreader and accelerate the overall malware spread, whereas a device that rarely comes online would hardly participate in transmission, effectively serving as a “firebreak”. Future work can investigate how different distributions of duty cycles affect outbreak outcomes, assessing the model’s sensitivity to heterogeneous node availability.
- Adaptive Link-Prediction Parameter: In our simulations, the link-prediction infection rate was treated as a constant. In reality, however, might not be fixed and could vary over time or with network conditions. Future extensions of the model could tie to network metrics, allowing it to adjust dynamically as the epidemic unfolds and the network structure evolves.
- Empirical Validation: At present, due to the lack of publicly available datasets on malware propagation in urban communication networks, the CLP-SEIRS-T model has only been evaluated via simulations on synthetic networks. In the future, we plan to validate the model against real-world data. One possible approach is to apply the model to network traffic traces from actual systems, or to conduct controlled experiments in a virtual machine-based network environment to simulate real malware spread scenarios.
- Stochastic Node Activation: We assumed periodic, deterministic online/offline patterns for nodes to facilitate analysis. In reality, however, node activity can be stochastic. The CLP-SEIRS-T framework can be extended to incorporate random activation times or use an “activity-driven” network model. We anticipate that randomizing node activation would, on average, still mitigate malware spread similar to having a duty cycle, but the detailed temporal dynamics may differ. Future work can explore how non-periodic, probabilistic connectivity patterns affect malware propagation.
- Weighted and Directed Networks: Our model assumed an undirected, unweighted network, but it can be extended to directed or weighted graphs. In a directed network, one can account for asymmetry in transmission (different infection probabilities in each direction of a link); in a weighted network, the infection rate of an edge can be made proportional to its weight. These extensions would introduce additional parameters and complexity, but would allow the model to apply to a wider range of communication systems. It will be valuable in future work to study how asymmetric or weighted connectivity influences malware spread.
- Scalability and Complexity: We recognize that simulating CLP-SEIRS-T on very large networks (with more than nodes) could be computationally demanding. At each time step the model must evaluate transmissions via direct and predicted links, leading to a complexity on the order of in the worst case. While our current implementation easily handles networks of size , scaling to much larger networks would require optimized algorithms or parallel computing. For real-time outbreak forecasting on extremely large networks, further simplifications might be necessary to reduce computational cost. In future work we will explore more efficient implementations to improve the model’s applicability to large-scale networks.
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviation
| CLP-SEIRS-T | Clustered Link-Prediction SEIRS Model with Temporal Node Activation |
References
- Ghafur, S.; Kristensen, S.; Honeyford, K.; Martin, G.; Darzi, A.; Aylin, P. A retrospective impact analysis of the WannaCry cyberattack on the NHS. NPJ Digit. Med. 2019, 2, 98. [Google Scholar] [CrossRef] [PubMed]
- Tsvetanov, T.; Slaria, S. The effect of the Colonial Pipeline shutdown on gasoline prices. Econ. Lett. 2021, 209, 110122. [Google Scholar] [CrossRef]
- Zhang, X.; Upton, O.; Beebe, N.L.; Choo, K.K.R. IoT Botnet Forensics: A Comprehensive Digital Forensic Case Study on Mirai Botnet Servers. Forensic Sci.-Int.-Digit. Investig. 2020, 32, 300926. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiang, L.; Ewe, H.T. A Novel Data-Driven Modeling Method for the Spatial Temporal Correlated Complex Sea Clutter. IEEE Trans. Geosci. Remote Sens. 2022, 60, 1–11. [Google Scholar] [CrossRef]
- Ding, Y.; Xiao, Y.; Xie, J.; Zhang, T. A time-varying transition channel model for air-ground communication. In Proceedings of the 2017 IEEE/AIAA 36th Digital Avionics Systems Conference (DASC), St. Petersburg, FL, USA, 17–21 September 2017; pp. 1–8. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiang, L. A Novel Data-Driven Analysis Method for Electromagnetic Radiations Based on Dynamic Mode Decomposition. IEEE Trans. Electromagn. Compat. 2020, 62, 1443–1450. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiang, L. A Novel Data-Driven Scheme for the Ship Wake Identification on the 2-D Dynamic Sea Surface. IEEE Access 2020, 8, 69593–69600. [Google Scholar] [CrossRef]
- Hethcote, H. The mathematics of infectious diseases. SIAM Rev. 2000, 42, 599–653. [Google Scholar] [CrossRef]
- Mishra, B.K.; Pandey, S.K. Dynamic model of worm propagation in computer network. Appl. Math. Model. 2014, 38, 2173–2179. [Google Scholar] [CrossRef]
- Kephart, J.; White, S. Measuring and modeling computer virus prevalence. In Proceedings of the 1993 IEEE Computer Society Symposium on Research in Security and Privacy, Oakland, CA, USA, 24–26 May 1993; pp. 2–15. [Google Scholar] [CrossRef]
- Staniford, S.; Paxson, V.; Weaver, N. How to Own the Internet in Your Spare Time. In Proceedings of the 11th USENIX Security Symposium, San Francisco, CA, USA, 5–9 August 2002; pp. 149–167. [Google Scholar]
- Zou, C.C.; Towsley, D.; Gong, W. Modeling and simulation study of the propagation and defense of internet e-mail worms. IEEE Trans. Dependable Secur. Comput. 2007, 4, 105–118. [Google Scholar] [CrossRef]
- Ing, J.; Pavlovskaia, E.; Wiercigroch, M.; Banerjee, S. Bifurcation analysis of an impact oscillator with a one-sided elastic constraint near grazing. Phys. D Nonlinear Phenom. 2010, 239, 312–321. [Google Scholar] [CrossRef]
- Rathie, P.N.; Coutinho, M.; Sousa, T.R.; Rodrigues, G.S.; Carrijo, T.B. Stable and generalized-t distributions and applications. Commun. Nonlinear Sci. Numer. Simul. 2012, 17, 5088–5096. [Google Scholar] [CrossRef]
- Xiong, S.; Yao, Y.; Berry, M.; Qi, H.; Cao, Q. Frequent traffic flow identification through probabilistic bloom filter and its GPU-based acceleration. J. Netw. Comput. Appl. 2017, 87, 60–72. [Google Scholar] [CrossRef]
- Musa, A.; Almohannadi, H.; Alhamar, J. Malware Propagation Modelling in Peer-to-Peer Networks: A Review. In Proceedings of the 2018 6th International Conference on Future Internet of Things and Cloud Workshops (FiCloudW), Barcelona, Spain, 6–8 August 2018; pp. 198–202. [Google Scholar] [CrossRef]
- Liu, W.; Zhong, S. Modeling and analyzing the dynamic spreading of epidemic malware by a network eigenvalue method. Appl. Math. Model. 2018, 63, 491–507. [Google Scholar] [CrossRef]
- Nguyen, B. Modelling Cyber Vulnerability using Epidemic Models. In Proceedings of the 7th International Conference on Simulation and Modeling Methodologies, Technologies and Applications, SIMULTECH 2017, Madrid, Spain, 26–28 July 2017; SCITEPRESS: Setubal, Portugal, 2017; pp. 232–239. [Google Scholar] [CrossRef]
- Boccaletti, S.; Latora, V.; Moreno, Y.; Chavez, M.; Hwang, D.U. Complex networks: Structure and dynamics. Phys. Rep. Rev. Sect. Phys. Lett. 2006, 424, 175–308. [Google Scholar] [CrossRef]
- Pastor-Satorras, R.; Vespignani, A. Epidemic spreading in scale-free networks. Phys. Rev. Lett. 2001, 86, 3200–3203. [Google Scholar] [CrossRef] [PubMed]
- Pastor-Satorras, R.; Castellano, C.; Van Mieghem, P.; Vespignani, A. Epidemic processes in complex networks. Rev. Mod. Phys. 2015, 87, 925–979. [Google Scholar] [CrossRef]
- Wang, Y.; Chakrabarti, D.; Wang, C.; Faloutsos, C. Epidemic spreading in real networks: An eigenvalue viewpoint. In Proceedings of the 22nd International Symposium on Reliable Distributed Systems, Florence, Italy, 6–8 October 2003; pp. 25–34. [Google Scholar] [CrossRef]
- Newman, M. Spread of epidemic disease on networks. Phys. Rev. E 2002, 66. [Google Scholar] [CrossRef]
- Cohen, R.; Havlin, S.; ben Avraham, D. Efficient immunization strategies for computer networks and populations. Phys. Rev. Lett. 2003, 91, 247901. [Google Scholar] [CrossRef]
- Newman, M. The structure and function of complex networks. SIAM Rev. 2003, 45, 167–256. [Google Scholar] [CrossRef]
- Dorogovtsev, S.; Goltsev, A.; Mendes, J.; Samukhin, A. Spectra of complex networks. Phys. Rev. E 2003, 68, 046109. [Google Scholar] [CrossRef]
- Chakrabarti, D.; Faloutsos, C. Graph mining: Laws, generators, and algorithms. ACM Comput. Surv. 2006, 38, A1–A69. [Google Scholar] [CrossRef]
- Nadini, M.; Sun, K.; Ubaldi, E.; Starnini, M.; Rizzo, A.; Perra, N. Epidemic spreading in modular time-varying networks. Sci. Rep. 2018, 8, 2352. [Google Scholar] [CrossRef] [PubMed]
- Nematzadeh, A.; Ferrara, E.; Flammini, A.; Ahn, Y.Y. Optimal Network Modularity for Information Diffusion. Phys. Rev. Lett. 2014, 113, 088701, Erratum in Phys. Rev. Lett. 2014, 113, 259901. [Google Scholar] [CrossRef] [PubMed]
- Granell, C.; Gomez, S.; Arenas, A. Dynamical Interplay between Awareness and Epidemic Spreading in Multiplex Networks. Phys. Rev. Lett. 2013, 111, 128701. [Google Scholar] [CrossRef]
- Holme, P.; Saramaki, J. Temporal networks. Phys. Rep. Rev. Sect. Phys. Lett. 2012, 519, 97–125. [Google Scholar] [CrossRef]
- Masuda, N.; Miller, J.C.; Holme, P. Concurrency measures in the era of temporal network epidemiology: A review. J. R. Soc. Interface 2021, 18. [Google Scholar] [CrossRef]
- Valdano, E.; Ferreri, L.; Poletto, C.; Colizza, V. Analytical Computation of the Epidemic Threshold on Temporal Networks. Phys. Rev. X 2015, 5, 021005. [Google Scholar] [CrossRef]
- Rocha, L.E.C.; Blondel, V.D. Bursts of Vertex Activation and Epidemics in Evolving Networks. PLoS Comput. Biol. 2013, 9, e1002974. [Google Scholar] [CrossRef]
- Williams, O.E.; Lino, F.; Latora, V. Effects of memory on spreading processes in non-Markovian temporal networks. New J. Phys. 2019, 21, 043028. [Google Scholar] [CrossRef]
- Perra, N.; Goncalves, B.; Pastor-Satorras, R.; Vespignani, A. Activity driven modeling of time varying networks. Sci. Rep. 2012, 2, 469. [Google Scholar] [CrossRef]
- Sejunti, M.I.; Taylor, D.; Masuda, N. A Parrondo paradox in susceptible-infectious-susceptible dynamics over periodic temporal networks. Math. Biosci. 2024, 378, 109336. [Google Scholar] [CrossRef] [PubMed]
- Liben-Nowell, D.; Kleinberg, J. The link-prediction problem for social networks. J. Am. Soc. Inf. Sci. Technol. 2007, 58, 1019–1031. [Google Scholar] [CrossRef]
- Lu, L.; Zhou, T. Link prediction in complex networks: A survey. Phys. A-Stat. Mech. Its Appl. 2011, 390, 1150–1170. [Google Scholar] [CrossRef]
- Moore, S.; Rogers, T. Predicting the Speed of Epidemics Spreading in Networks. Phys. Rev. Lett. 2020, 124, 068301. [Google Scholar] [CrossRef]
- Gross, T.; D’Lima, C.J.D.; Blasius, B. Epidemic dynamics on an adaptive network. Phys. Rev. Lett. 2006, 96, 208701. [Google Scholar] [CrossRef]
- Shaw, L.B.; Schwartz, I.B. Enhanced vaccine control of epidemics in adaptive networks. Phys. Rev. E 2010, 81, 046120. [Google Scholar] [CrossRef]
- Li, R.; Guo, X. Dynamics of a Stochastic SEIR Epidemic Model with Vertical Transmission and Standard Incidence. Mathematics 2024, 12, 359. [Google Scholar] [CrossRef]
- Hernandez Guillen, J.D.; Martin del Rey, A.; Hernandez Encinas, L. Study of the stability of a SEIRS model for computer worm propagation. Phys. A-Stat. Mech. Its Appl. 2017, 479, 411–421. [Google Scholar] [CrossRef]
- Hosseini, S.; Zandvakili, A. The SEIRS-C model of information diffusion based on rumour spreading with fuzzy logic in social networks. Int. J. Comput. Math. 2022, 99, 1918–1940. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, H. Stability and Hopf Bifurcation in a Delayed SEIRS Worm Model in Computer Network. Math. Probl. Eng. 2013, 2013, 319174. [Google Scholar] [CrossRef]
- Asadi, E.; Hosseini, S. A clustered-LPSEIRS malware propagation model in complex networks. Appl. Netw. Sci. 2024, 9, 62. [Google Scholar] [CrossRef]
| Symbol | Description |
|---|---|
| N | Total number of nodes (individuals) in the network. |
| Adjacency matrix entry (1 if edge between exists, 0 otherwise). | |
| Infection rate for intra-community contacts. | |
| Infection rate for inter-community contacts. | |
| Infection rate for predicted links | |
| Progression rate from Exposed (E) to Infectious (I) | |
| Recovery rate from Infectious (I) to Recovered (R) | |
| Immunity waning rate from Recovered (R) back to Susceptible (S). | |
| Period of node i’s activity cycle (in discrete time steps). | |
| Periodic online status function for node i at time t. | |
| Phase offset of node i’s activity cycle (phase of initial activation). | |
| Duty cycle (fraction of each period that node i is online). |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, G.; Shi, Q.; Liu, Y. A Clustered Link-Prediction SEIRS Model with Temporal Node Activation for Modeling Computer Virus Propagation in Urban Communication Systems. AppliedMath 2025, 5, 128. https://doi.org/10.3390/appliedmath5040128
Chen G, Shi Q, Liu Y. A Clustered Link-Prediction SEIRS Model with Temporal Node Activation for Modeling Computer Virus Propagation in Urban Communication Systems. AppliedMath. 2025; 5(4):128. https://doi.org/10.3390/appliedmath5040128
Chicago/Turabian StyleChen, Guiqiang, Qian Shi, and Yijun Liu. 2025. "A Clustered Link-Prediction SEIRS Model with Temporal Node Activation for Modeling Computer Virus Propagation in Urban Communication Systems" AppliedMath 5, no. 4: 128. https://doi.org/10.3390/appliedmath5040128
APA StyleChen, G., Shi, Q., & Liu, Y. (2025). A Clustered Link-Prediction SEIRS Model with Temporal Node Activation for Modeling Computer Virus Propagation in Urban Communication Systems. AppliedMath, 5(4), 128. https://doi.org/10.3390/appliedmath5040128






