Erythrocyte Folyl Polyglutamate Synthetase Activity Profiling as a Potential Tool for the Prediction of Methotrexate Efficacy and Toxicity in Rheumatoid Arthritis
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Patient Information
2.3. Clinical Samples
2.4. FPGS Assay
2.5. UHPLC-MS/MS
2.5.1. Sample Preparation
2.5.2. Assay Conditions Tested for Optimization
2.5.3. MTX + Glu2 Detection and FPGS Activity Calculations
2.5.4. Enzyme Kinetics
2.5.5. Statistical Analysis
3. Results
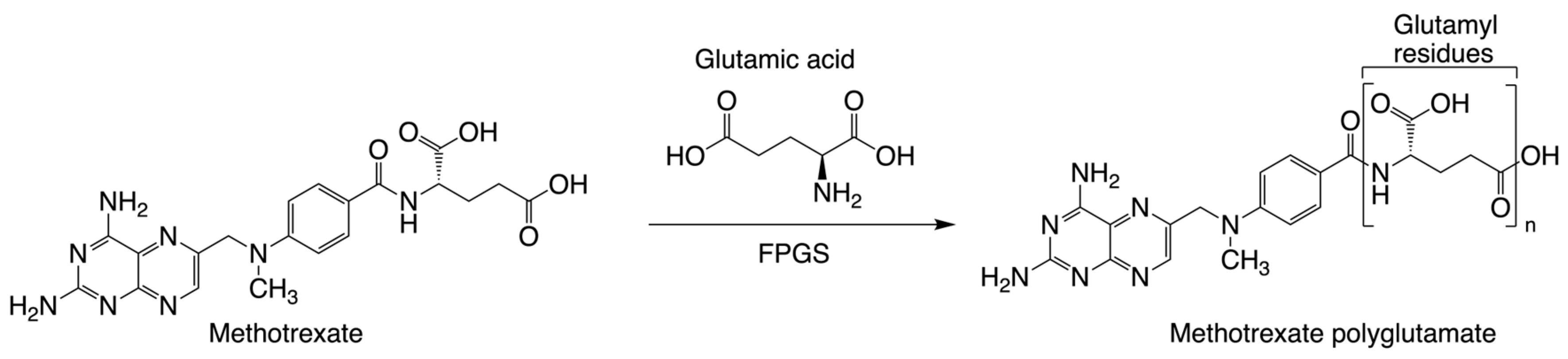
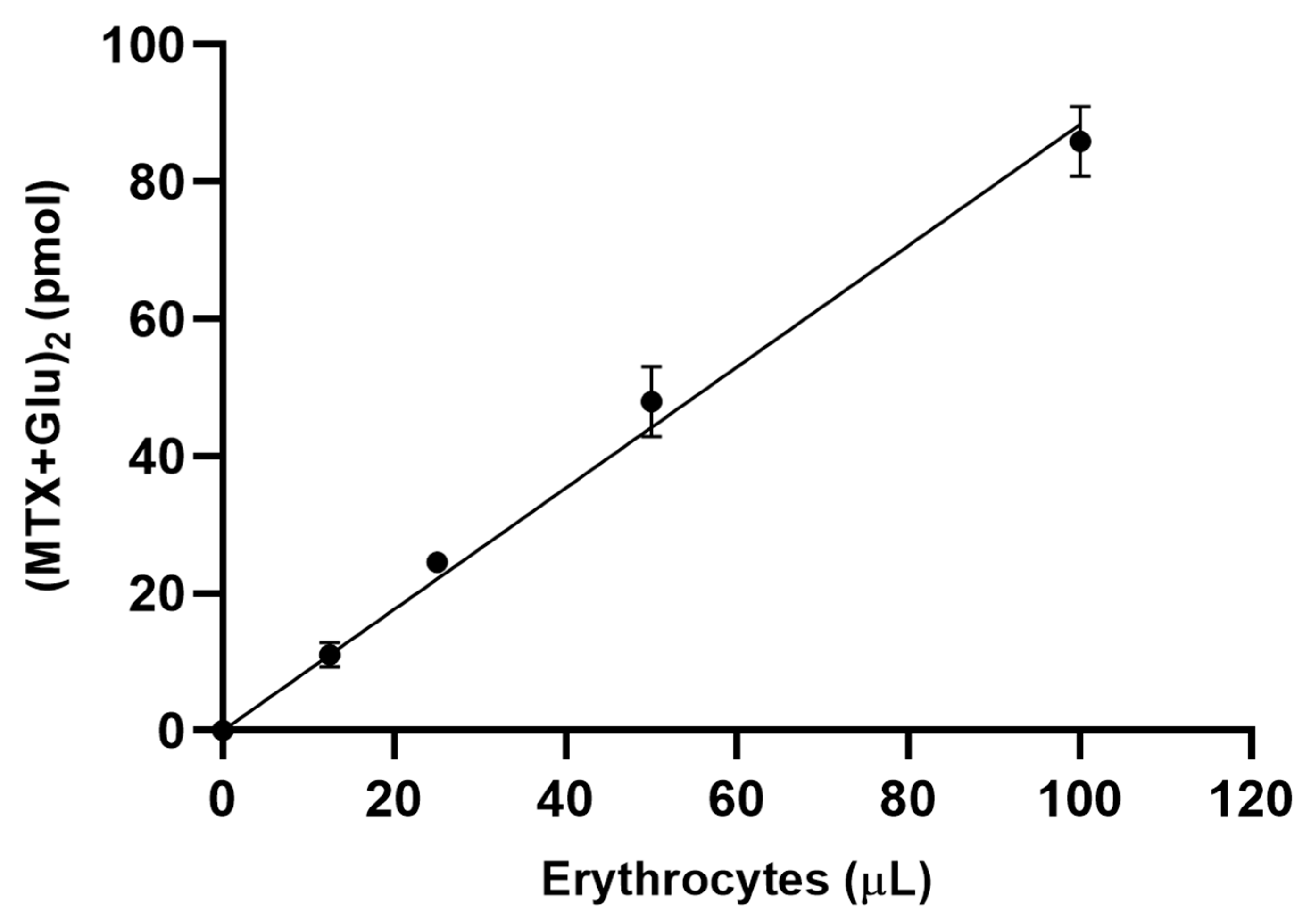
3.1. Assay Development/Optimization/Validation
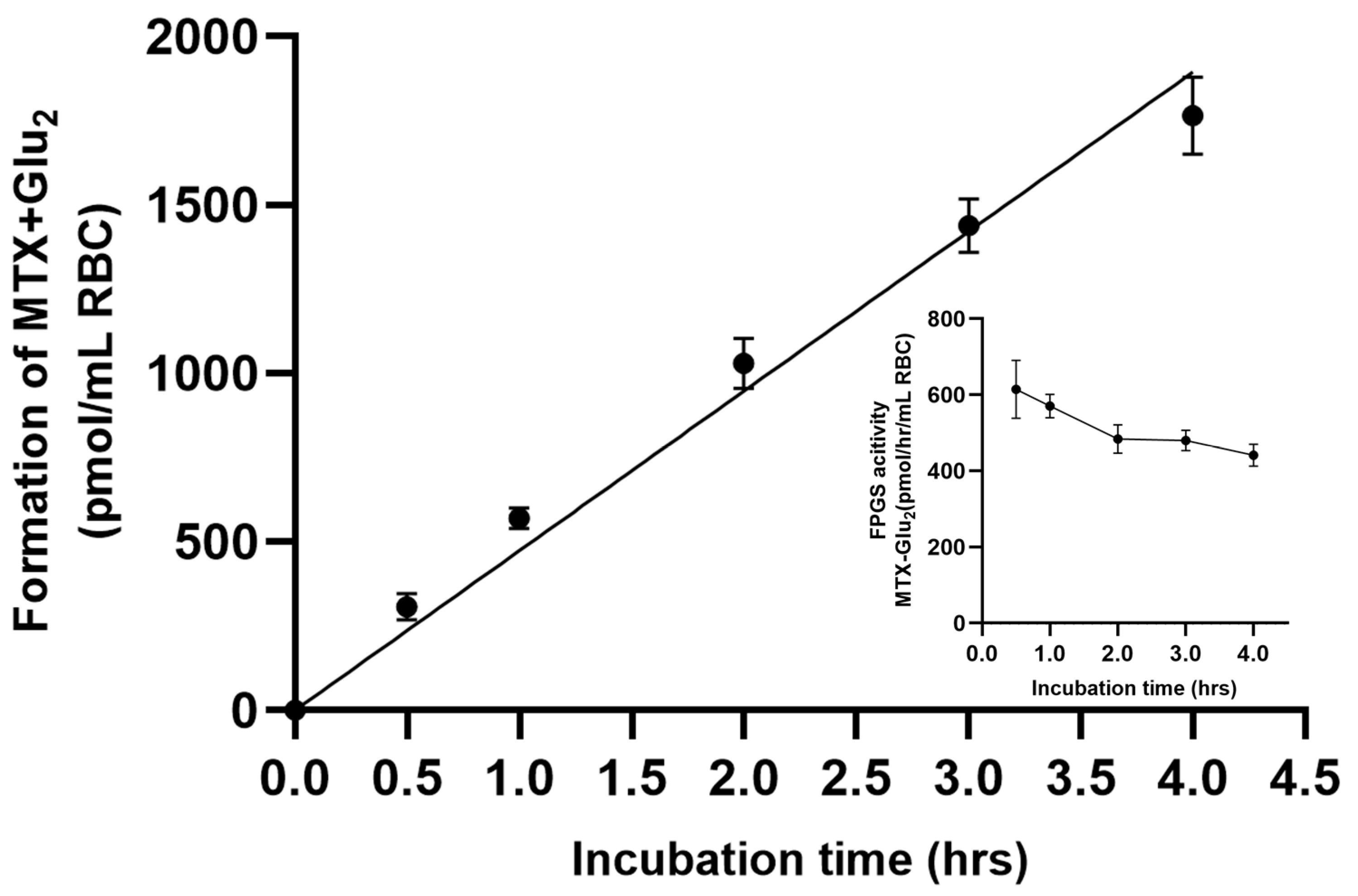
3.2. Enzyme Kinetics Analysis
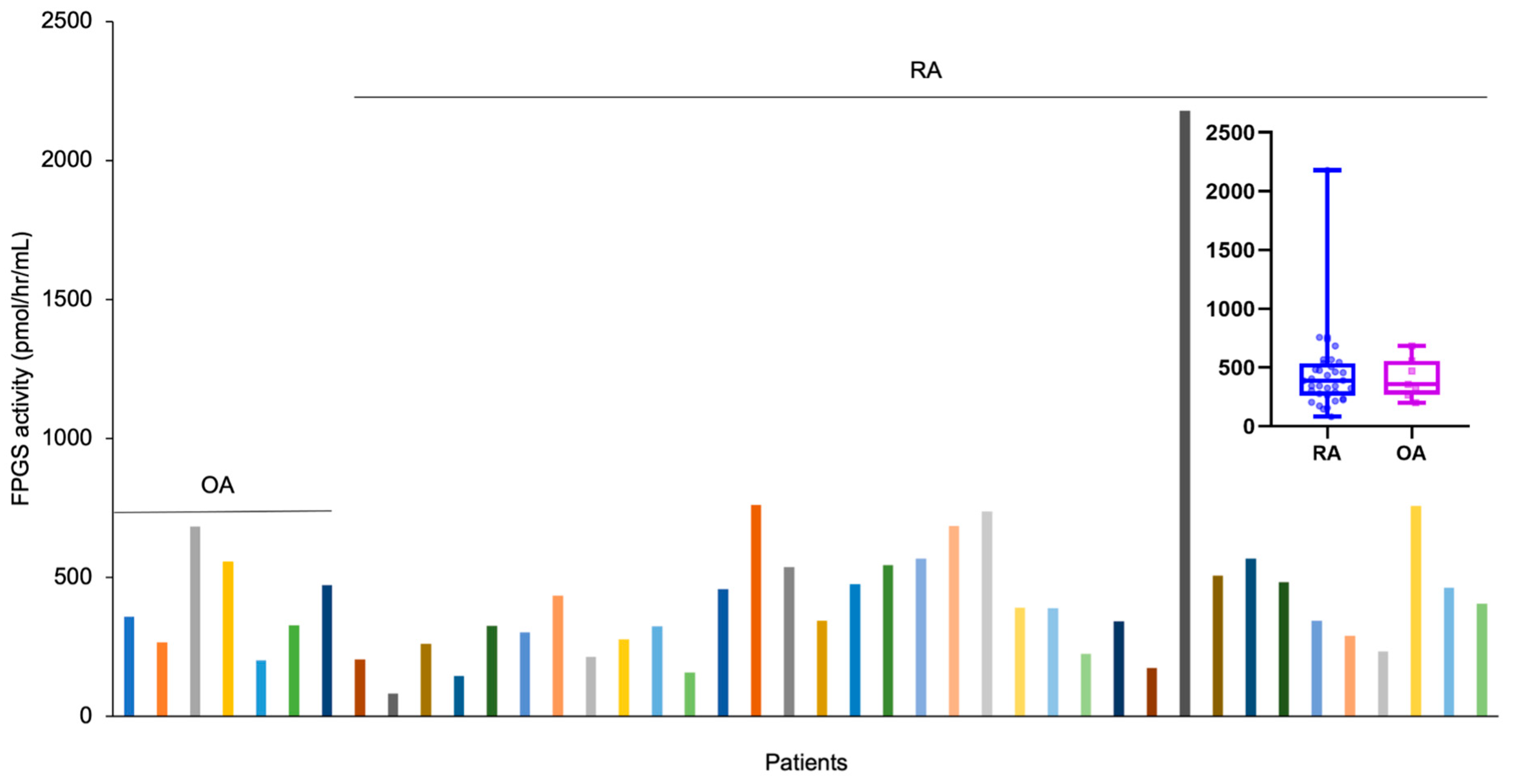
3.3. FPGS Activity in Arthritis Patients
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
References
- Visentin, M.; Zhao, R.; Goldman, I.D. The Antifolates. Hematol. Oncol. Clin. N. Am. 2012, 26, 629–648. [Google Scholar] [CrossRef] [PubMed]
- Kremer, J.M. Methotrexate and Leflunomide: Biochemical Basis for Combination Therapy in the Treatment of Rheumatoid Arthritis. Semin. Arthritis Rheum. 1999, 29, 14–26. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, O.; Goyal, A.; Lappin, S.L. Disease-Modifying Antirheumatic Drugs (DMARD). In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Swierkot, J.; Szechiński, J. Methotrexate in Rheumatoid Arthritis. Pharmacol. Rep. PR 2006, 58, 473–492. [Google Scholar] [PubMed]
- Taylor, P.C.; Balsa Criado, A.; Mongey, A.-B.; Avouac, J.; Marotte, H.; Mueller, R.B. How to Get the Most from Methotrexate (MTX) Treatment for Your Rheumatoid Arthritis Patient?-MTX in the Treat-to-Target Strategy. J. Clin. Med. 2019, 8, 515. [Google Scholar] [CrossRef]
- Lopez-Olivo, M.A.; Siddhanamatha, H.R.; Shea, B.; Tugwell, P.; Wells, G.A.; Suarez-Almazor, M.E. Methotrexate for Treating Rheumatoid Arthritis. Cochrane Database Syst. Rev. 2014, 2014, CD000957. [Google Scholar] [CrossRef]
- Dogra, S.; Singh, N.; Kumar, S.; Narang, T.; Handa, S. Comparison of Overall Efficacy and Safety of Oral versus Subcutaneous Methotrexate in Severe Psoriasis. Dermatol. Ther. 2022, 35, e15656. [Google Scholar] [CrossRef] [PubMed]
- Raza, K.; Buckley, C.E.; Salmon, M.; Buckley, C.D. Treating Very Early Rheumatoid Arthritis. Best Pract. Res. Clin. Rheumatol. 2006, 20, 849–863. [Google Scholar] [CrossRef] [PubMed]
- Bedoui, Y.; Guillot, X.; Sélambarom, J.; Guiraud, P.; Giry, C.; Jaffar-Bandjee, M.C.; Ralandison, S.; Gasque, P. Methotrexate an Old Drug with New Tricks. Int. J. Mol. Sci. 2019, 20, 5023. [Google Scholar] [CrossRef]
- Plant, D.; Maciejewski, M.; Smith, S.; Nair, N.; Maximising Therapeutic Utility in Rheumatoid Arthritis Consortium, the RAMS Study Group; Hyrich, K.; Ziemek, D.; Barton, A.; Verstappen, S. Profiling of Gene Expression Biomarkers as a Classifier of Methotrexate Nonresponse in Patients With Rheumatoid Arthritis. Arthritis Rheumatol. Hoboken NJ 2019, 71, 678–684. [Google Scholar] [CrossRef]
- Emery, P.; Pope, J.E.; Kruger, K.; Lippe, R.; DeMasi, R.; Lula, S.; Kola, B. Efficacy of Monotherapy with Biologics and JAK Inhibitors for the Treatment of Rheumatoid Arthritis: A Systematic Review. Adv. Ther. 2018, 35, 1535–1563. [Google Scholar] [CrossRef]
- Weinblatt, M.E. Methotrexate in Rheumatoid Arthritis: A Quarter Century of Development. Trans. Am. Clin. Climatol. Assoc. 2013, 124, 16–25. [Google Scholar]
- Fleischmann, R.; Schiff, M.; van der Heijde, D.; Ramos-Remus, C.; Spindler, A.; Stanislav, M.; Zerbini, C.A.F.; Gurbuz, S.; Dickson, C.; de Bono, S.; et al. Baricitinib, Methotrexate, or Combination in Patients With Rheumatoid Arthritis and No or Limited Prior Disease-Modifying Antirheumatic Drug Treatment. Arthritis Rheumatol. Hoboken NJ 2017, 69, 506–517. [Google Scholar] [CrossRef]
- de Rotte, M.C.F.J.; den Boer, E.; de Jong, P.H.P.; Pluijm, S.M.F.; Ćalasan, M.B.; Weel, A.E.; Huisman, A.M.; Gerards, A.H.; van Schaeybroeck, B.; Wulffraat, N.M.; et al. Methotrexate Polyglutamates in Erythrocytes Are Associated with Lower Disease Activity in Patients with Rheumatoid Arthritis. Ann. Rheum. Dis. 2015, 74, 408–414. [Google Scholar] [CrossRef]
- Pan, S.; Stamp, L.K.; Duffull, S.B.; Barclay, M.L.; Dalrymple, J.M.; Drake, J.; Zhang, M.; Korell, J. Assessment of the Relationship between Methotrexate Polyglutamates in Red Blood Cells and Clinical Response in Patients Commencing Methotrexate for Rheumatoid Arthritis. Clin. Pharmacokinet. 2014, 53, 1161–1170. [Google Scholar] [CrossRef]
- Daraghmeh, D.N.; Moghaddami, M.; Bobrovskaya, L.; Proudman, S.M.; Wiese, M.D. Quantitation of Methotrexate Polyglutamates in Human Whole Blood, Erythrocytes and Leukocytes Collected via Venepuncture and Volumetric Absorptive Micro-Sampling: A Green LC-MS/MS-Based Method. Anal. Bioanal. Chem. 2022, 414, 6029–6046. [Google Scholar] [CrossRef]
- Becker, M.L.; van Haandel, L.; Gaedigk, R.; Lasky, A.; Hoeltzel, M.; Stobaugh, J.; Leeder, J.S. Analysis of Intracellular Methotrexate Polyglutamates in Patients with Juvenile Idiopathic Arthritis: Effect of Route of Administration on Variability in Intracellular Methotrexate Polyglutamate Concentrations. Arthritis Rheum. 2010, 62, 1803–1812. [Google Scholar] [CrossRef]
- Hawwa, A.F.; AlBawab, A.; Rooney, M.; Wedderburn, L.R.; Beresford, M.W.; McElnay, J.C. Methotrexate Polyglutamates as a Potential Marker of Adherence to Long-Term Therapy in Children with Juvenile Idiopathic Arthritis and Juvenile Dermatomyositis: An Observational, Cross-Sectional Study. Arthritis Res. Ther. 2015, 17, 295. [Google Scholar] [CrossRef]
- van de Meeberg, M.M.; Hebing, R.C.F.; Nurmohamed, M.T.; Fidder, H.H.; Heymans, M.W.; Bouma, G.; de Bruin-Weller, M.S.; Tekstra, J.; van den Bemt, B.; de Jonge, R.; et al. A Meta-Analysis of Methotrexate Polyglutamates in Relation to Efficacy and Toxicity of Methotrexate in Inflammatory Arthritis, Colitis and Dermatitis. Br. J. Clin. Pharmacol. 2023, 89, 61–79. [Google Scholar] [CrossRef]
- Dervieux, T.; Zablocki, R.; Kremer, J. Red Blood Cell Methotrexate Polyglutamates Emerge as a Function of Dosage Intensity and Route of Administration during Pulse Methotrexate Therapy in Rheumatoid Arthritis. Rheumatol. Oxf. Engl. 2010, 49, 2337–2345. [Google Scholar] [CrossRef]
- Halilova, K.I.; Brown, E.E.; Morgan, S.L.; Bridges, S.L.; Hwang, M.-H.; Arnett, D.K.; Danila, M.I. Markers of Treatment Response to Methotrexate in Rheumatoid Arthritis: Where Do We Stand? Int. J. Rheumatol. 2012, 2012, 978396. [Google Scholar] [CrossRef]
- Yan, H.; Su, R.; Xue, H.; Gao, C.; Li, X.; Wang, C. Pharmacomicrobiology of Methotrexate in Rheumatoid Arthritis: Gut Microbiome as Predictor of Therapeutic Response. Front. Immunol. 2021, 12, 789334. [Google Scholar] [CrossRef]
- Takahashi, C.; Kaneko, Y.; Okano, Y.; Taguchi, H.; Oshima, H.; Izumi, K.; Yamaoka, K.; Takeuchi, T. Association of Erythrocyte Methotrexate-Polyglutamate Levels with the Efficacy and Hepatotoxicity of Methotrexate in Patients with Rheumatoid Arthritis: A 76-Week Prospective Study. RMD Open 2017, 3, e000363. [Google Scholar] [CrossRef]
- Wei, K.; Jiang, P.; Zhao, J.; Jin, Y.; Zhang, R.; Chang, C.; Xu, L.; Xu, L.; Shi, Y.; Guo, S.; et al. Biomarkers to Predict DMARDs Efficacy and Adverse Effect in Rheumatoid Arthritis. Front. Immunol. 2022, 13, 865267. [Google Scholar] [CrossRef]
- Becker, M.L. Using Methotrexate Metabolites to Make Clinical Decisions in JIA. Curr. Treat. Options Rheumatol. 2015, 1, 381–395. [Google Scholar] [CrossRef]
- Castaldo, P.; Magi, S.; Assunta Nasti, A.; Arcangeli, S.; Lariccia, V.; Alesi, N.; Tocchini, M.; Amoroso, S. Clinical Pharmacogenetics of Methotrexate. Curr. Drug Metab. 2011, 12, 278–286. [Google Scholar] [CrossRef]
- Brown, P.M.; Pratt, A.G.; Isaacs, J.D. Mechanism of Action of Methotrexate in Rheumatoid Arthritis, and the Search for Biomarkers. Nat. Rev. Rheumatol. 2016, 12, 731–742. [Google Scholar] [CrossRef]
- Stamp, L.K.; Roberts, R.L. Effect of Genetic Polymorphisms in the Folate Pathway on Methotrexate Therapy in Rheumatic Diseases. Pharmacogenomics 2011, 12, 1449–1463. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Cantley, L.C. Toward a Better Understanding of Folate Metabolism in Health and Disease. J. Exp. Med. 2019, 216, 253–266. [Google Scholar] [CrossRef]
- Raz, S.; Stark, M.; Assaraf, Y.G. Folylpoly-γ-Glutamate Synthetase: A Key Determinant of Folate Homeostasis and Antifolate Resistance in Cancer. Drug Resist. Updat. 2016, 28, 43–64. [Google Scholar] [CrossRef]
- Shane, B. Folate chemistry and metabolism*. Clin. Res. Regul. Aff. 2001, 18, 137–159. [Google Scholar] [CrossRef]
- Moran, R.G. Roles of Folylpoly-Gamma-Glutamate Synthetase in Therapeutics with Tetrahydrofolate Antimetabolites: An Overview. Semin. Oncol. 1999, 26 (Suppl. S6), 24–32. [Google Scholar] [PubMed]
- Stranzl, T.; Wolf, J.; Leeb, B.F.; Smolen, J.S.; Pirker, R.; Filipits, M. Expression of Folylpolyglutamyl Synthetase Predicts Poor Response to Methotrexate Therapy in Patients with Rheumatoid Arthritis. Clin. Exp. Rheumatol. 2003, 21, 27–32. [Google Scholar] [PubMed]
- Muller, I.B.; Lin, M.; Lems, W.F.; Ter Wee, M.M.; Wojtuszkiewicz, A.; Nurmohamed, M.T.; Cloos, J.; Assaraf, Y.G.; Jansen, G.; de Jonge, R. Association of Altered Folylpolyglutamate Synthetase Pre-mRNA Splicing with Methotrexate Unresponsiveness in Early Rheumatoid Arthritis. Rheumatol. Oxf. Engl. 2021, 60, 1273–1281. [Google Scholar] [CrossRef] [PubMed]
- Muller, I.B.; Lin, M.; Struys, E.A.; Heydari, P.; Hebing, R.C.F.; Nurmohamed, M.T.; Van Der Laken, C.; Lems, W.F.; Cloos, J.; Jansen, G.; et al. Development and Validation of a Sensitive UHPLC-MS/MS–Based Method for the Analysis of Folylpolyglutamate Synthetase Enzymatic Activity in Peripheral Blood Mononuclear Cells: Application in Rheumatoid Arthritis and Leukemia Patients. Ther. Drug Monit. 2019, 41, 598–606. [Google Scholar] [CrossRef]
- den Boer, E.; Meesters, R.J.W.; Van Zelst, B.D.; Luider, T.M.; Hazes, J.M.W.; Heil, S.G.; de Jonge, R. Measuring Methotrexate Polyglutamates in Red Blood Cells: A New LC-MS/MS-Based Method. Anal. Bioanal. Chem. 2012, 405, 1673–1681. [Google Scholar] [CrossRef]
- Liani, E.; Rothem, L.; Bunni, M.A.; Smith, C.A.; Jansen, G.; Assaraf, Y.G. Loss of Folylpoly-?-Glutamate Synthetase Activity Is a Dominant Mechanism of Resistance to Polyglutamylation-Dependent Novel Antifolates in Multiple Human Leukemia Sublines. Int. J. Cancer 2003, 103, 587–599. [Google Scholar] [CrossRef]
- Leil, T.A.; Endo, C.; Adjei, A.A.; Dy, G.K.; Salavaggione, O.E.; Reid, J.R.; Ames, M.M.; Adjei, A.A. Identification and Characterization of Genetic Variation in the Folylpolyglutamate Synthase Gene. Cancer Res. 2007, 67, 8772–8782. [Google Scholar] [CrossRef]
- Chen, L.; Qi, H.; Korenberg, J.; Garrow, T.A.; Choi, Y.-J.; Shane, B. Purification and Properties of Human Cytosolic Folylpoly-γ-Glutamate Synthetase and Organization, Localization, and Differential Splicing of Its Gene. J. Biol. Chem. 1996, 271, 13077–13087. [Google Scholar] [CrossRef]
- Kovarik, M.L.; Allbritton, N.L. Measuring Enzyme Activity in Single Cells. Trends Biotechnol. 2011, 29, 222–230. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, O.J.; Kim, S.-J.; Sohn, Y.B.; Park, H.-D.; Lee, S.-Y.; Kim, C.-H.; Ko, A.-R.; Yook, Y.-J.; Lee, S.-J.; Park, S.W.; et al. A Study of the Relationship between Clinical Phenotypes and Plasma Iduronate-2-Sulfatase Enzyme Activities in Hunter Syndrome Patients. Korean J. Pediatr. 2012, 55, 88. [Google Scholar] [CrossRef]
- Huen, K.; Richter, R.; Furlong, C.; Eskenazi, B.; Holland, N. Validation of PON1 Enzyme Activity Assays for Longitudinal Studies. Clin. Chim. Acta 2009, 402, 67–74. [Google Scholar] [CrossRef]
- Wang, S.; Ramamurthy, D.; Tan, J.; Liu, J.; Yip, J.; Chua, A.; Yu, Z.; Lim, T.K.; Lin, Q.; Pines, O.; et al. Post-Translational Modifications of Fumarase Regulate Its Enzyme Activity and Function in Respiration and the DNA Damage Response. J. Mol. Biol. 2020, 432, 6108–6126. [Google Scholar] [CrossRef]
- Härmä, H.; Tong-Ochoa, N.; Van Adrichem, A.J.; Jelesarov, I.; Wennerberg, K.; Kopra, K. Toward Universal Protein Post-Translational Modification Detection in High Throughput Format. Chem. Commun. 2018, 54, 2910–2913. [Google Scholar] [CrossRef] [PubMed]
- Bisswanger, H. Enzyme Assays. Perspect. Sci. 2014, 1, 41–55. [Google Scholar] [CrossRef]
- Angelis-Stoforidis, P.; Vajda, F.J.; Christophidis, N. Methotrexate Polyglutamate Levels in Circulating Erythrocytes and Polymorphs Correlate with Clinical Efficacy in Rheumatoid Arthritis. Clin. Exp. Rheumatol. 1999, 17, 313–320. [Google Scholar] [PubMed]
- Van De Meeberg, M.M.; Seinen, M.L.; Fidder, H.H.; Lin, M.; Oldenburg, B.; De Boer, N.K.; Bouma, G.; De Jonge, R.; Bulatović Ćalasan, M., on behalf of the Dutch Initiative on Crohn and Colitis (ICC). Subcutaneous Administration, Higher Age and Lower Renal Function Are Associated with Erythrocyte Methotrexate Accumulation in Crohn’s Disease: A Cross-Sectional Study. BMC Gastroenterol. 2022, 22, 365. [Google Scholar] [CrossRef]
- Barredo, J.; Moran, R.G. Determinants of Antifolate Cytotoxicity: Folylpolyglutamate Synthetase Activity during Cellular Proliferation and Development. Mol. Pharmacol. 1992, 42, 687–694. [Google Scholar] [PubMed]
- Dervieux, T.; Furst, D.; Lein, D.O.; Capps, R.; Smith, K.; Walsh, M.; Kremer, J. Polyglutamation of Methotrexate with Common Polymorphisms in Reduced Folate Carrier, Aminoimidazole Carboxamide Ribonucleotide Transformylase, and Thymidylate Synthase Are Associated with Methotrexate Effects in Rheumatoid Arthritis. Arthritis Rheum. 2004, 50, 2766–2774. [Google Scholar] [CrossRef] [PubMed]
- Dervieux, T. Pharmacogenetic and Metabolite Measurements Are Associated with Clinical Status in Patients with Rheumatoid Arthritis Treated with Methotrexate: Results of a Multicentred Cross Sectional Observational Study. Ann. Rheum. Dis. 2005, 64, 1180–1185. [Google Scholar] [CrossRef]
- Hornung, N.; Ellingsen, T.; Attermann, J.; Stengaard-Pedersen, K.; Poulsen, J.H. Patients with Rheumatoid Arthritis Treated with Methotrexate (MTX): Concentrations of Steady-State Erythrocyte MTX Correlate to Plasma Concentrations and Clinical Efficacy. J. Rheumatol. 2008, 35, 1709–1715. [Google Scholar] [PubMed]
- Chládek, J.; Simková, M.; Vanecková, J.; Hroch, M.; Chládkova, J.; Martínková, J.; Vávrová, J.; Beránek, M. The Effect of Folic Acid Supplementation on the Pharmacokinetics and Pharmacodynamics of Oral Methotrexate during the Remission-Induction Period of Treatment for Moderate-to-Severe Plaque Psoriasis. Eur. J. Clin. Pharmacol. 2008, 64, 347–355. [Google Scholar] [CrossRef]
- Schmiegelow, K.; Schrøder, H.; Gustafsson, G.; Kristinsson, J.; Glomstein, A.; Salmi, T.; Wranne, L. Risk of Relapse in Childhood Acute Lymphoblastic Leukemia Is Related to RBC Methotrexate and Mercaptopurine Metabolites during Maintenance Chemotherapy. Nordic Society for Pediatric Hematology and Oncology. J. Clin. Oncol. 1995, 13, 345–351. [Google Scholar] [CrossRef] [PubMed]
- TPMT Testing before Azathioprine Therapy? Drug Ther. Bull. 2009, 47, 9–12. [CrossRef]
- Danila, M.I.; Hughes, L.B.; Brown, E.E.; Morgan, S.L.; Baggott, J.E.; Arnett, D.K.; Bridges, S.L. Measurement of Erythrocyte Methotrexate Polyglutamate Levels: Ready for Clinical Use in Rheumatoid Arthritis? Curr. Rheumatol. Rep. 2010, 12, 342–347. [Google Scholar] [CrossRef][Green Version]
- Muller, I.B.; Hebing, R.F.; Jansen, G.; Nurmohamed, M.T.; Lems, W.F.; Peters, G.J.; De Jonge, R. Personalized Medicine in Rheumatoid Arthritis: Methotrexate Polyglutamylation Revisited. Expert Rev. Precis. Med. Drug Dev. 2018, 3, 331–334. [Google Scholar] [CrossRef]
- den Boer, E. Therapeutic Drug-Monitoring of Methotrexate-Polyglutamates in Rheumatoid Arthritis; Erasmus University Rotterdam: Rotterdam, The Netherlands, 2014. [Google Scholar]
- Dalrymple, J.M.; Stamp, L.K.; O’Donnell, J.L.; Chapman, P.T.; Zhang, M.; Barclay, M.L. Pharmacokinetics of Oral Methotrexate in Patients with Rheumatoid Arthritis. Arthritis Rheum. 2008, 58, 3299–3308. [Google Scholar] [CrossRef]
- Yamamoto, T.; Shikano, K.; Nanki, T.; Kawai, S. Folylpolyglutamate Synthase Is a Major Determinant of Intracellular Methotrexate Polyglutamates in Patients with Rheumatoid Arthritis. Sci. Rep. 2016, 6, 35615. [Google Scholar] [CrossRef]
- Owen, S.A.; Hider, S.L.; Martin, P.; Bruce, I.N.; Barton, A.; Thomson, W. Genetic Polymorphisms in Key Methotrexate Pathway Genes Are Associated with Response to Treatment in Rheumatoid Arthritis Patients. Pharmacogenom. J. 2013, 13, 227–234. [Google Scholar] [CrossRef]
- Pasnoor, M.; Heim, A.J.; Herbelin, L.; Statland, J.; Dimachkie, M.M.; Becker, M.; Barohn, R.J.; Methotrexate in MG Investigators of the Muscle Group Study. Methotrexate Polyglutamation in a Myasthenia Gravis Clinical Trial. Kans. J. Med. 2020, 13 (Suppl. S2), 10–13. [Google Scholar] [CrossRef]
- Zarou, M.M.; Vazquez, A.; Vignir Helgason, G. Folate Metabolism: A Re-Emerging Therapeutic Target in Haematological Cancers. Leukemia 2021, 35, 1539–1551. [Google Scholar] [CrossRef]
- Foldesi, B. Guide to Enzyme Unit Definitions and Assay Design. Biomol GmbH—Life Science Shop. Available online: https://www.biomol.com/resources/biomol-blog/guide-to-enzyme-unit-definitions-and-assay-design (accessed on 13 June 2023).
- Restriction Enzyme Resource Guide. Promega.com. Available online: https://www.promega.com/resources/guides/nucleic-acid-analysis/restriction-enzyme-resource/ (accessed on 13 June 2023).
- New England Biolabs. Optimizing Restriction Endonuclease Reactions|NEB. Neb.com. Available online: https://www.neb.com/tools-and-resources/usage-guidelines/optimizing-restriction-endonuclease-reactions (accessed on 13 June 2023).
- Cox, K.L.; Devanarayan, V.; Kriauciunas, A.; Manetta, J.; Montrose, C. Immunoassay Methods. Nih.gov. Available online: https://www.ncbi.nlm.nih.gov/books/NBK92434/ (accessed on 13 June 2023).
- Cox, K.L.; Devanarayan, V.; Kriauciunas, A.; Manetta, J.; Montrose, C.; Sittampalam, S. Immunoassay Methods. In Assay Guidance Manual; Markossian, S., Grossman, A., Brimacombe, K., Arkin, M., Auld, D., Austin, C., Baell, J., Chung, T.D.Y., Coussens, N.P., Dahlin, J.L., et al., Eds.; Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda, MD, USA, 2004. [Google Scholar]
- Voet, D.; Voet, J.G.; Pratt, C.W. Fundamentals of Biochemistry: Life at the Molecular Level, 5th ed.; Wiley: Hoboken, NJ, USA, 2016. [Google Scholar]
- Boyer, R.F. Concepts in Biochemistry, 2nd ed.; John Wiley & Sons, Inc.: New York, NY, USA, 2002. [Google Scholar]
- Punekar, N.S. Enzymes: Catalysis, Kinetics and Mechanisms; Springer: Singapore, 2018. [Google Scholar]
- CTG Labs—NCBI. clinicaltrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT00695188 (accessed on 2 October 2023).
- Becker, M.L. Role of Methotrexate in Juvenile Idiopathic Arthritis: Where We Have Been and Where We Are Going. Int. J. Clin. Rheumatol. 2013, 8, 123–135. [Google Scholar] [CrossRef]
- Den Boer, E.; De Rotte, M.C.J.F.; Pluijm, S.M.F.; Heil, S.G.; Hazes, J.M.; De Jonge, R. Determinants of Erythrocyte Methotrexate Polyglutamate Levels in Rheumatoid Arthritis. J. Rheumatol. 2014, 41, 2167–2178. [Google Scholar] [CrossRef]
- Jansen, G. Folate Supplementations for Methotrexate Therapies in Cancer and Arthritis: Rationales Revisited. J. Mol. Clin. Med. 2022, 5, 1. [Google Scholar] [CrossRef]
- Qiu, Q.; Huang, J.; Shu, X.; Fan, H.; Zhou, Y.; Xiao, C. Polymorphisms and Pharmacogenomics for the Clinical Efficacy of Methotrexate in Patients with Rheumatoid Arthritis: A Systematic Review and Meta-Analysis. Sci. Rep. 2017, 7, 44015. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, A.; Alshamarri, T.; Adeyeye, T.; Lazariu, V.; McNutt, L.-A.; Carpenter, D.O. A Comparison of Risk Factors for Osteo- and Rheumatoid Arthritis Using NHANES Data. Prev. Med. Rep. 2020, 20, 101242. [Google Scholar] [CrossRef]
- Stamp, L.K.; O’Donnell, J.L.; Chapman, P.T.; Zhang, M.; Frampton, C.; James, J.; Barclay, M.L. Determinants of Red Blood Cell Methotrexate Polyglutamate Concentrations in Rheumatoid Arthritis Patients Receiving Long-Term Methotrexate Treatment. Arthritis Rheum. 2009, 60, 2248–2256. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Yuasa, H. Molecular Basis for Pharmacokinetics and Pharmacodynamics of Methotrexate in Rheumatoid Arthritis Therapy. Drug Metab. Pharmacokinet. 2014, 29, 12–19. [Google Scholar] [CrossRef]
- Intriago, M.; Maldonado, G.; Cárdenas, J.; Ríos, C. Clinical Characteristics in Patients with Rheumatoid Arthritis: Differences between Genders. Sci. World J. 2019, 2019, 8103812. [Google Scholar] [CrossRef]
- van Vollenhoven, R.F. Sex Differences in Rheumatoid Arthritis: More than Meets the Eye. BMC Med. 2009, 7, 12. [Google Scholar] [CrossRef]

| Kinetic Parameters | Mean | Standard Deviation |
|---|---|---|
| Vmax (pmol/h/mL)−1 | 611.95 | 193.36 |
| Km (µM)−1 | 30.29 | 4.81 |
| Intra-Day | Inter-Day | |||
|---|---|---|---|---|
| Day-1 | Day-2 | Day-3 | Combined | |
| Average (pmol/h/mL) | 754.9 | 599.6 | 642.2 | 665.6 |
| Standard deviation (%) | 3.03 | 7.90 | 3.10 | 12.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, A.; Arain, M.I.; Bhadbhade, P.; Funk, R. Erythrocyte Folyl Polyglutamate Synthetase Activity Profiling as a Potential Tool for the Prediction of Methotrexate Efficacy and Toxicity in Rheumatoid Arthritis. Future Pharmacol. 2023, 3, 819-833. https://doi.org/10.3390/futurepharmacol3040049
Kumar A, Arain MI, Bhadbhade P, Funk R. Erythrocyte Folyl Polyglutamate Synthetase Activity Profiling as a Potential Tool for the Prediction of Methotrexate Efficacy and Toxicity in Rheumatoid Arthritis. Future Pharmacology. 2023; 3(4):819-833. https://doi.org/10.3390/futurepharmacol3040049
Chicago/Turabian StyleKumar, Amar, Mudassar Iqbal Arain, Pooja Bhadbhade, and Ryan Funk. 2023. "Erythrocyte Folyl Polyglutamate Synthetase Activity Profiling as a Potential Tool for the Prediction of Methotrexate Efficacy and Toxicity in Rheumatoid Arthritis" Future Pharmacology 3, no. 4: 819-833. https://doi.org/10.3390/futurepharmacol3040049
APA StyleKumar, A., Arain, M. I., Bhadbhade, P., & Funk, R. (2023). Erythrocyte Folyl Polyglutamate Synthetase Activity Profiling as a Potential Tool for the Prediction of Methotrexate Efficacy and Toxicity in Rheumatoid Arthritis. Future Pharmacology, 3(4), 819-833. https://doi.org/10.3390/futurepharmacol3040049







