Abstract
Although methotrexate (MTX) is the first line disease-modifying therapy used in the treatment of autoimmune arthritis, it is limited by its unpredictable and variable response profile and lack of therapeutic biomarkers to predict or monitor therapeutic response. The purpose of this work is to evaluate the utility of red blood cell (RBC) metabolite profiles to screen for molecular biomarkers associated with MTX response. Methods: Utilizing the collagen-induced arthritis mouse model, DBA/1J mice were treated with subcutaneous MTX (20 mg/kg/week) and RBC samples were collected and analyzed by semi-targeted global metabolomic profiling and analyzed by univariate analysis. Results: MTX treatment normalized the following RBC metabolite levels that were found to be altered by disease induction: N-methylisoleucine, nudifloramide, phenylacetylglycine, 1-methyl-L-histidine, PC 42:1, PE 36:4e, PC 42:3, PE 36:4e (16:0e/20:4), and SM d34:0. Changes in the RBC metabolome weakly but significantly correlated with changes in the plasma metabolome following MTX treatment (ρ = 0.24, p = 1.1 × 10−13). The RBC metabolome resulted in the detection of nine significant discriminatory biomarkers, whereas the plasma metabolome resulted in two. Overall, the RBC metabolome yielded more highly sensitive and specific biomarkers of MTX response compared to the plasma metabolome. N-methylisoleucine was found to be highly discriminatory in both plasma and RBCs. Conclusions: Our results suggest that RBCs represent a promising biological matrix for metabolomics and future studies should consider the RBC metabolome in their biomarker discovery strategy.
1. Introduction
Methotrexate (MTX) is a first-line therapy in the treatment of chronic autoimmune conditions, such as rheumatoid arthritis (RA), and is an antimetabolite that targets folate metabolism [1,2,3,4,5,6]. However, MTX therapy is characterized by a widely variable response rate with approximately two-thirds of patients failing to achieve satisfactory response within six months [7,8]. To date, no established pretreatment or early treatment clinical biomarkers exist to stratify RA patients based on their likelihood to adequately respond to MTX [3,9]. Consequentially, the focus of this work is to evaluate the potential utility of red blood cell (RBC) metabolite profiles in screening for molecular biomarkers associated with MTX response in an autoimmune arthritis mouse model.
The collagen-induced arthritis (CIA) mouse model is the most extensively utilized animal model to study the development, progression, and therapeutic response of autoimmune arthritis. The model’s systemic inflammatory response mimics the physiologic and pathogenic characteristics of RA and demonstrably responds to disease modifying therapies, such as MTX [7,10,11,12].
Metabolomics has become particularly useful in precision medicine to identify biomarkers of disease and therapeutic response. Metabolomics is an emerging “omics” science concerned with the comprehensive analysis of small molecule metabolites within a biological system [12,13,14]. The human metabolome is comprised of intermediates, products, and side-products resulting from interactions between the genome, exposome (i.e., environmental exposures), and gut microbiome. Thus, the metabolome (sum of all metabolites in an organism) is a highly sensitive measure of an organism’s phenotype and provides insight into metabolic changes associated with a physiologic state, such as development of disease or exposure/response to drug therapy [15,16,17,18,19].
Initially, urine was the favored biofluid used in metabolomics for its non-invasive collection method and ease of processing. However, urine is a biological waste material containing mostly breakdown products in which metabolite concentrations can drastically vary based on timing of collection, limiting its applicability and practicality in biomarker discovery [13,20]. More recently, plasma or serum has become the preferred biofluid for metabolomic studies [21]. While arguably superior to urine, plasma and serum still exhibits a low level of metabolic activity [22]. Previous metabolomic studies of MTX response in autoimmune arthritis have primarily focused on analysis of synovial fluid, plasma, serum, and urine [23].
RBCs have previously been overlooked as a biofluid source in MTX metabolomic studies. RBCs are anucleated cells lacking multiple organelles, including mitochondria and ribosomes. In recent works, the understanding of RBC metabolism has been furthered through the application of the “-omic” sciences, predominately lipidomics and proteomics, in an effort to improve preservation and storage conditions of these cells for transfusion therapy [24]. As a result, RBCs are now known to be metabolically active cellular compartments which express a sophisticated metabolism [25,26]; this makes RBCs an intriguing prospect for metabolomic studies of MTX [26,27]. Therefore, the biochemical composition of RBCs and related biochemical pathways represents a cellular phenotype that may be informative about disease pathology and treatment response [22,25,28]. Furthermore, RBC folate concentration is considered by some to be the most reliable indicator of body folate status [29,30], which is highly pertinent when studying a folate antimetabolite drug like MTX and further supports the potential of the RBC metabolome as a relevant matrix for the identification of metabolomic markers of MTX response in autoimmune arthritis.
Altogether, RBCs could feasibly provide a more robust analysis for MTX response in autoimmune arthritis metabolomic studies, while being equally practical to obtain and process as plasma. As a result, this work utilizes a CIA mouse model to evaluate the performance of a semi-targeted metabolomic profiling approach to identify RBC metabolomic biomarkers of MTX response, and to compare their performance to metabolomic biomarkers identified using corresponding plasma samples.
2. Materials and Methods
2.1. Animals
The methodology of this study is described in depth in our previous publication [3]. Briefly, this study included 40 male DBA/1J mice at six to eight weeks of age from Jackson Laboratory (Bar Harbor, ME, USA). Mice were randomly assigned to one of four groups, including healthy control mice (n = 10), healthy control mice treated with MTX (n = 10), untreated CIA disease mice (n = 10), and CIA disease mice treated with MTX (n = 10). Mice were kept under pathogen-free conditions and all experimental procedures were conducted under a protocol reviewed and approved by the Institutional Animal Care and Use Committee at The University of Kansas Medical Center and are consistent with the Guide for the Care and Use of Laboratory Animals (Protocol ID: 2018-2481).
2.2. Disease Induction and Treatment
Mice (7 to 9 weeks old) were allowed a one-week acclimation period and then underwent the CIA disease induction protocol. This protocol included intradermal tail injections of a commercially prepared collagen emulsion in complete Freund’s adjuvant containing 1 mg/mL chicken type II collagen and 2 mg/mL killed mycobacterium tuberculosis H37Ra (Hooke Laboratories, Lawrence, MA, USA) given on experimental day 0. A subsequent booster dose on day 19 was given containing collagen emulsion in incomplete Freund’s adjuvant [7].
The final analysis included data from 37 mice that completed the study, with three mice expiring (two from the disease group and one from the MTX treated disease group). All three mice died following the booster injection and likely resulted from inadvertent injection into the tail vein. MTX treatment was initiated in all mice on day 14 upon signs of measurable disease activity scores. MTX injections were administered every seven days subcutaneously at a dose of 20 mg/kg for a total of six doses administered over a 54-day experimental period.
On experimental day 54, all mice were euthanized by CO2 asphyxiation, after which plasma and RBC samples were collected for analysis. Blood samples were collected in potassium EDTA-containing tubes and centrifuged at 1600× g for 10 min at room temperature, separating the blood samples into RBCs and plasma fractions which were stored at −80 °C prior to analysis.
2.3. Metabolomics Analysis
RBC and plasma samples were submitted for metabolomic analysis to the National Institutes of Health (NIH) West Coast Metabolomics Center at the University of California, Davis (Davis, CA, USA) and has been previously described [3]. Briefly, samples were prepared using standardized protocols established by the NIH West Coast Metabolomics Center and included biphasic liquid-liquid extraction with subsequent analysis using three independent standardized analytical methods that have been optimized for the detection and semi-quantitative analysis of intermediates of primary metabolism, biogenic amines, and lipids [31,32]. Metabolite identification was guided by retention times and mass spectral data accessed using MassBank of North America on 18 September 2019 (http://mona.fiehnlab.ucdavis.edu). Curated raw peak intensity data were provided and underwent a standardized normalization procedure to integrate the metabolomic data across the three analytical platforms. Normalized peak intensities for each identified metabolite was determined for each RBC and plasma sample as previously described [3]. Metabolites with duplicate detection across the analytical methods were combined by mean normalization of normalized peak intensities and averaged to preserve equal weighting between methods. Data analysis and visualization was conducted using MetaboAnalyst 5.0. Normalized peak intensities for multivariate analysis underwent logarithmic transformation and Pareto scaling. Changes in metabolite levels were represented based on fold-change and visualized using volcano plots.
2.4. Statistical Analysis
Following normalization of peak metabolite concentrations, statistical analyses were conducted using MetaboAnalyst 5.0. Peak intensities for each metabolite were compared between groups by non-parametric unpaired comparisons assuming unequal group variance. Significantly altered metabolites were defined based on a false-discovery rate (FDR) corrected p-value (q-value) threshold of 0.25. Spearman’s rank correlation analysis was used to determine correlation coefficients (ρ) and evaluate associations between continuous variables using JMP Pro 16 (SAS Institute, Cary, NC, USA). Classical univariate receiver operating characteristic curves were generated and the area under the curve (AUC) and p-values were determined for each metabolite to evaluate the performance of each of the identified biomarkers. Optimal cut-off points were determined based on the Youden index.
3. Results
3.1. RBC Metabolomic Differences Associated with CIA Disease Induction and the Effect of MTX
RBC samples were collected from mice at the end of a 54-day experimental period and included healthy control mice (n = 10), healthy control mice treated with MTX (n = 10), untreated CIA disease mice (n = 8), and CIA disease mice treated with MTX (n = 9). The results of CIA disease induction and MTX treatment on disease activity scores (DAS) and paw volumes (PV) for these mice have been previously published [3]. The median [IQR] DAS increased from 0 [0,0] in healthy control mice to 10 [6,11] in disease mice (p = 0.003), and was reduced to 1 [0,2] in disease mice treated with MTX (p = 0.0006). Similarly, median [IQR] paw volume increased from 0.36 [0.32,0.38] mL in healthy control mice to 0.48 [0.44,0.52] mL in disease mice (p = 0.0007), and was reduced to 0.37 [0.36,0.39] mL in disease mice treated with MTX (p = 0.0006). Albeit significantly reduced, MTX treated disease mice did have higher DAS scores compared to healthy control mice (p = 0.003). However, paw volume measurements did not significantly differ between healthy control mice and disease mice treated with MTX (p = 0.20). RBC and plasma datasets containing normalized peak intensities as well as DAS and PV are available in Supplementary Files S1 and S2, respectively.
RBC samples were analyzed by semi-targeted global metabolomic profiling and included intermediates of primary metabolism, lipids, and biogenic amines. RBC metabolomic data identified 1022 metabolites and were analyzed by unpaired univariate analysis, the results of which are presented as volcano plots (Figure 1). Metabolites significantly altered between healthy control and disease mice (Figure 1a) and between disease mice and disease mice treated with MTX (Figure 1b) were identified. Significant metabolites were defined based on an FDR-adjusted p-value (i.e., q-value) of less than 0.25; these metabolites are labeled by color, with blue dots indicating a decrease in metabolite concentrations and red dots indicating an increase. Thirty-three total metabolites were found to significantly differ between healthy control and disease mice (Figure 1a), of which 19 metabolites were decreased and 13 were increased. A total of 197 metabolites were found to differ between disease mice and disease mice treated with MTX (Figure 1b), where 21 metabolites decreased and 176 increased.
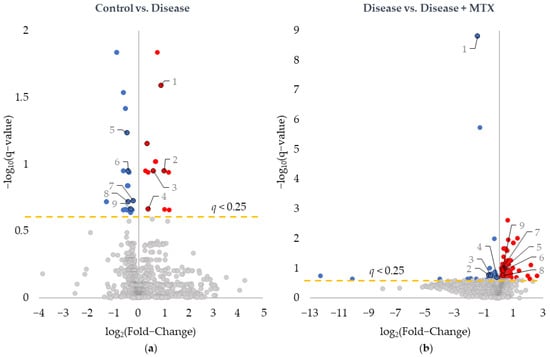
Figure 1.
Identification of metabolites of interest associated with CIA disease induction and MTX treatment. Univariate analysis of differences in the RBC metabolome resulted in volcano plots comparing (a) healthy control mice and disease mice and (b) disease mice and MTX treated disease mice. Metabolites were plotted based on log base 2 fold-change and negative log base 10 of the q-value. Red-colored metabolites were found to increase, and blue-colored metabolites were found to decrease (q-value < 0.25). Labeled metabolites were found to be significantly altered following disease induction and subsequently trend towards healthy control levels with MTX treatment.
Of the metabolites identified, nine were found to be altered upon disease induction in the healthy control mice and subsequently trended towards baseline levels in the disease mice treated with MTX. These metabolites are labeled on Figure 1. For each of the metabolites identified in Figure 1, the corresponding fold-change and q-values are provided for both comparisons (Table 1). With regard to raw p-values, all nine metabolites had raw p-values less than 0.005 for the comparison of healthy control and disease mice, and less than 0.02 in the comparison of disease and disease mice treated with MTX.

Table 1.
Metabolites altered in RBCs from CIA disease mice that are at least partially corrected in disease mice treated with MTX.
Spearman’s regression analyses were conducted to further evaluate the relationship of the identified RBC metabolites with disease activity and MTX response. Normalized peak intensities for each of the nine identified metabolites were evaluated for their relationship with DAS and PV measurements (Table 2). The Spearman’s correlation coefficient, or Spearman’s rho (ρ), and corresponding p-value for each comparison is provided. All of the identified metabolites demonstrated a statistically significant correlation with both DAS and PV. This observation further supports the relationship of the identified RBC metabolites with arthritis disease activity and response to MTX therapy in the CIA mouse model.

Table 2.
Spearman’s regression analysis evaluating the correlation between RBC metabolite levels and disease activity measurements.
3.2. Comparison of RBC and Plasma Metabolomic Differences Associated with MTX Treatment in the CIA Mouse Model
Having identified RBC metabolites associated with disease induction and MTX response in the CIA mouse model, metabolomic differences between disease mice and disease mice treated with MTX were compared in RBCs and plasma (Figure 2). Using a q-value threshold of 0.25, a total of 70 plasma metabolites were found to be altered with MTX treatment. The p-values for these metabolites ranged from 0.017 to 1.1 × 10−8. Meanwhile, 197 metabolites were significantly altered in RBCs with p-values ranging from 0.048 to 1.5 × 10−12. The increased number of metabolites altered in RBCs compared to plasma suggests that the RBC metabolome may be more sensitive and responsive to the effects of MTX therapy resulting in a broader metabolic change compared to what is observed in the plasma. A comparison between all metabolites appearing on both RBC and plasma metabolomes resulted in 940 compounds (i.e., 92.0% of the RBC metabolites were also present in the plasma metabolome) and found a weak but statistically significant correlation between the fold-changes between the two metabolomes (ρ = 0.24, p = 1.1 × 10−13). A total of 37 overlapping metabolites were found to be significantly altered in both the RBC and plasma analysis, which represented 18.8% and 52.9% of significantly altered metabolites, respectively for each of the matrices (Table 3). All of the overlapping metabolites but one (i.e., PC 38:8e) were altered in the same direction (i.e., increased or decreased) in both RBCs and plasma in response to MTX treatment. Altered metabolites associated with MTX treatment included reduction in the hydrophilic metabolites N,N-diethyl-2-aminoethanol and N-methylisoleucine, as well as increases in a variety of phospholipid species including phosphatidylcholines (PC), lyso-PCs (LPC), phosphatidylethanolamines (PE), and phosphatidylinositols (PI).
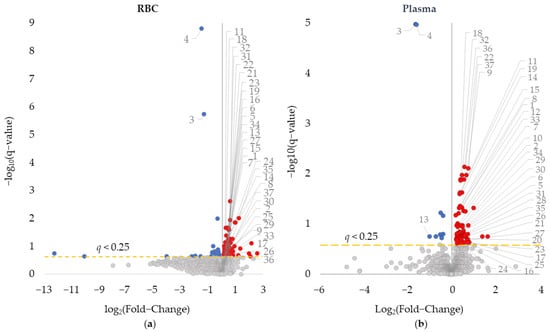
Figure 2.
Comparison of (a) RBC and (b) plasma metabolomics in detecting metabolic changes associated with MTX therapy in CIA disease mice. Univariate analysis of RBC and plasma metabolic data comparing the metabolome in disease and MTX treated disease mice was plotted based on log base 2 fold-change and negative log base 10 of the q-value. Red-colored metabolites were found to increase, and blue-colored metabolites were found to decrease (q value < 0.25). Overlapping metabolites that appeared on both RBC and plasma analysis are labeled.

Table 3.
Metabolites altered in both RBCs and plasma in CIA disease mice treated with MTX.
3.3. Comparison of MTX’s Effect on the RBC Metabolome in Healthy Control and CIA Disease Mice
After comparing the plasma and RBC metabolomic changes, a comparison of MTX’s effect on the RBC metabolome of healthy control mice and CIA disease mice was conducted. This comparison was used to differentiate changes in the metabolome resulting directly from the effect of MTX on metabolism compared to those metabolic changes that occur secondary to the effect of MTX on CIA disease activity. Differences in the RBC metabolome of healthy control mice and healthy control mice treated with MTX identified 138 significantly altered metabolites with 121 increased and 27 decreased (Figure 3). Significantly altered metabolites were compared to those previously identified in the comparison of disease mice and disease mice treated with MTX (Figure 1b and Figure 2a). A total of 38 metabolites were found to overlap between the two comparisons and are labeled in Figure 3 and presented on Table 4. This represented 19% of metabolites identified as altered in CIA disease mice treated with MTX. With the exception of four metabolites, the majority of directional changes in metabolite levels were consistent across the healthy control and CIA disease mice. Overall, these results suggest that MTX directly induced changes in the metabolome that may represent the direct effect of MTX on metabolism, however, the majority of identified metabolites are likely secondary to the effect of MTX on disease activity. Metabolic changes associated with MTX treatment include increases in several quaternary ammonium compounds (i.e., 3-Carboxypropyltrimethylammonium and 4-Trimethylammoniobutanoic acid), ceramides, sphingolipids, PCs, PEs, PIs, phosphatidylglycerol (PG), methotrexate, the acetylated aminoglycan nucleotide donor of N-acetylglucosamine (i.e., Uridine-5-diphosphoacetylglucosamine) and reductions in several amino acids including S-adenosyl-homocysteine and N-acetyl-leucine.
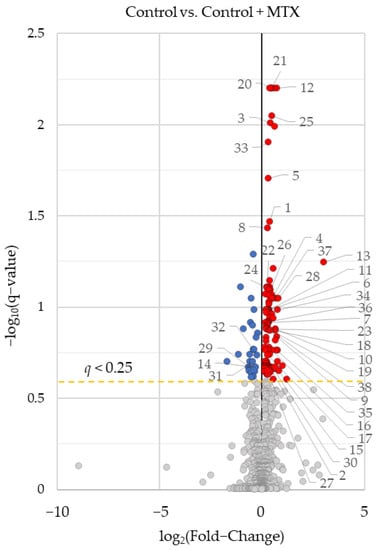
Figure 3.
Identification of metabolites associated with MTX’s effect on the RBC metabolome in healthy control mice. Univariate analysis of differences in the RBC metabolome comparing healthy control mice and healthy control mice treated with MTX. Results of this analysis were graphed as volcano plots. Labeled metabolites were significantly altered by MTX treatment in control and CIA disease mice. Metabolites were plotted based on log base 2 fold-change and negative log base 10 of the q-value. Red-colored metabolites were found to increase with MTX treatment, and blue-colored metabolites were found to decrease (q-value < 0.25).

Table 4.
Overlapping metabolites which were significantly altered by MTX treatment in both healthy control and CIA disease mice.
3.4. Evaluation of Metabolomic Markers as Biomarkers of MTX Treatment in RBCs and Plasma
Following the identification of the nine metabolites of interest from the RBC metabolome, receiver-operating characteristic (ROC) curve analyses were conducted for each of the metabolites. The performance of metabolites identified via RBC metabolomic analysis were compared to those identified by plasma metabolomic analysis to determine their ability to discriminate between untreated disease mice and those treated with MTX. ROC curve analysis is often employed to determine the accuracy of a diagnostic test or biomarker by plotting the false positive rate (i.e., 1-specificity) versus the true positive rate (i.e., sensitivity). The resulting area under the curve (AUC) is commonly used to describe the quality of the biomarker in discriminating between the two groups. In a previous publication by this author group, N-methylisoleucine (NMI) and quinolone were found to be discriminatory biomarkers in the plasma metabolome [3]. In total, all 11 metabolites were found to be strongly discriminatory between disease and MTX treated disease mice with AUCs for the ROC curves ranging from 0.79 to 1.0. The RBC metabolomic analysis resulted in the identification of nine potential biomarkers (Figure 4) whereas the plasma metabolome only identified two potential biomarkers (Figure 5). N-methylisoleucine was the only metabolite identified in both the RBC and plasma metabolomic analysis and was found to strongly discriminate between disease mice and disease mice treated with MTX.
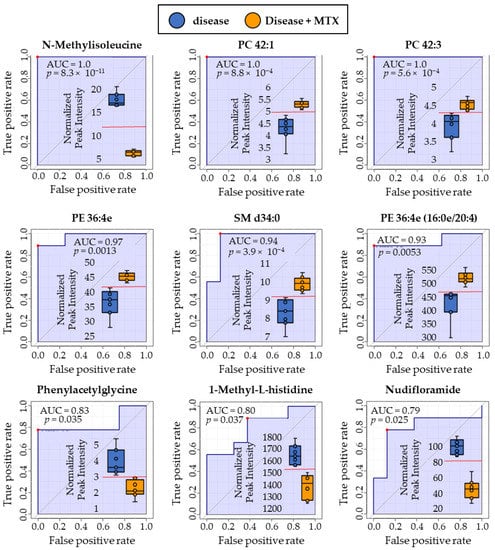
Figure 4.
ROC curve analyses were conducted using RBC metabolites identified by metabolomic profiling to compare the discriminatory ability of each biomarker. Red dots on each plot indicate the optimal cut-off based on the Youden Index. Comparison is based on metabolite levels in RBC for disease mice and disease mice treated with MTX. Box and whisker plots are inlayed within each ROC curve depicting the separation between disease (blue, left) and MTX treated disease mice (right, orange). Normalized peak intensities (y-axis) are labeled in thousands (×1000).
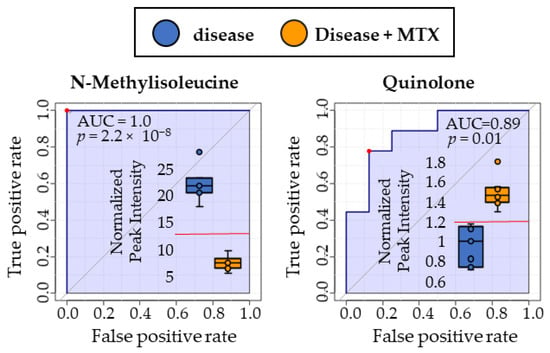
Figure 5.
ROC curve analyses were conducted using plasma metabolites identified by metabolomic profiling to compare the discriminatory ability of each biomarker. Red dots on each plot indicate the optimal cut-off based on the Youden Index. Comparison is based on metabolite levels in plasma for disease mice and disease mice treated with MTX. Box and whisker plots are inlayed within each ROC curve depicting the separation between disease (blue, left) and MTX treated disease mice (right, orange). Normalized peak intensities (y-axis) are labeled in thousands (×1000).
4. Discussion
In this study, semi-targeted metabolomic profiling of RBCs was evaluated by screening for metabolites associated with disease induction and MTX treatment in the CIA mouse model to identify putative biomarkers of MTX response in autoimmune arthritis and to establish the plausibility of using RBCs as a matrix for metabolomic biomarker discovery.
First, the RBC metabolome was compared between healthy control and disease mice, as well as disease and MTX treated disease mice. RBC metabolites of interest were identified based on three criteria: (1) the metabolite was significantly altered in disease mice compared to healthy control mice, (2) the metabolite was significantly altered in disease mice treated with MTX compared to untreated disease mice, and (3) MTX treatment resulted in at least a partial correction in metabolite levels towards those observed in the healthy control mice. The RBC biomarkers that met these criteria included: N-methylisoleucine, nudifloramide, phenylacetylglycine, 1-methyl-L-histidine, PC 42:1, PE 36:4e, PC 42:3, PE 36:4e (16:0e/20:4), and SM d34:0. In a separate regression analysis RBC levels of all of the identified metabolites were found to significantly correlate with both DAS and PV measurements in these mice. Together, these findings supported these RBC metabolites as associated with disease activity and response to MTX treatment in the CIA mouse model.
Next, a comparison of RBC and plasma metabolomic changes in disease mice treated with MTX demonstrated a high-level of similarity in metabolites and metabolite classes identified in each of the separate metabolomic studies. The validity of our results on MTX’s effect on the metabolome are strengthened by the high level of overlap (37 overlapping compounds) detected between the RBC and plasma metabolomic analyses. In particular, the findings indicate that MTX treatment is associated with a highly significant reduction in two hydrophilic metabolites in both plasma and RBCs (i.e., N,N-diethyl-2-aminoethanol and N-methylisoleucine). In addition, MTX treatment was associated with a general increase in numerous phospholipid species in both RBCs and plasma that include PCs, lyso-PCs, LPCs, PEs, and PIs. Some literature suggests that plasma and especially serum may contain higher concentrations of extracellular vesicles (EVs) which may contain microvesicle particles (ectosomes) from other blood cells (i.e., white and red blood cells, platelets) which could potentially confound the results obtained by these comparisons, especially with regard to phospholipids [33]. A weak, but highly statistically significant correlation was observed between changes in the RBC and plasma metabolomes in CIA disease mice treated with MTX. This observation suggests a relationship between the plasma and RBC metabolome that may be expected since metabolites, depending on their physicochemical properties, likely readily partition between the plasma and RBC compartment. However, the observation of only a weak correlation also suggests that many metabolites are differentially represented and impacted in RBCs compared to plasma and represent discrete biological compartment.
To differentiate the effects of MTX and CIA disease activity on the RBC metabolome, metabolomic effects of MTX treatment in healthy control mice was evaluated and compared to the effect of MTX observed in the CIA disease mice. Approximately 20% of metabolites found to be altered in the CIA disease mice following treatment with MTX were also observed to be altered in the healthy control mice treated with MTX. These finding support distinct metabolic changes that are likely the direct impact of MTX on metabolism but support the majority of metabolic changes in the CIA disease mice as secondary to the effect of MTX on disease activity.
Finally, ROC curve analyses were conducted on the nine identified metabolites of interest in both RBCs and plasma to evaluate and compare their ability to potentially serve as discriminatory biomarkers of MTX treatment response in autoimmune arthritis. N-methylisoleucine was highly discriminatory between disease mice and disease mice treated with MTX in both plasma and RBCs, with an AUC of 1.0. RBC levels of nudifloramide, phenylacetylglycine, 1-methyl-L-histidine, PC 42:1, PE 36:4e, PE 36:4e, PC 42:3, PE 36:4e (16:0e/20:4), and SM d34:0 were also found to be highly discriminatory between the two groups of mice with AUCs ranging from 0.79 to 1.0. In contrast, the plasma metabolomic study only identified one additional biomarker (i.e., quinolone) which was also found to be highly discriminatory with an AUC of 0.89.
To maximize applicability and relevance to clinical practice, translational research using animal models must accurately model disease processes. Using this rationale, the CIA mouse model was selected to model RA because it produces consistent systemic inflammation that resembles RA and has been demonstrated to be responsive to MTX [3,10,11,30]. The current clinical standard of care consists of once weekly subcutaneous MTX injections, a regimen thought to be superior to oral MTX administration [34,35]. This study employed this MTX administration strategy to reduce the risk of MTX-related toxicities and to avoid the variation in oral bioavailability from MTX’s saturable transporter-mediated absorption across the gastrointestinal lumen [36,37,38]. In this study, MTX was dosed at 20 mg/kg based on previous dose-escalation studies done by this lab [2]. Despite the pharmacokinetic differences between the two routes of administration, observations made in this study are expected to similarly apply to the use of oral MTX [39].
Several observations can be derived from interpreting the biomarkers of MTX response, many of which are related to the drug’s known pharmacology. The concentrations of N-methylisoleucine, 1-methyl-L-histidine, and phenylacetylglycine were all increased in CIA disease mice compared to healthy control mice and were decreased in response to MTX treatment. All three of these metabolites can be categorized as modified amino acids. Both N-methylisoleucine and 1-methyl-L-histidine are methylated products of their essential amino acid counterparts, whereas phenylacetylglycine is a product of gut microbial phenylalanine metabolism to phenylacetic acid that is conjugated with glycine in the mouse liver [40]. Increased methylated products in disease mice with reductions following MTX treatment can potentially be explained by MTX therapy’s effect on folates, which serve as the primary methyl donors and regulators of one-carbon metabolism. Previous work in our lab has found that RBC folate levels are increased in CIA disease mice and that MTX treatment results in a dose-dependent depletion of folates that corresponds with MTX treatment and response [3]. These results reinforce the hypothesis that MTX induced folate depletion results in a reduction in methyltransferase activity and thus the methylation of various biomolecules, including amino acids [3,41,42,43,44,45]. However, it remains unclear whether these methylated amino acid biomarkers are a direct result of endogenous metabolism, or a product of exogenous metabolites generated from the gut microbiome [3,46,47]. However, these changes in methylation were not observed in our healthy control mice treated with MTX, which may suggest that the antagonism of folates by MTX may not be responsible for these changes. In the case of phenylacetylglycine, it has been demonstrated to be a gut microbiota-dependent metabolite that promotes platelet responsiveness and thrombosis through activation of G-protein coupled adrenergic receptors [40]. Although phenylacetylglycine levels changed in both healthy control and CIA disease mice treated with MTX, the changes were in opposite directions (i.e., increased in healthy control mice and decreased in CIA disease mice) and suggest the effect of MTX on phenylacetylglycine likely is not a direct effect of MTX on this metabolite. However, these findings support previous findings that the gut microbiome may play a significant role in autoimmune arthritis disease activity and MTX response [48,49]. Of these compounds, N-methylisoleucine appears to be the most promising biomarker of MTX response as it has been demonstrated to be a discriminatory plasma metabolite of MTX response in patients with RA [46] and in CIA studies using both RBCs and plasma analyses, as described here. N-methylisoleucine also represented the greatest absolute difference of any of the proposed biomarker with mean RBC levels of N-methylisoleucine three-fold lower in MTX treated disease mice compared to disease mice, a finding that was consistent in both plasma and RBC analyses.
Our results also indicate that disease induction in the CIA mouse model is associated with an increase in RBC nudifloramide that is reduced in response to MTX therapy. Nudifloramide is an end product of nicotinamide-adenine dinucleotide (NAD) degradation and is an inhibitor of poly(ADP-ribose) [50]. Increased plasma nudifloramide levels have been observed in patients with chronic renal failure as a result of reduced renal excretion and has been proposed to function as a uremic toxin [51,52]. It is unclear if CIA disease induction causes an increase in nudifloramide production or a reduction in clearance. Reduced clearance may represent reduced kidney function in the disease mice that is reversed with MTX treatment. However, increased NAD metabolism is known to be a component of the immunoinflammatory response and MTX has been found to inhibit cellular NAD production via inhibition of de novo purine biosynthesis [53,54]. The observation that MTX treatment in the healthy control mice resulted in a significant, but opposite impact on nudifloramide levels compared to the CIA disease mice suggests that reductions in nudifloramide is not a direct effect of MTX therapy, but may represent a downstream effect of MTX on CIA disease activity.
RBC and plasma phospholipid levels were found to be decreased in CIA disease mice and normalized with MTX therapy. Previous studies have found reduced plasma and RBC phospholipid levels in patients with RA that has been attributed to increased lipid peroxidation related to increased oxidative stress [55,56]. As a result, our data supports a dyslipidemia associated with disease activity in the CIA mouse model that is responsive to treatment with MTX. The effect of MTX on lipid levels may not represent a direct effect of MTX on lipid metabolism and may represent a downstream effect that is secondary to MTX’s effect on disease activity in the mouse model. However, the observation that several phospholipid species, in addition to ceramides and sphingolipids, are increased with MTX in healthy control mice may suggest a direct effect of MTX on lipid metabolism.
In our analysis, the RBC metabolome appeared to be more sensitive in detecting metabolic changes associated with disease induction and MTX response than the plasma metabolome. Our findings support the value of the RBC metabolome as a potentially useful biological matrix for screening for biomarkers of MTX response in autoimmune arthritis. Future metabolomic studies are encouraged to explore this underutilized and highly accessible biofluid as a method to categorize the biochemical effects of various disease processes and treatment responses. As with all metabolomic studies, this study is hypothesis generating in nature and is limited by relatively small sample sizes and lack of independent validation metabolomic studies. As a result, future studies with a larger sample size are warranted. In addition, future functional metabolomic and metabolic flux analysis studies may be useful in understanding and validating the effect of disease and MTX treatment on the identified metabolites. Future studies will need to translate metabolomic findings from pre-clinical models to the autoimmune arthritis disease population, including metabolomic studies in humans using RBC as the biofluid sample source.
5. Conclusions
Nine RBC metabolites were identified that were altered in the CIA mouse model and normalized with MTX treatment. All identified RBC metabolites were found to be highly discriminatory between disease mice and mice treated with MTX and supported their potential as biomarkers of MTX response. When compared to a previous plasma metabolomic analysis, RBCs were more sensitive at detecting changes in the metabolome associated with MTX response. N-methylisoleucine appeared as a highly discriminatory biomarker in both the plasma and RBC metabolomic analysis, and supports previous plasma metabolomic findings in RA. These findings further support N-methylisoleucine as a promising therapeutic biomarker of MTX response in RA. The results of this study suggest that RBCs may be a useful sample source for future metabolomic works in the discovery of therapeutic and disease biomarkers.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/futurepharmacol2040038/s1, File S1: RBC metabolomics dataset containing normalized peak intensities for each identified metabolite along with disease activity scores and paw volumes for the mice included in this study. File S2: Plasma metabolomics dataset containing normalized peak intensities for each identified metabolite along with disease activity scores and paw volumes for the mice included in this study.
Author Contributions
Conceptualization, R.S.F.; methodology, R.S.F. and K.P.; formal analysis, Y.M.S. and Y.K.C.; data curation, Y.M.S.; writing—original draft preparation, Y.M.S.; writing—review and editing, R.S.F.; visualization, Y.M.S.; supervision, R.S.F.; project administration, R.S.F.; funding acquisition, R.S.F. and Y.M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the University of Kansas, Frontiers: University of Kansas Clinical and Translational Science Institute supported by a CTSA grant from NCATS, KL2TR002367 (R.F.), the Kansas Institute of Precision Medicine Centers of Biomedical Research Excellence grant from NIGMS awarded to the University of Kansas Medical Center, P20GM130423 (R.F.), and the American Foundation for Pharmaceutical Education Gateway to Research Award (Y.M.S.).
Institutional Review Board Statement
The study was conducted in accordance with the Guide for the Care and Use of Laboratory Animals as adopted and promulgated by the U.S. National Institutes of Health, and were conducted under a protocol approved by the Institutional Animal Care and Use Committee at The University of Kansas (Protocol ID: 2018-2481).
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article or supplementary material. The data presented in this study are available in Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fraenkel, L.; Bathon, J.M.; England, B.R.; St Clair, E.W.; Arayssi, T.; Carandang, K.; Deane, K.D.; Genovese, M.; Huston, K.K.; Kerr, G.; et al. 2021 American College of Rheumatology Guideline for the Treatment of Rheumatoid Arthritis. Arthritis Rheumatol. 2021, 73, 1108–1123. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; van Haandel, L.; Kiptoo, P.; Becker, M.L.; Siahaan, T.J.; Funk, R.S. Methotrexate disposition, anti-folate activity and efficacy in the collagen-induced arthritis mouse model. Eur. J. Pharmacol. 2019, 853, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Salamoun, Y.M.; Polireddy, K.; Cho, Y.K.; Medcalf, M.R.; Funk, R.S. Methotrexate Disposition, Anti-Folate Activity, and Metabolomic Profiling to Identify Molecular Markers of Disease Activity and Drug Response in the Collagen-Induced Arthritis Mouse Model. Metabolites 2022, 12, 24. [Google Scholar] [CrossRef] [PubMed]
- Luan, H.; Gu, W.; Li, H.; Wang, Z.; Lu, L.; Ke, M.; Lu, J.; Chen, W.; Lan, Z.; Xiao, Y.; et al. Serum Metabolomic and Lipidomic Profiling Identifies Diagnostic Biomarkers for Seropositive and Seronegative Rheumatoid Arthritis Patients. J. Transl. Med. 2021, 19, 500. [Google Scholar] [CrossRef]
- Friedman, B.; Cronstein, B. Methotrexate mechanism in treatment of rheumatoid arthritis. Jt. Bone Spine 2019, 86, 301–307. [Google Scholar] [CrossRef]
- Cronstein, B.N. Molecular therapeutics. Methotrexate and its mechanism of action. Arthritis Rheum. 1996, 39, 1951–1960. [Google Scholar] [CrossRef]
- Brand, D.D.; Latham, K.A.; Rosloniec, E.F. Collagen-induced arthritis. Nat. Protoc. 2007, 2, 1269–1275. [Google Scholar] [CrossRef]
- Medcalf, M.R.; Bhadbhade, P.; Mikuls, T.R.; O’Dell, J.R.; Gundry, R.L.; Funk, R.S. Plasma Metabolome Normalization in Rheumatoid Arthritis Following Initiation of Methotrexate and the Identification of Metabolic Biomarkers of Efficacy. Metabolites 2021, 11, 824. [Google Scholar] [CrossRef]
- Funk, R.S.; Becker, M.L. Disease modifying anti-rheumatic drugs in juvenile idiopathic arthritis: Striving for individualized therapy. Expert Rev. Precis. Med. Drug Dev. 2016, 1, 53–68. [Google Scholar] [CrossRef]
- Holmdahl, R.; Bockermann, R.; Bäcklund, J.; Yamada, H. The molecular pathogenesis of collagen-induced arthritis in mice—A model for rheumatoid arthritis. Ageing Res. Rev. 2002, 1, 135–147. [Google Scholar] [CrossRef]
- Lindqvist, E.; Saxne, T.; Geborek, P.; Eberhardt, K. Ten year outcome in a cohort of patients with early rheumatoid arthritis: Health status, disease process, and damage. Ann. Rheum. Dis. 2002, 61, 1055–1059. [Google Scholar] [CrossRef] [PubMed]
- Kremer, J.M.; Galivan, J.; Streckfuss, A.; Kamen, B. Methotrexate metabolism analysis in blood and liver of rheumatoid arthritis patients: Association with hepatic folate deficiency and formation of polyglutamates. Arthritis Rheum. Off. J. Am. Coll. Rheumatol. 1986, 29, 832–835. [Google Scholar] [CrossRef] [PubMed]
- Bouatra, S.; Aziat, F.; Mandal, R.; Guo, A.C.; Wilson, M.R.; Knox, C.; Bjorndahl, T.C.; Krishnamurthy, R.; Saleem, F.; Liu, P.; et al. The human urine metabolome. PLoS ONE 2013, 8, e73076. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chen, B.; Fang, Z.; Leng, Y.-F.; Wang, D.-W.; Chen, F.-Q.; Xu, X.; Sun, Z.-L. Metabolomics in the development and progression of rheumatoid arthritis: A systematic review. Jt. Bone Spine 2020, 87, 425–430. [Google Scholar] [CrossRef]
- Ng, D.J.Y.; Pasikanti, K.K.; Chan, E.C.Y. Trend analysis of metabonomics and systematic review of metabonomics-derived cancer marker metabolites. Metabolomics 2011, 7, 155–178. [Google Scholar] [CrossRef]
- Mendrick, D.L.; Schnackenberg, L. Genomic and metabolomic advances in the identification of disease and adverse event biomarkers. Biomark. Med. 2009, 3, 605–615. [Google Scholar] [CrossRef]
- Kaddurah-Daouk, R.; Beecher, C.; Kristal, B.S.; Matson, W.R.; Bogdanov, M.; Asa, D.J. Bioanalytical advances for metabolomics and metabolic profiling. Pharmagenomics 2004, 4, 46–51. [Google Scholar]
- Dunne, V.G.; Bhattachayya, S.; Besser, M.; Rae, C.; Griffin, J.L. Metabolites from cerebrospinal fluid in aneurysmal subarachnoid haemorrhage correlate with vasospasm and clinical outcome: A pattern-recognition 1H NMR study. NMR Biomed. 2005, 18, 24–33. [Google Scholar] [CrossRef]
- Brindle, J.T.; Antti, H.; Holmes, E.; Tranter, G.; Nicholson, J.K.; Bethell, H.W.; Clarke, S.; Schofield, P.M.; McKilligin, E.; Mosedale, D.E.; et al. Rapid and noninvasive diagnosis of the presence and severity of coronary heart disease using 1H-NMR-based metabonomics. Nat. Med. 2002, 8, 1439–1444. [Google Scholar] [CrossRef]
- Atzori, L.; Antonucci, R.; Barberini, L.; Griffin, J.L.; Fanos, V. Metabolomics: A new tool for the neonatologist. J. Matern.-Fetal Neonatal Med. 2009, 22, 50–53. [Google Scholar] [CrossRef]
- Wawrzyniak, R.; Kosnowska, A.; Macioszek, S.; Bartoszewski, R.; Jan Markuszewski, M. New plasma preparation approach to enrich metabolome coverage in untargeted metabolomics: Plasma protein bound hydrophobic metabolite release with proteinase K. Sci. Rep. 2018, 8, 9541. [Google Scholar] [CrossRef] [PubMed]
- Mill, J.; Patel, V.; Okonkwo, O.; Li, L.; Raife, T. Erythrocyte sphingolipid species as biomarkers of Alzheimer’s disease. J. Pharm. Anal. 2022, 12, 178–185. [Google Scholar] [CrossRef]
- Xu, L.; Chang, C.; Jiang, P.; Wei, K.; Zhang, R.; Jin, Y.; Zhao, J.; Xu, L.; Shi, Y.; Guo, S.; et al. Metabolomics in rheumatoid arthritis: Advances and review. Front. Immunol. 2022, 13, 961708. [Google Scholar] [CrossRef] [PubMed]
- Simon, T.L.; McCullough, J.; Snyder, E.L.; Solheim, B.G.; Strauss, R.G. Rossi’s Principles of Transfusion Medicine; John Wiley & Sons, Incorporated: Hoboken, NJ, USA, 2016. [Google Scholar]
- Hess, J.R.; Beyer, G.M. Chapter 13—Red Blood Cell Metabolism during Storage: Basic Principles and Practical Aspects. In Blood Banking and Transfusion Medicine (Second Edition); Hillyer, C.D., Silberstein, L.E., Ness, P.M., Anderson, K.C., Roback, J.D., Eds.; Churchill Livingstone: Philadelphia, PA, USA, 2007; pp. 205–211. [Google Scholar]
- Hess, J.R.; D’Alessandro, A. Red blood cell metabolism and preservation. In Rossi’s Principles of Transfusion Medicine; John Wiley & Sons, Incorporated: Hoboken, NJ, USA, 2022; pp. 143–157. [Google Scholar]
- Yurkovich, J.T.; Zielinski, D.C.; Yang, L.; Paglia, G.; Rolfsson, O.; Sigurjónsson, Ó.E.; Broddrick, J.T.; Bordbar, A.; Wichuk, K.; Brynjólfsson, S.; et al. Quantitative time-course metabolomics in human red blood cells reveal the temperature dependence of human metabolic networks. J. Biol. Chem. 2017, 292, 19556–19564. [Google Scholar] [CrossRef] [PubMed]
- Greenwalt, T.J. The Ernest Witebsky memorial lecture. Red but not dead: Not a hapless sac of hemoglobin. Immunol. Investig. 1995, 24, 3–21. [Google Scholar] [CrossRef]
- Galloway, M.; Rushworth, L. Red cell or serum folate? Results from the National Pathology Alliance benchmarking review. J. Clin. Pathol. 2003, 56, 924–926. [Google Scholar] [CrossRef]
- Guidelines on the investigation and diagnosis of cobalamin and folate deficiencies. A publication of the British Committee for Standards in Haematology. BCSH General Haematology Test Force. Clin. Lab. Haematol. 1994, 16, 101–115.
- Cajka, T.; Fiehn, O. Toward Merging Untargeted and Targeted Methods in Mass Spectrometry-Based Metabolomics and Lipidomics. Anal. Chem. 2016, 88, 524–545. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics by gas chromatography–mass spectrometry: Combined targeted and untargeted profiling. Curr. Protoc. Mol. Biol. 2016, 114, 30.4.1–30.4.32. [Google Scholar] [CrossRef]
- Zhang, X.; Takeuchi, T.; Takeda, A.; Mochizuki, H.; Nagai, Y. Comparison of serum and plasma as a source of blood extracellular vesicles: Increased levels of platelet-derived particles in serum extracellular vesicle fractions alter content profiles from plasma extracellular vesicle fractions. PLoS ONE 2022, 17, e0270634. [Google Scholar] [CrossRef]
- Hazlewood, G.S.; Thorne, J.C.; Pope, J.E.; Lin, D.; Tin, D.; Boire, G.; Haraoui, B.; Hitchon, C.A.; Keystone, E.C.; Jamal, S. The comparative effectiveness of oral versus subcutaneous methotrexate for the treatment of early rheumatoid arthritis. Ann. Rheum. Dis. 2016, 75, 1003–1008. [Google Scholar] [CrossRef] [PubMed]
- Vena, G.A.; Cassano, N.; Iannone, F. Update on subcutaneous methotrexate for inflammatory arthritis and psoriasis. Ther. Clin. Risk Manag. 2018, 14, 105. [Google Scholar] [CrossRef] [PubMed]
- Hoekstra, M.; Haagsma, C.; Neef, C.; Proost, J.; Knuif, A.; van de Laar, M. Bioavailability of higher dose methotrexate comparing oral and subcutaneous administration in patients with rheumatoid arthritis. J. Rheumatol. 2004, 31, 645–648. [Google Scholar] [PubMed]
- Radmanesh, M.; Rafiei, B.; Moosavi, Z.B.; Sina, N. Weekly vs. daily administration of oral methotrexate (MTX) for generalized plaque psoriasis: A randomized controlled clinical trial. Int. J. Dermatol. 2011, 50, 1291–1293. [Google Scholar] [CrossRef]
- Weinstein, G.; Roenigk, H.; Maibach, H.; Cosmides, J.; Halprin, K.; Millard, M.; Almeyda, J.; Auerbach, R.; Tobias, H.; Bergfeld, W. Psoriasis-liver-methotrexate interactions. Arch. Dermatol. 1973, 108, 36–42. [Google Scholar] [CrossRef]
- Giannini, E.H.; Brewer, E.J.; Kuzmina, N.; Shaikov, A.; Maximov, A.; Vorontsov, I.; Fink, C.W.; Newman, A.J.; Cassidy, J.T.; Zemel, L.S. Methotrexate in resistant juvenile rheumatoid arthritis: Results of the USA–USSR double-blind, placebo-controlled trial. N. Engl. J. Med. 1992, 326, 1043–1049. [Google Scholar] [CrossRef]
- Nemet, I.; Saha, P.P.; Gupta, N.; Zhu, W.; Romano, K.A.; Skye, S.M.; Cajka, T.; Mohan, M.L.; Li, L.; Wu, Y.; et al. A Cardiovascular Disease-Linked Gut Microbial Metabolite Acts via Adrenergic Receptors. Cell 2020, 180, 862–877.e22. [Google Scholar] [CrossRef]
- Emmerson, J.T.; Murray, L.K.; Jadavji, N.M. Impact of dietary supplementation of one-carbon metabolism on neural recovery. Neural Regen. Res. 2017, 12, 1075. [Google Scholar]
- Abbenhardt, C.; Miller, J.W.; Song, X.; Brown, E.C.; Cheng, T.-Y.D.; Wener, M.H.; Zheng, Y.; Toriola, A.T.; Neuhouser, M.L.; Beresford, S.A. Biomarkers of one-carbon metabolism are associated with biomarkers of inflammation in women. J. Nutr. 2014, 144, 714–721. [Google Scholar] [CrossRef]
- Borovikova, L.V.; Ivanova, S.; Zhang, M.; Yang, H.; Botchkina, G.I.; Watkins, L.R.; Wang, H.; Abumrad, N.; Eaton, J.W.; Tracey, K.J. Vagus nerve stimulation attenuates the systemic inflammatory response to endotoxin. Nature 2000, 405, 458–462. [Google Scholar] [CrossRef]
- Fox, J.T.; Stover, P.J. Folate-mediated one-carbon metabolism. Vitam. Horm. 2008, 79, 1–44. [Google Scholar] [PubMed]
- Annibal, A.; Tharyan, R.G.; Schonewolff, M.F.; Tam, H.; Latza, C.; Auler, M.M.K.; Grönke, S.; Partridge, L.; Antebi, A. Regulation of the one carbon folate cycle as a shared metabolic signature of longevity. Nat. Commun. 2021, 12, 3486. [Google Scholar] [CrossRef] [PubMed]
- Medcalf, M.R.; Bantis, L.E.; Shi, P.; Bhadbhade, P.; Gundry, R.L.; Mikuls, T.R.; England, B.R.; O’Dell, J.R.; Funk, R.S. Plasma metabolomic profiling as a tool to identify predictive biomarkers of methotrexate efficacy in rheumatoid arthritis. Semin. Arthritis Rheum. 2022, 56, 152056. [Google Scholar] [CrossRef] [PubMed]
- Yajima, T.; Mason, K.; Katz, E. Biogenetic origin of the D-isoleucine and N-methyl-L-alloisoleucine residues in the actinomycins. Antimicrob. Agents Chemother. 1976, 9, 224–232. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Artacho, A.; Isaac, S.; Nayak, R.; Flor-Duro, A.; Alexander, M.; Koo, I.; Manasson, J.; Smith, P.B.; Rosenthal, P.; Homsi, Y.; et al. The Pretreatment Gut Microbiome Is Associated With Lack of Response to Methotrexate in New-Onset Rheumatoid Arthritis. Arthritis Rheumatol. 2021, 73, 931–942. [Google Scholar] [CrossRef] [PubMed]
- Nayak, R.R.; Alexander, M.; Deshpande, I.; Stapleton-Gray, K.; Rimal, B.; Patterson, A.D.; Ubeda, C.; Scher, J.U.; Turnbaugh, P.J. Methotrexate impacts conserved pathways in diverse human gut bacteria leading to decreased host immune activation. Cell Host Microbe 2021, 29, 362–377.e11. [Google Scholar] [CrossRef] [PubMed]
- Giner, M.P.; Christen, S.; Bartova, S.; Makarov, M.V.; Migaud, M.E.; Canto, C.; Moco, S. A Method to Monitor the NAD+ Metabolome-From Mechanistic to Clinical Applications. Int. J. Mol. Sci. 2021, 22, 10598. [Google Scholar] [CrossRef] [PubMed]
- Slominska, E.M.; Smolenski, R.T.; Szolkiewicz, M.; Leaver, N.; Rutkowski, B.; Simmonds, H.A.; Swierczynski, J. Accumulation of plasma N-methyl-2-pyridone-5-carboxamide in patients with chronic renal failure. Mol. Cell. Biochem. 2002, 231, 83–88. [Google Scholar] [CrossRef]
- Rutkowski, B.; Slominska, E.; Szolkiewicz, M.; Smolenski, R.T.; Striley, C.; Rutkowski, P.; Swierczynski, J. N-methyl-2-pyridone-5-carboxamide: A novel uremic toxin? Kidney Int Suppl. 2003, 63, S19–S21. [Google Scholar] [CrossRef]
- Singh, R.K.; van Haandel, L.; Heruth, D.P.; Ye, S.Q.; Leeder, J.S.; Becker, M.L.; Funk, R.S. Nicotinamide Phosphoribosyltransferase Deficiency Potentiates the Antiproliferative Activity of Methotrexate through Enhanced Depletion of Intracellular ATP. J. Pharmacol. Exp. Ther. 2018, 365, 96–106. [Google Scholar] [CrossRef]
- Funk, R.S.; Singh, R.; Pramann, L.; Gigliotti, N.; Islam, S.; Heruth, D.P.; Ye, S.Q.; Chan, M.A.; Leeder, J.S.; Becker, M.L. Nicotinamide Phosphoribosyltransferase Attenuates Methotrexate Response in Juvenile Idiopathic Arthritis and In Vitro. Clin. Transl. Sci. 2016, 9, 149–157. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lazarevic, M.B.; Vitic, J.; Mladenovic, V.; Myones, B.L.; Skosey, J.L.; Swedler, W.I. Dyslipoproteinemia in the course of active rheumatoid arthritis. Semin. Arthritis Rheum. 1992, 22, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Vijayakumar, D.; Suresh, K.; Manoharan, S. Altered pattern of lipids in plasma and erythrocyte membranes of rheumatoid arthritis patients. Indian J. Clin. Biochem. 2005, 20, 52–55. [Google Scholar] [CrossRef] [PubMed][Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).