Abstract
SARS-CoV-2 vaccination is a life-saving strategy for the entire population living in this pandemic. Several vaccines were developed using different platforms such as nucleic acids, viral vectors recombinant proteins, live attenuated, and inactivated virus modalities, etc. Although immunogenicity and efficacy of these COVID vaccines were investigated, Covaxin (a vaccine code-named BBV152), an inactivated COVID-19 vaccine, has not been well studied yet. This study aimed to explore the interactions between biomolecules with vaccine adjuvants by analyzing molecular and protein–protein interactions of S protein, angiotensin-converting enzyme 2 (ACE2), and human serum albumin (HSA) with the ingredients of Covaxin (2-phenoxyethanol and imidazoquinolinone) by computational methods using Autodock Vina, Cluspro, and Swiss ADME. In addition, its drug-likeness property was investigated. The binding energies using Autodock Vina showed stronger interactions of 2-phenoxyethanol and imidazoquinolinone with viral surface protein, S protein, human cell membrane receptor ACE2, and drug carrier plasma HSA (−5.2, −5.3 and −5.3 kcal/mol; −8.5, −8.5 and −9.1 kcal/mol, respectively). The interaction between S protein with ACE2 in the presence of 2-phenoxyethanol and imidazoquinolinone hindered the S protein function by reducing the binding energy between these proteins. In addition, imidazoquinolinone may have the drug-likeness property based on pharmacokinetic and physicochemical parameters. These results suggest that the Covaxin vaccine, owing to these ingredients, may impart greater efficacy in averting the virus and thus it may be more effective in producing herd immunity. In conclusion, for the first time, this computational study predicts the possible useful effects of these two adjuvants of Covaxin in therapeutic and drug-likeness strategies against SARS-CoV-2.
Keywords:
SARS-CoV-2; coronavirus; Covaxin; ACE2; HSA; drug–protein interaction; 2-Phenoxyethanol; imidazoquinolinone 1. Introduction
The prevention of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic is a major international public health priority due to the high infection rate and associated mortality worldwide. Coronavirus disease 2019 (COVID-19) created humankind’s most significant catastrophe event in 2020 [1]. Despite wearing face masks, sanitization, and antiviral drugs, there is a need to develop vaccines against COVID-19. Several vaccines have been developed using different platforms, including nucleic acids, viral vectors, virus-like particles, peptide-based, recombinant proteins, live attenuated, and inactivated virus modalities. Several large clinical trials produced positive findings on the clinical efficacy and safety of COVID-19 vaccines. The Covaxin (code-named BBV152), an inactivated COVID-19 vaccine, was developed and assessed by Bharat Biotech, India [2]. Covaxin showed promising efficacy and immunogenicity towards SARS-CoV-2, which is available in a double dose [3]. Covaxin is included along with immune-potentiators, also known as vaccine adjuvants, which are added to the vaccine to increase and boost its immunogenicity. The manufacturer developed a formulation containing the whole virion inactivated SARS-CoV-2 antigen (strain: NIV-2020-770), adjuvant aluminium hydroxide gel, immunostimulatory imidazoquinolinone, and the preservative 2-phenoxyethanol (2-PE) (Figure 1) [4]. Imidazoquinoline (IMDG) molecule is also a toll-like receptor (TLR) 7/8 agonist and thus can have an immunomodulatory effect [5]. On 3 November 2021, the Technical Advisory Group for Emergency Use Listing, WHO listed the Covaxin vaccine against COVID-19 for emergency use.
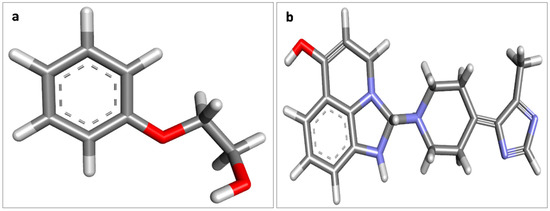
Figure 1.
Structure of 2-Phenoxyethanol and imidazoquinolinone, (a) 2-Phenoxyethanol, (b) imidazoquinolinone.
However, there are concerns regarding the formulation, safety, tolerability, and immunogenicity of Covaxin despite its efficacy [2]. The toxicity and adverse side-effects of most adjuvant formulations are the most serious concerns with adjuvant usage in human vaccinations, particularly regular childhood vaccines. Therefore, adjuvants for human vaccination are now chosen to balance the need for adjuvanticity and a modest amount of adverse effects [6]. 2-PE and imidazoquinolinone are used as adjuvants in Covaxin. Although several non-COVID19 vaccines such asPCV13 [7], PCV10 [7], IPV [8], and influenza vaccines [9] use these two adjuvants, there is no information available on their impacts on SARS-CoV-2 and its interactions with the host. Among these adjuvants, 2-PE is a highly effective vaccine preservative as it can kill bacteriabydecouplingoxidativephosphorylationfromrespirationandcompetitively inhibitingmalatedehydrogenase [10,11].
Human serum albumin (HSA) reversibly binds a wide range of drugs and small compounds, thus modulating biochemical pathways. The SARS-CoV-2 inhibits the HSA-mediated transport of compounds by altering the formation of endothelial glycocalyx [12]. The physiological functions of HSA include maintaining osmotic pressure, anti-inflammation, and transport of plasma molecules [12,13,14]. Therefore, it is essential to know whether imidazoquinolinone and 2-PE interact with HSA and modulate the symptoms of COVID-19.
SARS-CoV-2 comprises a genome size of ~30 kilobases of different structural and accessory proteins [15,16]. In coronavirus morphology, there are four structural proteins: membrane (M) protein, spike (S) protein, nucleocapsid (N) protein, and envelope (E) protein [17]. The S1 subunit of the N terminal contains the receptor-binding domain (RBD), and the C terminal S2 subunit induces membrane fusion. The S2 subunit, characterized by Heptad Repeats (HR) regions that assemble into an intra-hairpin helical structure with a six-helix bundle following viral endocytosis, facilitates the membrane fusion process inside the host cell [18,19]. The binding of S1 and S2 subunits causes fusion in the cell membrane, which induces viral invasion into the human cell by attaching to the angiotensin-converting enzyme 2 (ACE2) [18,20,21], the coronavirus receptor, which can be a specific target to avert viral entry [22].
The molecular interaction, protein–protein interactions, ligand-based binding affinities, and drug-likeness studies can be assessed using various bioinformatics tools. Computational approaches are essential tools to predict apparent binding modes and affinities of ligands for macromolecules before experimental studies, which are expensive and time-consuming [18]. Furthermore, advances in computational docking techniques’ speed, dependability, and accuracy over the past few years have made them an excellent option for developing pharmaceuticals with a structure-based structure. Here, we employed computational approaches to study the interaction between Covaxin, 2-PE, and imidazoquinolinone ingredients of Covaxin with HSA, ACE2, and S protein.
Here we report that we studied Covaxin safety, immunogenicity, hesitance, and resistance against the SARS-CoV-2 using a new methodology. Our data suggest that this new methodology may help develop future vaccines and other targeted therapies.
2. Materials and Methods
2.1. Sequence Analysis
The Cryo-EM structures of COVID-19 S protein (PDB ID-6vsb), X-ray crystal structures of ACE2 (PDB ID-1r42), and HSA (PDB ID-1e78) were retrieved from the PDB database at resolutions of 3.46 Å, 2.2 Å, and 2.6 Å, respectively, which were used for the computational study. Before molecular interaction analysis, the FASTA sequence of the above PDB structures was used to analyze its physicochemical property and secondary structure prediction in ExPASy ProtParam [23] and SOPMA programs [24].
2.2. Investigation of S Protein-ACE2 Interaction in the Presence of 2-PE and Imidazoquinolinone
A computerized rigid-body docking tool, ClusPro 2.0 (https://cluspro.bu.edu/publications.php, accessed on 8 August 2021), was used for S protein-ACE2 protein-protein docking analysis in the presence or absence of 2-PE and imidazoquinolinone. This program aids in screening docked conformations for clustering features based on various protein parameters. The filtered conformations were selected based on empirical estimation of free energy. Free energy was calculated by taking desolvation and electrostatic energies into account. The ClusPro clustering program detects native sites with the help of Piper’s rigid docking tool based on FFT [25]. The native site is assumed to possess a wide range of free energies to draw a more significant number of results. Initially, the sample was taken for about 109 positions of the ligand for the receptor. Only the top 103 positions were selected among all relative ligand positions corresponding to the receptor [18,26,27].
2.3. Docking Analysis between ACE2, HSA, and S Protein with 2-PE and Imidazoquinolinone
The binding affinities of HSA, ACE2, and S protein with 2-PE and imidazoquinolinone were evaluated through the molecular docking program AutoDock Tools 1.5.6, which is a free graphic user interface (GUI). The canonical SMILES id of 2-PE is acquired from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). However, a canonical SMILES id of imidazoquinolinone molecule was not available in the PubChem database. However, its 2D structure was designed by using ACD/ChemSketch. The CHIMERA 1.11.2 program [28] converted 2D into 3D structures and energy minimization of the ligands. The grid box was constructed using 112, 114, and 126, pointing in x, y, and z directions, respectively, with a grid point spacing of 0.508 Å. The centre grid box of 49.108 Å, 28.216 Å, and 59.46 Å around the selected amino acids was based on the CASTp result for HSA. Similarly, for S protein and the ACE2 grid box, it was 126, 126, 126 and 96, 88, 95 pointing in x, y, and z directions, respectively, with the centre grid box being 225.616 Å, 226.49 Å, and 243.662 Å and 53.829 Å, 59.942 Å, and 29.379 Å.
The binding affinity of HSA, ACE2, and S protein with 2-PE and imidazoquinolinone was estimated using AutoDock Vina 1.1.2 [29]. The binding sites of the above receptors are used to recognize the binding affinity, receptor pocket atom, receptor-interacting atom, atomic contact energy, receptor–ligand interaction site, and side amino acid residues. Discovery Studio 2017 R2 was used to analyze the Pictorial depiction of docking results [30].
2.4. Drug-Likeness Analysis of 2-PE and Imidazoquinolinone
Swiss ADME is a web-based tool that links physicochemical, pharmacokinetics, and molecular chemistry of drug-likeliness to determine proficiency. Bioavailability Radar (solubility, size, polarity, lipophilicity, saturation, and flexibility) has been used to assess drug-likeness. For drug development, the analysis of ADME (abstraction, distribution, metabolism, and excretion) is critically important. The SwissADME (http://www.swissadme.ch/, accessed on 8 August 2021) program was used to predict ADME properties using the 2-PE Canonical SMILES ID obtained from the PubChem database. Similarly, the structure of imidazoquinolinone was used for ADME prediction.
3. Results and Discussion
Adjuvants are vaccine ingredients that boost the potency, breadth, and longevity of the immune response. The traditional procedure of developing new vaccine adjuvants has been recognized as one of the most time-consuming in medical history. Since their original licencing in the 1920s, insoluble aluminium salts (alum) have been the only adjuvant used in approved products such as hepatitis B, diphtheria, tetanus, and pertussis vaccinations, as well as vaccines against the human papillomavirus [31]. Many vaccine adjuvants have been developed in recent years with proven significant effectiveness in preclinical models; however, most have yet to be approved for use in humans, typically due to safety or tolerability issues [31].
Covaxin vaccine contains a number of adjuvants to improve the potency, breadth, and durability of the immune response, including 2-PE and imidazoquinolinone. In this study, we primarily analyzed the molecular interactions of S protein, ACE2, and HSA with 2-PE and imidazoquinolinone through an insilico approach. Inspecting the drug-likeness and protein–protein interaction through various computational tools enhances our understanding of vaccine adjuvants and its impact on SARS-CoV2.
3.1. Physicochemical Properties and Structural Analysis of S Protein, ACE2, and HSA
The predicted secondary structures of S protein, ACE2, and HSA using SOPMA revealed that S protein, ACE2, and HSA contain α-Helix was 26.86%, 58.05%, and 69.57%, respectively. The higher proportion of α-helices in the structures of HSA and ACE2 makes them more stable proteins [32]. Physicochemical data also indicate that the percentage of the total number of negatively charged residues (Asp + Glu) was higher (1.53%) compared to the total number of positively charged residues (Arg + Lys) in ACE2 molecules. In contrast, the total numbers of negatively charged residues in S protein and HSA were 1.12% and 1.18%, respectively negatively charged amino acids are thermostable due to the formation of salt bridges throughout the protein structure, preventing protein denaturation at high temperatures [33]. The half-life of a protein is defined as the time is taken for its complete disintegration since its synthesis in a cell. The maximum half-life of the S protein of 2019-nCoV and ACE2 was estimated as 30 h in mammalian reticulocytes, whereas HSA has only 1.1 h. The instability index of S protein, ACE2, and HSA was 31.58, 43.59, and 38.85, respectively. A smaller instability index (<40) of a protein depicts its stability [33]. The instability index of ACE2 is a little higher than 40, which makes it unstable. Similarly, S protein, ACE2, and HSA have an aliphatic index of 81.58, 76.76, and 76.92, respectively. The aliphatic index is the relative volume occupied by its aliphatic side chains (alanine, valine, isoleucine, and leucine). Thus, higher aliphatic index values represent the withstanding potential of protein at high temperatures.
3.2. Molecular Interactions between Covaxin Adjuvant with S Protein, ACE2, and HSA
Covaxin is demonstrated to produce antibodies and show a good immune response against the COVID-19 virus [34]; however, the information on the molecular interactions of it ingredients with COVID virus and host cell proteins are not known.
The binding energies of the molecular interaction study were determined as ΔGb-5.3 Kcal/mol and ΔGb-8.5 Kcal/mol when ACE2 interacted with 2-PE and imidazoquinolinone, respectively (Figure 2 and Figure 3). In contrast, ΔGb-5.3 Kcal/mol and ΔGb-9.1 Kcal/mol were the energies, respectively, when HSA interacted with 2-PE and imidazoquinolinone (Figure 4 and Figure 5). On the contrary, the binding affinities scored were ΔGb-5.2 Kcal/mol and ΔGb-8.5 Kcal/mol when S protein interacts with these molecules (Figure 6 and Figure 7). Imidazoquinolinone showed the higher affinity forS protein, ACE2, and HSA compared withthose of 2-PE.
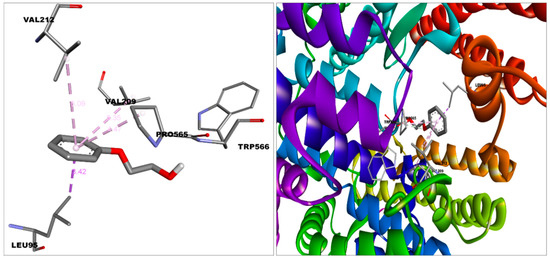
Figure 2.
Docked pose of 2-phenoxyethanol in the binding pocket of ACE2. This figure was produced using Discovery Studio Visualizer (http://accelrys.com/products/collaborative-science/biovia-discovery-studio/visualization-download.php).
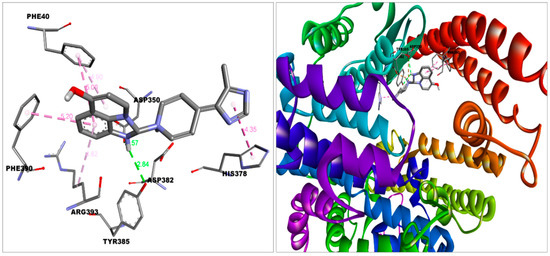
Figure 3.
Docking of imidazoquinolinone in the binding pocket of ACE2.Discovery Studio Visualizer was used to create this figure. (http://accelrys.com/products/collaborative-science/biovia-discovery-studio/visualization-download.php).
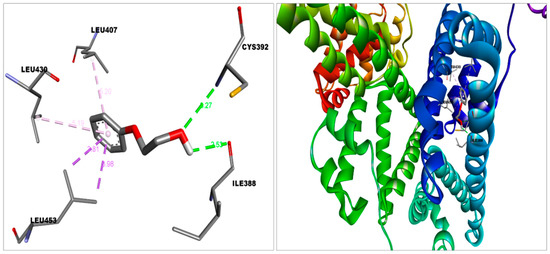
Figure 4.
2-phenoxyethanol docked in HSA’s binding bag.Discovery Studio Visualizer was used to create this figure. (http://accelrys.com/products/collaborative-science/biovia-discovery-studio/visualization-download.php).
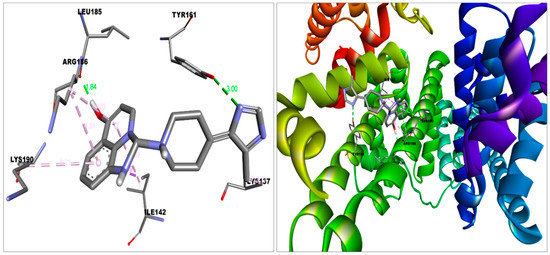
Figure 5.
Imidazoquinolinone in a docked position in the binding pocket of HSA.This figure was produced using Discovery Studio Visualizer (http://accelrys.com/products/collaborative-science/biovia-discovery-studio/visualization-download.php).
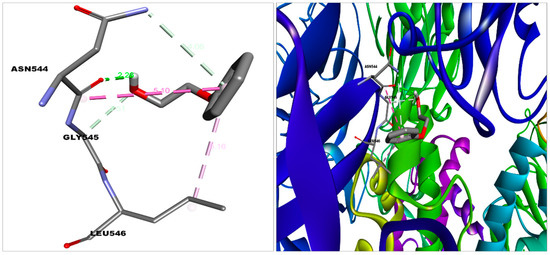
Figure 6.
2-phenoxyethanol docked in the binding pocket of S protein.A Discovery studio visualizer was used to build this diagram. (http://accelrys.com/products/collaborative-science/biovia-discovery-studio/visualization-download.php).
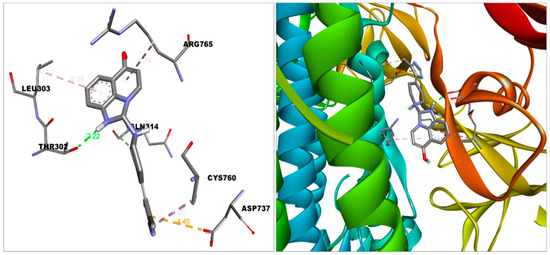
Figure 7.
In the binding pocket of S protein, imidazoquinolinone is docked. Discovery Studio visualizer was used to build this figure. (http://accelrys.com/products/collaborative-science/biovia-discovery-studio/visualization-download.php).
The imidazoquinolinone bound S protein through multiple bonds and interactions, including Vander Waal’s (Asn317, Ser316, Thr315, Thr761, Thr302, Tyr313, Thr768, Gln314, Asn764, Thr739); Conventional hydrogen bond (Thr302); Pi-Anion (Asp-737); Pi-alkyl (Cys760, Leu303) and Alkyl (Arg765) bond (Figure 7). Imidazoquinolinone engaged with ACE2 with Vander Waal’s (Ala348, Thr347, Glu402, His401, Trp69); Conventional hydrogen bond (Tyr385); Pi-Pi T-shaped (His378); Pi-Pi stacked (Phe390, Phe40); Pi-alkyl (Arg393), and Salt bridge (Asp350, Asp382) (Figure 3). Furthermore, imidazoquinolinone engages with HSA by Van der Waal’s (Met123, Phe134, Glu141, Tyr138, Phe157, Gly189, His146, Leu115); Conventional hydrogen bond (Tyr161, Leu185); Pi-sigma (Ile142); Pi-alkyl (Arg186, Lys190); and Alkyl (Lys137) bonds (Figure 5).
2-PE bound with the S protein by Van der Waal’s (Gln564, Phe565, Val576, Phe543, Leu517, Cys391, Ala522, Leu518, Pro521); Carbon hydrogen bond (Asn544); Conventional Hydrogen Bond (Asn544); and Pi-Alkyl (Leu546) bonds (Figure 6), with ACE2 by Van der Waal’s (Leu91, Asn210, Lys562, Ala396, Glu564); Carbon hydrogen bond (Pro565); Unfavorable donor (Trp566); Pi-alkyl (Val212, Val209); and Pi-sigma (Leu95) bonds and with HSA by Van der Waal’s (Asn391, Ala449, Leu387, Val433, Phe403, Tyr411); Conventional hydrogen bond (Cys392,Ile388); Pi-sigma (Leu453); and Pi-alkyl (Leu430, Leu407) bonds (Figure 2 and Figure 4).
The above molecular interaction data indicated that 2-PE bound with RBD (Receptor binding domain) of S protein, which spans from 319 to 591 amino acid residues in S protein [35], whereas imidazoquinolinone strongly interacted with S protein compared to 2-PE with the proximity of the RBD site. The number of hydrogen bonds formed during the interaction of 2-PE and imidazoquinolinone with S protein is 2 and 1, respectively. Such H-bonds are formed between the amino acid residues of the S protein and OH groups of the adjuvant. The binding affinity and position of the above two compounds suggest that these molecules may cause hindering in S protein function, which was also reflected in the protein–protein interaction study.
Imidazoquinolinone strongly interacted with the IB domain (Span from 108–196) of HSA with a binding affinity ΔGb-9.1 Kcal/mol, which plays (IB domain) a vital role in drug delivery [13,14]. Similarly, 2-PE bound with the IIIA domain (Span from 384–497) of HSA with a binding affinity ΔGb-5.3 Kcal/mol [36].
Covaxin ingredients (2-PE, imidazoquinolinone) showed good binding affinity towards ACE2, S protein, and HSA (Table 1). Furthermore, imidazoquinolinone showed the highest affinity with S protein, ACE2, and HSA with energies ΔGb-8.5 Kcal/mol, ΔGb-8.5 Kcal/mol, and ΔGb-9.1 Kcal/mol, respectively.

Table 1.
The binding energy, interaction type, and amino acid involved in the interaction of S protein ACE2 of 2019-nCoV and HSA with 2-PE and imidazoquinolinone.
3.3. Protein–Protein Interaction Study in the Presence of Covaxin Adjuvants with S Protein, ACE2, and HSA
There are 10 best docking models listed on the ClusPro Web server with various free energies. A grouping criterion was used based on the overall RMSD [26]. We used five ClusPro docking models chosen based on binding affinity, S protein complex with 2-PE, and imidazoquinolinone to engage with the anticipated sites ACE2 and the lowest binding energy during such interactions. For the S protein-ACE2 interaction, the average binding energy for all five binding sites is −901.2 kJ/mol. Nevertheless, the average binding energy for S protein-ACE2 in the presence of 2-PE is −696.64 kJ/mol, and −589.46 kJ/mol in the presence of imidazoquinolinone (Figure 8, Figure 9 and Figure 10; Table 2).
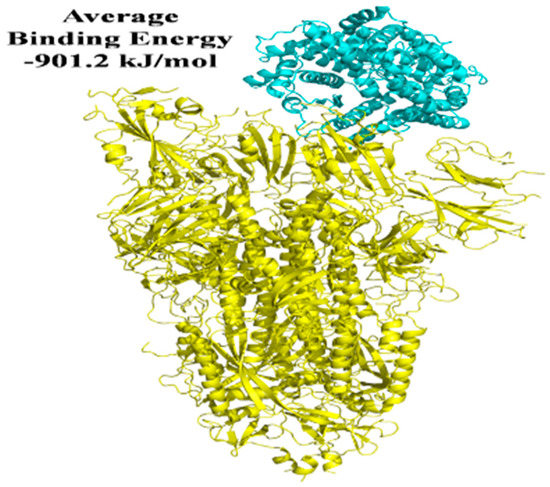
Figure 8.
In the absence of Covaxin−adjuvant, a docked model revealed the interaction of S protein with the ACE2 receptor.
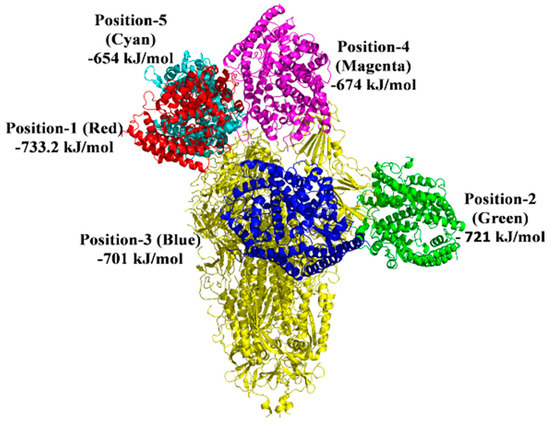
Figure 9.
Docked model depicting the interaction of S protein with the ACE2 receptor in the presence of 2−phenoxyethanol.
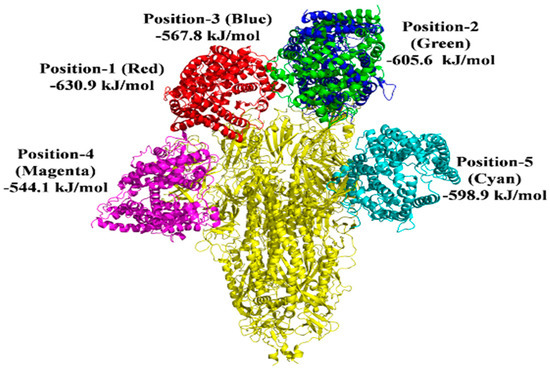
Figure 10.
In the presence of imidazoquinolinone, the docked model depicts the S protein’s interaction with the ACE2 receptor.

Table 2.
Protein–protein interaction depicting five lowest binding energies for S protein-ACE2 complex in the presence or absence of Covaxin adjuvant.
Protein–protein interactions are mediated via electrostatic forces, hydrogen bonding, and hydrophobic interactions between two or more protein molecules [18,36]. An analysis of protein–protein interactions can provide important information about the molecular networks of a living cell [37]. Furthermore, protein–protein interaction plays a significant role in predicting the protein’s function of molecules that target protein and drug’ stability [38]. In the presence of 2-PE and imidazoquinolinone, the binding energy between S protein and ACE2 was reduced. In comparison to direct binding, S protein interaction with ACE2 in the presence of 2-PE resulted in a substantial decrease in the binding energy of 204.56 kJ/mol and 311.74 kJ/mol in the presence of Imidazoquinolinone (Figure 8, Figure 9 and Figure 10). These data suggest that 2-PE or imidazoquinolinone may prevent the RBD site of S protein from attaching to the ACE2 receptor protein of the host cells.
3.4. Drug Likeliness Analysis of Covaxin Ingredients
Imidazoquinoline is a TLR7/8 selective agonist and thus can modulate the immune reaction via cytokine production [5]; however, the drug-likeness of 2-PE is not well known. The SwissADME tool was used to assess the drug-likeness of 2-PE and imidazoquinolinone. This tool determined the connection between the pharmacokinetics and physicochemical properties of the molecules. The physicochemical properties of 2-PE (C8H10O2) were determined, i.e., 138.16 g/mol molecular weight, molar refractivity of 38.90, 10 heavy atoms, 3 rotatable bonds, 2 hydrogen bond acceptors, 1 hydrogen bond donor, and topological polar surface area of 29.46 Ų. Similarly, physicochemical properties for imidazoquinolinone (C19H21N5O) showed 335.40 g/mol molecular weight, molar refractivity of 100.12, 25 heavy atoms, 2 rotatable bonds, 3 hydrogen bond acceptors, 1 hydrogen bond donor, and a topological polar surface area of 66.81 Ų.
2-PE has a very high solubility in water. SwissADME uses five distinct criteria to predict drug-likeness (Lipinski, Ghose, Veber, Egan, and Muegge). Lipinski, Veber, and Egan obey the drug-likeness property of 2-PE, whereas Ghose and Muegge are not. Similarly, for a lipophilicity score of imidazoquinolinone, iLOGP, XLOGP3, WLOGP, MLOGP, and SILICON-IT models compute to 2.27. The bioavailability score for both molecules is 0.55. All five models render imidazoquinolinone competent as an adequate drug molecule (Tables S1 and S2). The BOILED-Egg model assumes that 2-PE and imidazoquinolinone can easily pass through the blood–brain barrier and absorb by human gastrointestinal absorption [38]. The 2-phenoxyethanol’s pharmacokinetics and drug-likeness properties have sparked debate about whether it should be considered a drug molecule. Swiss Target prediction, a webserver, plays a critical role in identifying ligand–target of known molecules [39,40]. It accurately predicts the targets to modulate their behavior, elucidating the molecular mechanism and predicting cross-reactivity in 2D and 3D similarity events with known ligands [41,42]. It also detects potential side effects and assists in repurposing molecules for new uses [39,43]. Using the SwissTarget method, 2-PE showed a high predictive performance level of interaction with A-G and C-G coupled receptors, kinases, enzymes, and nuclear receptors. The primary drug-likeness targets from this prediction are A-G coupled receptors and enzymes. In addition (Figure S4), imidazoquinolinone showed a variety of A-G and B-G coupled receptors, enzymes, and histone-modifying enzymes without affecting the vaccine ingredients’ function (Figure S3).
Molecular interaction data suggested that imidazoquinolinone had a robust binding affinity compared to 2-PE, corroborating the protein–protein interaction data. Compared to 2-PE, imidazoquinolinone had the maximum hindering effects on the S protein, ACE2 interaction. Molecular interaction data also suggest that imidazoquinolinone binds to the RBD site of S protein, which may cause hindering in S protein-ACE2 complex formation. Protein–protein interactions regulate various biological functions, including cell to cell interactions, metabolic regulation, and developmental control [44]. This could open the door to repurposing/designing appropriate treatment to deter viral penetration using 2-PE and imidazoquinolinone in Covaxin. A molecule must achieve the target in optimum concentration and be usable in the bioactive form before the necessary biological events initiated to be an effective drug. The SwissADME technology reduces the time and resources required for drug development. To be an oral drug candidate, development products’ structural or physicochemical properties must be evaluated for drug-likeness. A molecule’s drug-likeness is determined for bioavailability by qualitatively assessing the likelihood that the molecule will be formed into an oral drug. Bioavailability radar defines the optimal set of properties like lipophilicity, saturation, solubility, polarity, size, and flexibility for the input molecule drug-likeness of 2-PE and imidazoquinolinone (Figures S1 and S2). These adjuvants are effective against virus entry but less effective against biological targets in humans.
4. Conclusions
Using various computational methods, we suggest that using 2-PE and imidazoquinolinone as ingredients in Covaxin may provide beneficial repurposing/designing effective therapy to prevent viral entry. Our computational data also showed the possibility of adjuvant of Covaxin for therapeutic strategy against SARS-CoV2. 2-PE and imidazoquinolinone exhibited good binding affinities for the aforementioned macromolecules, as well as hindering the S protein function by reducing the binding energy towards the human cell receptor protein ACE2. Bioavailability radar and drug-likeness properties of these adjuvants suggest that it may be an effective adjuvant for vaccines. However, there are several weaknesses in this study, including the lack of a molecular dynamic simulation, in vitro and in vivo investigations. Since several issues still remain unaddressed, a future extension of this research can lead to a comprehensive understanding of system vaccinology to predict Covaxin vaccine immunogenicity and reactogenicity.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/futurepharmacol2030021/s1. Figure S1. the Bioavailability Radar depicts glimpse of the drug-likeness of a molecule. The pink area represents the optimal range for each properties of Imidazoquinolinone. Figure S2. the Bioavailability Radar depicts glimpse of the drug-likeness of a molecule. The pink area represents the optimal range for each properties of 2-phenoxyethanol. Figure S3. Imidazoquinolinone targeted biomolecules. Figure S4. 2-phenoxyethanol targeted biomolecules. Table S1. Lipophilicity, Pharmacokinetics and Drug likeness property of 2 Phenoxyethanol. Table S2. Lipophilicity, Pharmacokinetics and Drug likeness property of Imidazoquinolinone.
Author Contributions
Conceptualization, A.B.J. and A.K.D.; writing—original draft preparation, methodology, A.B.J.; supervision, writing review and editing, A.K.D. All authors have read and agreed to the published version of the manuscript.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Seitz, B.M.; Aktipis, A.; Buss, D.M.; Alcock, J.; Bloom, P.; Gelfand, M.; Haselton, M.G. The pandemic exposes human nature: 10 evolutionary insights. Proc. Natl. Acad. Sci. USA 2020, 117, 27767–27776. [Google Scholar] [CrossRef] [PubMed]
- Talukder, A.; Kalita, C.; Neog, N.; Goswami, C.; Sarma, M.K.; Hazarika, I. A comparative analysis on the safety and efficacy of Covaxin versus other vaccines against COVID-19: A review. Z. Für Nat. C 2022, 77, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Darbar, S.; Agarwal, S.; Saha, S. COVID-19 Vaccine: COVAXIN®-India’s First Indigenous Effective Weapon to Fight against Coronavirus (A Review). Parana J. Sci. Educ. 2021, 7, 1–9. [Google Scholar]
- Ganneru, B.; Jogdand, H.; Daram, V.K.; Das, D.; Molugu, N.R.; Prasad, S.D.; Kannappa, S.V.; Ella, K.M.; Ravikrishnan, R.; Vadrevu, K.M. Th1 skewed immune response of whole virion inactivated SARS CoV 2 vaccine and its safety evaluation. iScience 2021, 24, 102298. [Google Scholar] [CrossRef] [PubMed]
- Ganapathi, L.; Van Haren, S.; Dowling, D.J.; Bergelson, I.; Shukla, N.M.; Malladi, S.S.; Balakrishna, R.; Tanji, H.; Ohto, U.; Shimizu, T.; et al. The Imidazoquinoline Toll-Like Receptor-7/8 Agonist Hybrid-2 Potently Induces Cytokine Production by Human Newborn and Adult Leukocytes. PLoS ONE 2015, 10, e0134640. [Google Scholar] [CrossRef]
- Gupta, R.K.; Siber, G.R. Adjuvants for human vaccines—Current status, problems and future prospects. Vaccine 1995, 13, 1263–1276. [Google Scholar] [CrossRef]
- Veve, M.P. Side Effects of Drugs Annual. Vaccines 2018, 42, 383–413. [Google Scholar] [CrossRef]
- Vidor, E. Poliovirus vaccine-inactivated. Vaccines 2013, 573–597. [Google Scholar] [CrossRef]
- Miller, S.M.; Cybulski, V.; Whitacre, M.; Bess, L.S.; Livesay, M.T.; Walsh, L.; Burkhart, D.; Bazin, G.; Evans, J.T. Novel Lipidated Imidazoquinoline TLR7/8 Adjuvants Elicit Influenza-Specific Th1 Immune Responses and Protect Against Heterologous H3N2 Influenza Challenge in Mice. Front. Immunol. 2020, 11, 406. [Google Scholar] [CrossRef]
- Dréno, B.; Zuberbier, T.; Gelmetti, C.; Gontijo, G.; Marinovich, M. Safety review of phenoxyethanol when used as a preservative in cosmetics. J. Eur. Acad. Derm. Venereol. 2019, 33, 15–24. [Google Scholar] [CrossRef]
- Hua, D.; Xu, P. Recent advances in biotechnological production of 2-phenyl ethanol. Biotechnol. Adv. 2011, 29, 654–660. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.S.; Fatemi, R.; Winlow, W. SARS-CoV-2 Bound Human Serum Albumin and Systemic Septic Shock. Front. Cardiovasc. Med. 2020, 7, 153. [Google Scholar] [CrossRef] [PubMed]
- Guido, T.C.; Louro, S.R.; Anteneodo, C. Hydration of hydrophobic biological porphyrins. J. Chem. Physics. 2011, 134, 02B608. [Google Scholar] [CrossRef]
- Guido, T.R.; Louro, S.R.; Pascutti, P.G.; Anteneodo, C. Solvation of anionic water—Soluble porphyrins: A computational study. Int. J. Quantum. Chem. 2010, 110, 2094–2100. [Google Scholar] [CrossRef]
- Ahmed, S.F.; Qadeer, A.A.; McKay, M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses 2020, 12, 254. [Google Scholar] [CrossRef]
- Gu, W.; Zhou, T.; Ma, J.; Sun, X.; Lu, Z. Analysis of synonymous codon usage in SARS Coronavirus and other Nidovirales. Virus Res. 2004, 101, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Bosch, B.J.; Van der Zee, R.; De Haan, C.A.; Rottier, P.J. The coronavirus spike protein is a class I virus fusion protein: Structural and functional characterization of the fusion core complex. J. Virol. 2003, 77, 8801–8811. [Google Scholar] [CrossRef]
- Jena, A.B.; Kanungo, N.; Nayak, V.; Chainy, G.B.N.; Dandapat, J. Catechin and curcumin interact with S protein of SARS-CoV2 and ACE2 of human cell membrane: Insights from computational studies. Sci. Rep. 2021, 11, 2043. [Google Scholar] [CrossRef]
- Mallick, R.; Duttaroy, A.K. Origin and Structural Biology of Novel Coronavirus (SARS-CoV-2). Adv. Exp. Med. Biol. 2021, 1352, 1–13. [Google Scholar] [CrossRef]
- Walls, A.C.; Tortorici, M.A.; Frenz, B.; Snijder, J.; Li, W.; Rey, F.A.; DiMaio, F.; Bosch, B.J.; Veesler, D. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2016, 23, 899–905. [Google Scholar] [CrossRef]
- Millet, J.K.; Whittaker, G.R. Host cell proteases: Critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015, 202, 120–134. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Liu, Y.; Yang, Y.; Zhang, P.; Zhong, W.; Wang, Y.; Wang, Q.; Xu, Y.; Li, M.; Li, X.; et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B 2020, 10, 766–788. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. Proteom. Protoc. Handb. 2005, 571–607. [Google Scholar]
- Geourjon, C.; Deleage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 1995, 11, 681–684. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein–protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef]
- Pi, M.; Kapoor, K.; Ye, R.; Nishimoto, S.K.; Smith, J.C.; Baudry, J.; Quarles, L.D. Evidence for Osteocalcin Binding and Activation of GPRC6A in β-Cells. Endocrinology 2016, 157, 1866–1880. [Google Scholar] [CrossRef]
- Jena, A.B.; Kanungo, N.; Chainy, G.B.N.; Devaraji, V.; Dandapat, J. 8-Hydroxydihydrosanguinarine (8-HDS), a pyridone containing analogue of sanguinarine, can be a potential inhibitor of S protein and M protease of SARS CoV2: Insights from computational studies. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Dassault Systemes BIOVIA. BIOVIA Discovery Studio—BIOVIA—Dassault Systèmes®, 2017. Available online: https://www.3ds.com/products-services/biovia/products/molecular-modeling-simulation/biovia-discovery-studio/(accessed on 29 September 2021).
- Pulendran, B.; Arunachalam, P.; O’Hagan, D.T. Emerging concepts in the science of vaccine adjuvants. Nat. Rev. Drug Discov. 2021, 20, 454–475. [Google Scholar] [CrossRef]
- Jena, A.B.; Samal, R.R.; Kumari, K.; Pradhan, J.; Chainy, G.B.; Subudhi, U.; Pal, S.; Dandapat, J. The benzene metabolite p-benzoquinone inhibits the catalytic activity of bovine liver catalase: A biophysical study. Int. J. Biol. Macromol. 2021, 167, 871–880. [Google Scholar] [CrossRef] [PubMed]
- Oprea, T.I.; Bauman, J.E.; Bologa, C.G.; Buranda, T.; Chagaev, A.; Edwards, B.S.; Jarvik, J.W.; Gresham, H.D.; Haynes, M.K.; Hjelle, B.; et al. Drug repurposing from an academic perspective. Drug Discov. Today Ther. Strat. 2011, 8, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Kyriakidis, N.C.; López-Cortés, A.; González, E.V.; Grimaldos, A.B.; Prado, E.O. SARS-CoV-2 vaccines strategies: A comprehensive review of phase 3 candidates. NPJ Vaccines 2021, 6, 28. [Google Scholar] [CrossRef] [PubMed]
- Wrap, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Lee, P.; Wu, X. Modifications of human serum albumin and their binding effect. Curr. Pharm. Des. 2015, 21, 1862–1865. [Google Scholar] [CrossRef] [PubMed]
- Sprinzak, E.; Sattath, S.; Margalit, H. How reliable are experimental protein-protein interaction data? J. Mol. Biol. 2003, 327, 919–923. [Google Scholar] [CrossRef]
- Daina, A.; Zoete, V. A BOILED-Egg To Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. Chem. Med. Chem. 2016, 11, 1117–1121. [Google Scholar] [CrossRef]
- Lounkine, E.; Keiser, M.J.; Whitebread, S.; Mikhailov, D.; Hamon, J.; Jenkins, J.L.; Lavan, P.; Weber, E.; Doak, A.K.; Côté, S.; et al. Large-scale prediction and testing of drug activity on side-effect targets. Nature 2012, 486, 361–367. [Google Scholar] [CrossRef]
- Keiser, M.J.; Setola, V.; Irwin, J.J.; Laggner, C.; Abbas, A.I.; Hufeisen, S.J.; Jensen, N.H.; Kuijer, M.B.; Matos, R.C.; Tran, T.B.; et al. Predicting new molecular targets for known drugs. Nature 2009, 462, 175–181. [Google Scholar] [CrossRef]
- Gfeller, D.; Grosdidier, A.; Wirth, M.; Daina, A.; Michielin, O.; Zoete, V. SwissTargetPrediction: A web server for target prediction of bioactive small molecules. Nucleic Acids. Res. 2014, 42, W32–W38. [Google Scholar] [CrossRef]
- Kola, I.; Landis, J. Can the pharmaceutical industry reduce attrition rates? Nat. Rev. Drug Discov. 2004, 3, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Braun, P.; Gingras, A.C. History of protein-protein interactions: From egg-white to complex networks. Proteomics 2012, 12, 1478–1498. [Google Scholar] [CrossRef] [PubMed]
- Rao, V.S.; Srinivas, K.; Sujini, G.N.; Kumar, G.N. Protein-protein interaction detection: Methods and analysis. Int. J. Proteom. 2014, 147648. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).