The Ins and Outs of Urea: Identification of Putative DUR3-like Urea Transporters in the Oligohaline Nerite Snail Theodoxus fluviatilis and Their Expression under Changing Salinities
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Transfer Experiments
2.3. Total RNA Extraction and cDNA Synthesis
2.4. Generation and Assembly of Transcriptome Data
2.5. Generation and Assembly of Genome Data
2.6. Identification and Characterization of DUR3-like Urea Transporter Homologues
2.7. Bioinformatic Tools
2.8. Primer Design for qRT-PCR
2.9. qRT-PCR Reactions and Gene Expression Analyses
2.10. Statistical Analyses
3. Results
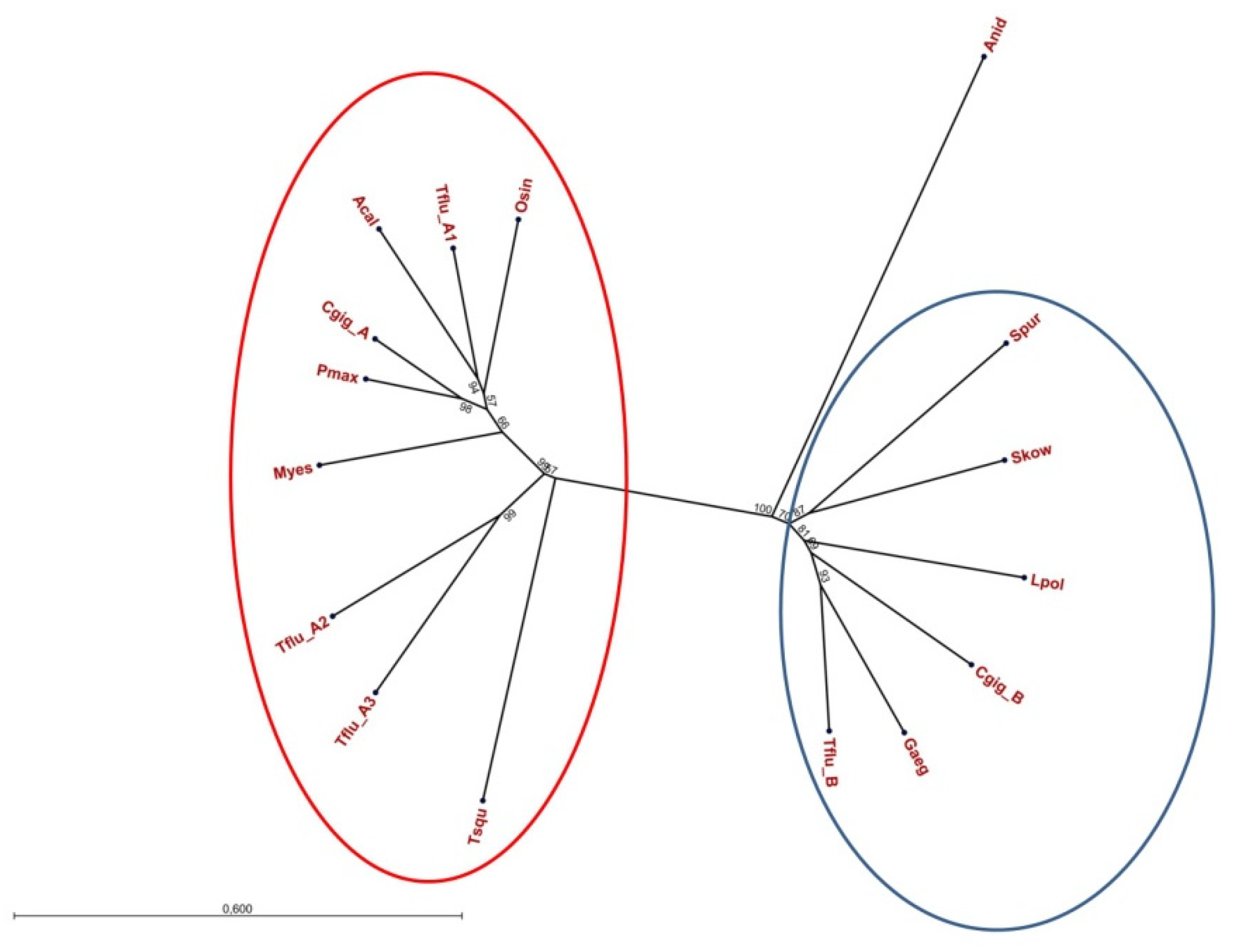
3.1. Identification of Putative DUR3-like Urea Transporter Sequences
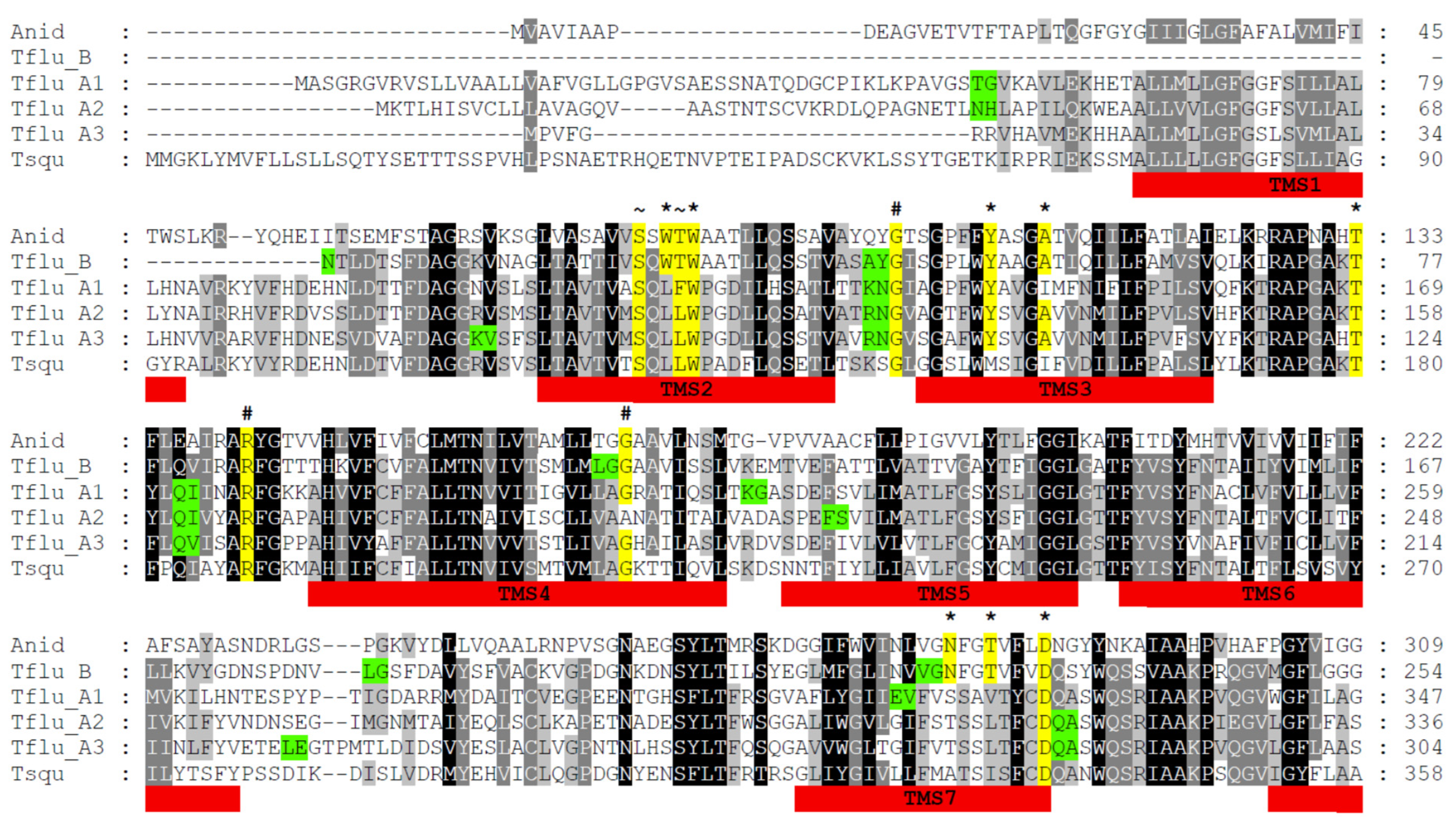
3.2. Structural Features of Putative DUR3-like Urea Transporters of T. fluviatilis
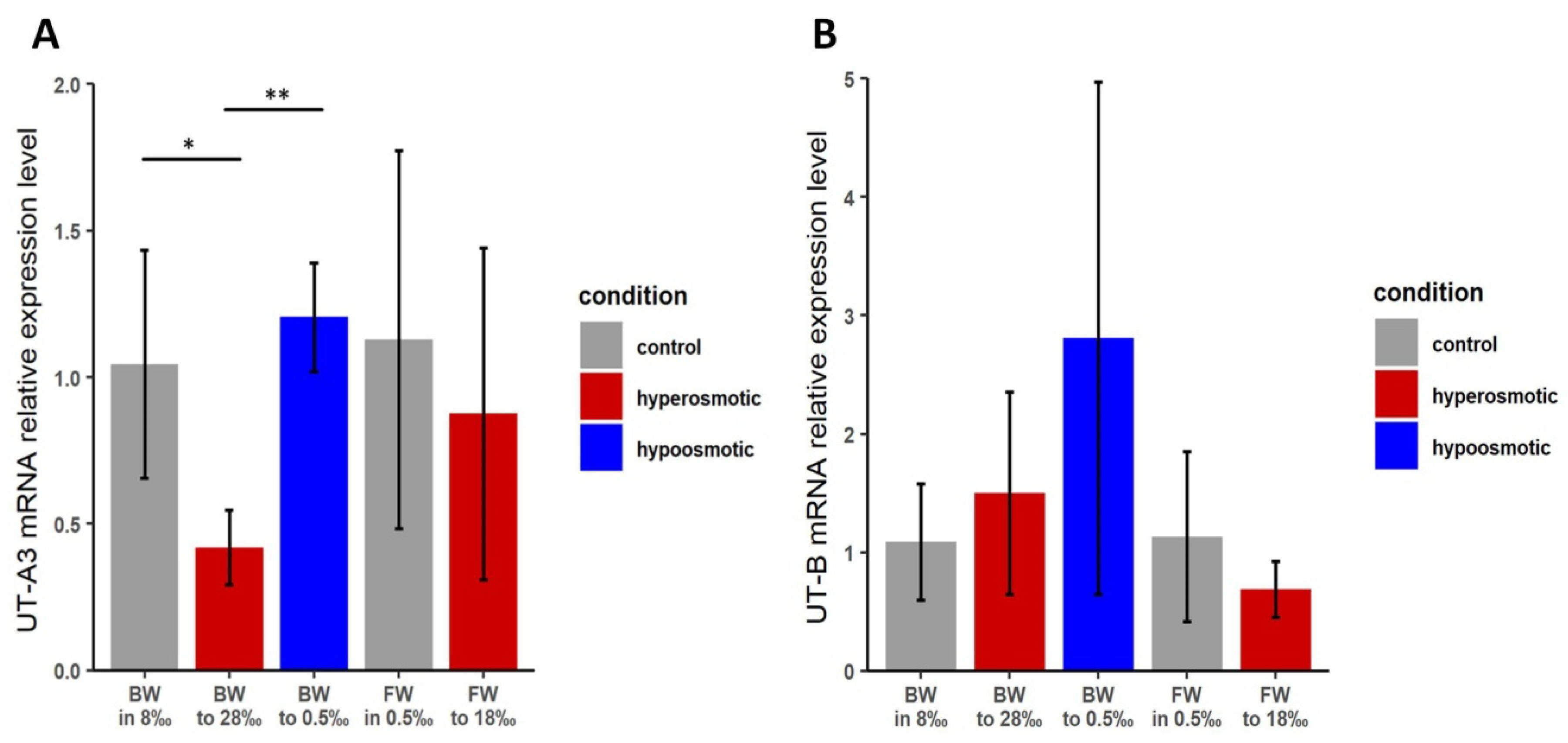
3.3. Effects of Osmotic Stress on Transcript Levels of TfUT-A3 and TfUT-B
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Ethical Approval
References
- Wang, H.; Ran, J.; Jiang, T. Urea. In Urea Transporters in Subcellular Biochemistry; Yang, B., Sands, J.M., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 7–29. [Google Scholar] [CrossRef]
- Oja, S.S.; Saransaari, P.; Korpi, E.R. Neurotoxicity of ammonia. Neurochem. Res. 2017, 42, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Pallone, T.L.; Turner, M.R.; Edwards, A.; Jamison, R.L. Countercurrent exchange in the renal medulla. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2003, 284, R1153–R1175. [Google Scholar] [CrossRef] [PubMed]
- Browning, J. Urea levels in plasma and erythrocytes of the southern fiddler skate, Trygonorhina fasciata guanerius. J. Exp. Biol. 1978, 203, 325–329. [Google Scholar] [CrossRef]
- Hazon, N.; Wells, A.; Pillans, R.D.; Good, J.P.; Anderson, W.G.; Franklin, C.E. Urea based osmoregulation and endocrine control in elasmobranch fish with special reference to euryhalinity. Comp. Biochem. Physiol. B 2003, 136, 685–700. [Google Scholar] [CrossRef]
- Li, C.; Wang, W. Urea transport mediated by aquaporin water channel proteins. In Urea Transporters in Subcellular Biochemistry; Yang, B., Sands, J.M., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 227–265. [Google Scholar] [CrossRef]
- McDonald, M.D.; Wood, C.M. Evidence for facilitated diffusion of urea across the gill basolateral membrane of the rainbow trout (Oncorhynchus mykiss). Biochim. Biophys. Acta 2004, 1663, 89–96. [Google Scholar] [CrossRef]
- Kato, A.; Sands, J.M. Active sodium-urea counter-transport is inducible in the basolateral membrane of rat renal initial inner medullary collecting ducts. J. Clin. Investig. 1998, 102, 1008–1015. [Google Scholar] [CrossRef] [PubMed]
- Morgan, R.L.; Wright, P.A.; Ballantyne, J.S. Urea transport in kidney brush-border membrane vesicles from an elasmobranch, Raja erinacea. J. Exp. Biol. 2003, 206, 3293–3302. [Google Scholar] [CrossRef] [PubMed]
- Fines, G.A.; Ballantyne, J.S.; Wright, P.A. Active urea transport and an unusual basolateral membrane composition in the gills of a marine elasmobranch. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2001, 280, R16–R24. [Google Scholar] [CrossRef]
- McDonald, M.D.; Smith, C.P.; Walsh, P.J. The physiology and evolution of urea transport in fishes. J. Membr. Biol. 2006, 212, 93–107. [Google Scholar] [CrossRef]
- Smith, C.P. Mammalian urea transporters. Exp. Physiol. 2009, 94, 180–185. [Google Scholar] [CrossRef]
- Sands, J.M.; Blount, M.A. Genes and proteins of urea transporters. In Urea Transporters in Subcellular Biochemistry; Yang, B., Sands, J.M., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 45–63. [Google Scholar] [CrossRef]
- Levin, E.J.; Cao, J.; Enkavi, G.; Quick, M.; Pan, Y.; Tajkhorshid, E.; Zhou, M. Structure and permeation mechanism of a mammalian urea transporter. Proc. Natl. Acad. Sci. USA 2012, 109, 11194–11199. [Google Scholar] [CrossRef] [PubMed]
- Sumrada, R.; Gorski, M.; Cooper, T. Urea transport-defective strains of Saccharomyces cerevisiae. J. Bacteriol. 1976, 125, 1048–1056. [Google Scholar] [CrossRef] [PubMed]
- ElBerry, H.M.; Majumdar, M.L.; Cunningham, T.S.; Sumrada, R.A.; Cooper, T.G. Regulation of the urea active transporter gene (DUR3) in Saccharomyces cerevisiae. J. Bacteriol. 1993, 175, 4688–4698. [Google Scholar] [CrossRef] [PubMed]
- Navarathna, D.H.M.L.P.; Das, A.; Morschhäuser, J.; Nickerson, K.W.; Roberts, D.D. Dur3 is the major urea transporter in Candida albicans and is co-regulated with the urea amidolyase Dur1,2. Microbiology 2011, 157, 270–279. [Google Scholar] [CrossRef]
- Abreu, C.; Sanguinetti, M.; Amillis, S.; Ramon, A. UreA, the major urea/H+ symporter in Aspergillus nidulans. Fungal Genet. Biol. 2010, 47, 1023–1033. [Google Scholar] [CrossRef]
- Kojima, S.; Bohner, A.; von Wirén, N. Molecular mechanisms of urea transport in plants. J. Membr. Biol. 2006, 212, 83–91. [Google Scholar] [CrossRef]
- Chan, C.Y.L.; Hiong, K.C.; Boo, M.V.; Choo, C.Y.L.; Wong, W.P.; Chew, S.F.; Ip, Y.K. Light exposure enhances urea absorption in the fluted giant clam, Tridacna squamosa, and up-regulates the protein abundance of a light-dependent urea active transporter, DUR3-like, in its ctenidium. J. Exp. Biol. 2018, 221, jeb176313. [Google Scholar] [CrossRef]
- Cooper, T.G.; Sumrada, R. Urea transport in Saccharomyces cerevisiae. J. Bacteriol. 1978, 121, 571–576. [Google Scholar] [CrossRef]
- Liu, L.H.; Ludewig, U.; Frommer, W.B.; von Wirén, N. AtDUR3 encodes a new type of high-affinity urea/H+ symporter in Arabidopsis. Plant Cell 2003, 15, 790–800. [Google Scholar] [CrossRef]
- Kojima, S.; Bohner, A.; Gassert, B.; Yuan, L.X.; von Wiren, N. AtDUR3 represents the major transporter for high-affinity urea transport across the plasma membrane of nitrogen-deficient Arabidopsis roots. Plant J. 2007, 52, 30–40. [Google Scholar] [CrossRef]
- Wang, W.H.; Köhler, B.; Cao, F.Q.; Liu, G.W.; Gong, Y.Y.; Sheng, S.; Song, Q.C.; Cheng, X.Y.; Garnett, T.; Okamoto, M.; et al. Rice DUR3 mediates high-affinity urea transport and plays an effective role in improvement of urea acquisition and utilization when expressed in Arabidopsis. New Phytol. 2011, 193, 432–444. [Google Scholar] [CrossRef]
- Hermida, M.; Robledo, D.; Diaz, S.; Costas, D.; Bruzos, A.L.; Blanco, A.; Pardo, B.G.; Martinez, P. The first high density genetic map of common cockle (Cerastoderma edule) reveals a major QTL controlling shell color variation. Sci. Rep. 2022, 12, 16971. [Google Scholar] [CrossRef]
- Shumway, S.E.; Freeman, R.F.H. Osmotic balance in a marine pulmonate, Amphibola crenata. Mar. Freshw. Behav. Physiol. 1984, 11, 157–183. [Google Scholar] [CrossRef]
- Henry, R.P.; McBride, C.J.; Williams, A.H. Responses of the marsh periwinkle, Littoraria (Littorina) irrorata to temperature, salinity and desiccation, and the potential physiological relationship to climbing behavior. Mar. Freshw. Behav. Physiol. 1993, 24, 45–54. [Google Scholar] [CrossRef]
- Jordan, P.J.; Deaton, L.E. Osmotic regulation and salinity tolerance in the freshwater snail Pomacea bridgesi and the freshwater clam Lampsilis teres. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 1999, 122, 199–205. [Google Scholar] [CrossRef]
- Symanowski, F.; Hildebrandt, J.-P. Differences in osmotolerance in freshwater and brackish water populations of Theodoxus fluviatilis (Gastropoda: Neritidae) are associated with differential protein expression. J. Comp. Physiol. B 2010, 180, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Wiesenthal, A.A.; Müller, C.; Harder, K.; Hildebrandt, J.-P. Alanine, proline and urea are major organic osmolytes in the snail Theodoxus fluviatilis under hyperosmotic stress. J. Exp. Biol. 2019, 222, 193557. [Google Scholar] [CrossRef] [PubMed]
- Knobloch, J.; Müller, C.; Hildebrandt, J.-P. Expression levels and activities of energy-yielding ATPases in the oligohaline neritid snail Theodoxus fluviatilis under changing environmental salinities. Biol. Open 2022, 11, bio059190. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. BUSCO update: Novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Anderson, J.B.; Cherukuri, P.F.; DeWeese-Scott, C.; Geer, L.Y.; Gwadz, M.; He, S.; Hurwitz, D.I.; Jackson, J.D.; Ke, Z.; et al. CDD: A Conserved Domain Database for protein classification. Nucleic Acids Res. 2005, 33, D192–D196. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Mizianty, M.J.; Kurgan, L. Prediction and analysis of nucleotide-binding residues using sequence and sequence-derived structural descriptors. Bioinformatics 2012, 28, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Nicholas, K.B.; Nicholas, H.B., Jr. GeneDoc: A Tool for Editing and Annotating Multiple Sequence Alignments. Distributed by the Author 1997. Available online: https://nrbsc.org/gfx/genedoc (accessed on 17 February 2023).
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Sanguinetti, M.; Santos, L.H.S.; Dourron, J.; Alamón, C.; Idiarte, J.; Amillis, S.; Pantano, S.; Ramón, A. Substrate recognition properties from an intermediate structural state of the UreA transporter. Int. J. Mol. Sci. 2022, 23, 16039. [Google Scholar] [CrossRef]
- Sanguinetti, M.; Amillis, S.; Pantano, S.; Scazzocchio, C.; Ramon, A. Modelling and mutational analysis of Aspergillus nidulans UreA, a member of the subfamily of urea/H+ transporters in fungi and plants. Open Biol. 2014, 4, 140070. [Google Scholar] [CrossRef]
- Smith, H.W. The retention and physiological role of urea in Elasmobranchii. Biol. Rev. 1936, 11, 49–82. [Google Scholar] [CrossRef]
- Trischitta, F.; Faggio, C.; Torre, A. Living with high concentrations of urea: They can! Open J. Anim. Sci. 2012, 2, 32–40. [Google Scholar] [CrossRef]
- Horne, F.R.; Barnes, G. Reevaluation of urea biosynthesis in prosobranch and pulmonate snails. J. Comp. Physiol. 1970, 69, 452–457. [Google Scholar] [CrossRef]
- Rees, B.B.; Hand, S.C. Biochemical correlates of estivation tolerance in the mountainsnail Oreohelix (Pulmonata: Oreohelicidae). Biol. Bull. 1993, 184, 230–242. [Google Scholar] [CrossRef]
- Hiong, K.C.; Loong, A.M.; Chew, S.F.; Ip, Y.K. Increases in urea synthesis and the ornithine-urea cycle capacity in the Giant African Snail, Achatina fulica, during fasting or aestivation, or after the injection with ammonium chloride. J. Exp. Zool. A 2005, 303, 1040–1053. [Google Scholar] [CrossRef]
- Lippe, C. Urea and thiourea permeabilities of phospholipid and cholesterol bilayer membranes. J. Mol. Biol. 1969, 39, 669–672. [Google Scholar] [CrossRef] [PubMed]
- Bankir, L. Active urea transport in lower vertebrates and mammals. In Urea Transporters in Subcellular Biochemistry; Yang, B., Sands, J.M., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 193–226. [Google Scholar] [CrossRef]
- LeMoine, C.M.R.; Walsh, P.J. Evolution of urea transporters in vertebrates: Adaptation to urea’s multiple roles and metabolic sources. J. Exp. Biol. 2015, 218, 1936–1945. [Google Scholar] [CrossRef] [PubMed]
- Kakinuma, M.; Suzuki, K.; Iwata, S.; Coury, D.A.; Iwade, S.; Mikami, K. Isolation and characterization of a new DUR3-like gene, PyDUR3.3, from the marine macroalga Pyropia yezoensis (Rhodophyta). Fish. Sci. 2016, 82, 171–184. [Google Scholar] [CrossRef]
- Valladares, A.; Montesinos, M.L.; Herrero, A.; Flores, E. An ABC-type, high-affinity urea permease identified in cyanobacteria. Mol. Microbiol. 2002, 43, 703–715. [Google Scholar] [CrossRef] [PubMed]


| Target Gene | Primer Sequence | Expected Amplicon Size | |
|---|---|---|---|
| GAPDH | fw | 5′-GACCTTGAAGGCAAGCTGAC-3′ | 117 bp |
| rev | 5′-GGCTTTCTTGATCTCGTCGT-3′ | ||
| TfUT-A3 | fw | 5′-GTGGCCCGCCACGATGAG-3′ | 122 bp |
| rev | 5′-GGGCACACACCTTCCTGCA-3′ | ||
| TfUT-B | fw | 5′-CAACGCGGGTCTGGTGCC-3′ | 127 bp |
| rev | 5′-GGTCACGGCCATGATCTCTGC-3′ | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Knobloch, J.; Gößeler, S.; Fuchs, L.I.R.; Fuß, J.; Torres-Oliva, M.; Müller, C.; Hildebrandt, J.-P. The Ins and Outs of Urea: Identification of Putative DUR3-like Urea Transporters in the Oligohaline Nerite Snail Theodoxus fluviatilis and Their Expression under Changing Salinities. Physiologia 2023, 3, 281-294. https://doi.org/10.3390/physiologia3020020
Knobloch J, Gößeler S, Fuchs LIR, Fuß J, Torres-Oliva M, Müller C, Hildebrandt J-P. The Ins and Outs of Urea: Identification of Putative DUR3-like Urea Transporters in the Oligohaline Nerite Snail Theodoxus fluviatilis and Their Expression under Changing Salinities. Physiologia. 2023; 3(2):281-294. https://doi.org/10.3390/physiologia3020020
Chicago/Turabian StyleKnobloch, Jan, Sarah Gößeler, Laura I. R. Fuchs, Janina Fuß, Montserrat Torres-Oliva, Christian Müller, and Jan-Peter Hildebrandt. 2023. "The Ins and Outs of Urea: Identification of Putative DUR3-like Urea Transporters in the Oligohaline Nerite Snail Theodoxus fluviatilis and Their Expression under Changing Salinities" Physiologia 3, no. 2: 281-294. https://doi.org/10.3390/physiologia3020020
APA StyleKnobloch, J., Gößeler, S., Fuchs, L. I. R., Fuß, J., Torres-Oliva, M., Müller, C., & Hildebrandt, J.-P. (2023). The Ins and Outs of Urea: Identification of Putative DUR3-like Urea Transporters in the Oligohaline Nerite Snail Theodoxus fluviatilis and Their Expression under Changing Salinities. Physiologia, 3(2), 281-294. https://doi.org/10.3390/physiologia3020020







