Human Sputum Proteomics: Advancing Non-Invasive Diagnosis of Respiratory Diseases with Enhanced Biomarker Analysis Methods
Abstract
1. Introduction
2. Methods and Systems Utilized to Study Sputum Proteomics
3. Considerations for the Selection of a Safe and Cost-Effective Lateral Flow Assay
4. Advanced Technologies to Enhance Lateral Flow Assay Analytical Sensitivities and Specificities
5. Various Technologies used to Study Proteins in Sputum
6. Potential Use of Point-of-Care Platforms for the Screening of Sputum Multi-Biomarker Proteins to Diagnose Infectious and Chronic Respiratory Diseases
7. Future Evolution and Benefits of a Point-of-Care Platform for the Determination of Multiple Biomarkers to Improve Early Detection and Diagnosis of Diseases
8. What Are the Advantages of Affinity-Capture–Separation Analytical Techniques When Compared with Traditional Methods?
9. Discussion and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Barberis, I.; Bragazzi, N.L.; Galluzo, L.; Martini, M. The history of tuberculosis: From the first historical records to the isolation of Koch’s bacillus. J. Prev. Med. Hyg. 2017, 58, E9–E12. [Google Scholar] [PubMed]
- Schluger, N.W. The acid-fast bacilli smear: Hail and farewell. Am. J. Resp. Crit. Care Med. 2019, 199, 692. [Google Scholar] [CrossRef] [PubMed]
- Singhal, R.; Myneedu, V.P. Microscopy as a diagnostic tool in pulmonary tuberculosis. Int. J. Mycobacteriol. 2015, 4, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Fireman, E. Induced sputum: Opening a new window to the lung. Sarcoidosis Vasc. Diffuse Lung Dis. 2001, 18, 263–271. [Google Scholar]
- Zhang, F.; Zhang, F.; Dong, Y.; Li, L.; Pang, Y. New insights into biomarkers for evaluating therapy efficacy in pulmonary tuberculosis: A narrative review. Infect. Dis. Ther. 2023, 12, 2665–2689. [Google Scholar] [CrossRef] [PubMed]
- Agustí, A.; Bafadhel, M.; Beasley, R.; Bel, E.H.; Faner, R.; Gibson, P.G.; Louis, R.; McDonald, V.M.; Sterk, P.J.; Thomas, M.; et al. Precision medicine in airway diseases: Moving to clinical practice. Eur. Respir. J. 2017, 50, 1701655. [Google Scholar] [CrossRef] [PubMed]
- de Souza, V.M.L.; Carvalho, L. Heterogeneity in lung cancer. Pathobiology 2018, 85, 96–107. [Google Scholar]
- Anjum, A.; Hosein, M. Diagnostic importance of saliva—An overview. J. Pak. Dent. Assoc. 2019, 28, 129–135. [Google Scholar] [CrossRef]
- Khurshid, Z.; Zafar, M.S.; Khan, R.S.; Najeeb, S.; Slowey, P.D.; Rehman, I.U. Role of salivary biomarkers in oral cancer detection. Adv. Clin. Chem. 2018, 86, 23–70. [Google Scholar]
- Goswami, Y.; Mishra, R.; Agrawal, A.P.; Agrawal, L.A. Salivary biomarkers-A review of powerful diagnostic tool. IOSR J. Dent. Med. Sci. 2015, 14, 80–87. [Google Scholar]
- Yoshizawa, J.M.; Schafer, C.A.; Schafer, J.J.; Farrell, J.J.; Paster, B.J.; Wong, D.T.W. Salivary biomarkers: Toward future clinical diagnostic utilities. Clin. Microbiol. Rev. 2013, 26, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Samara, M.; Ramachandran, R.A.; Gosh, S.; George, H.; Wang, R.; Pesavento, R.P.; Mathew, M.T. A review on saliva-based health diagnostics: Biomarker selection and future directions. Biomed. Mater. Dev. 2024, 2, 121–138. [Google Scholar] [CrossRef] [PubMed]
- Dosedělová, V.; Laštovičková, M.; Ayala-Cabrera, J.F.; Dolina, J.; Konečný, Š.; Schmitz, O.J.; Kubáň, P. Quantification and identification of bile acids in saliva by liquid chromatography-mass spectrometry: Possible non-invasive diagnostics of Barret’s esophagus? J. Chromatogr. A 2022, 1676, 463287. [Google Scholar] [CrossRef] [PubMed]
- Esther, C.R.; O’Neal, W.K.; Anderson, W.H.; Kesimer, M.; Ceppe, A.; Doerschuk, C.M.; Alexis, N.E.; Hastie, A.T.; Barr, R.G.; Bowler, R.P.; et al. Identification of sputum biomarkers predictive of pulmonary exacerbations in COPD. Chest 2022, 161, 1239–1249. [Google Scholar] [CrossRef] [PubMed]
- Correnti, S.; Prejanò, M.; Gamboni, F.; Stephenson, D.; Pelaia, C.; Pelaia, G.; Savino, R.; D’Alessandro, A.; Terraciano, R. An integrated metabo-lipidomics profile of induced sputum for the identification of novel biomarkers in the differential diagnosis of asthma and COPD. J. Transl. Med. 2024, 22, 301. [Google Scholar] [CrossRef] [PubMed]
- Bonnesen, B.; Jensen, J.-U.S.; Mathioudakis, A.G.; Corlateanu, A.; Sivapalan, P. Promising treatment biomarkers in asthma. Front. Drug Saf. Regul. 2023, 3, 1291471. [Google Scholar] [CrossRef]
- Cao, C.; Li, W.; Hua, W.; Yan, F.; Zhang, F.; Huang, H.; Ying, Y.; Li, N.; Lan, F.; Wang, S.; et al. Proteomic analysis of sputum reveals novel biomarkers for various presentations of asthma. J. Transl. Med. 2017, 15, 171. [Google Scholar] [CrossRef]
- D’Amato, M.; Iadarola, P.; Viglio, S. Proteomic analysis of human sputum for the diagnosis of lung disorders: Where are we today? Int. J. Mol. Sci. 2022, 23, 5692. [Google Scholar] [CrossRef] [PubMed]
- Hull, R.C.; Huang, J.T.J.; Barton, A.K.; Keir, H.R.; Ellis, H.; Cookson, W.O.C.; Moffatt, M.F.; Loebinger, M.R.; Chalmers, J.D. Sputum proteomics in nontuberculous mycobacterial lung disease. Chest 2022, 161, 1180–1191. [Google Scholar] [CrossRef]
- Bishwal, S.C.; Das, M.K.; Badireddy, V.K.; Dabral, D.; Das, A.; Mahapatra, A.R.; Sahu, S.; Malakar, D.; Singh, I.I.; Mazumdar, H.; et al. Sputum proteomics reveals a shift in Vitamin D-binding protein and antimicrobial protein axis in tuberculosis patients. Sci. Rep. 2019, 9, 1036. [Google Scholar] [CrossRef]
- Kesimer, M.; Sheehan, J.K. Mass spectrometry analysis of mucin core proteins. Methods Mol. Biol. 2012, 842, 67–79. [Google Scholar]
- Burg, D.; Schofield, J.P.R.; Brandsma, J.; Staykova, D.; Folisi, C.; Bansal, A.; Nicholas, B.; Xian, Y.; Rowe, A.; Corfield, J.; et al. Large-scale label-free quantitative mapping of the sputum proteome. J. Proteome Res. 2018, 17, 2072–2091. [Google Scholar] [CrossRef] [PubMed]
- Maher, R.E.; Barrett, E.; Beynon, R.J.; Harman, V.M.; Jones, A.M.; McNamara, P.S.; Smith, J.A.; Lord, R.W. The relationship between lung disease severity and the sputum proteome in cystic fibrosis. Respir. Med. 2022, 204, 107002. [Google Scholar] [CrossRef]
- Moermans, C.; Brion, C.; Bock, G.; Graff, S.; Gerday, S.; Nekoee, H.; Poulet, C.; Bricmont, N.; Henket, M.; Paulus, V.; et al. Sputum Type 2 markers could predict remission in severe asthma treated with anti-IL-5. Chest 2023, 163, 1368–1379. [Google Scholar] [CrossRef] [PubMed]
- Nolasco, S.; Crimi, C.; Campisi, R. Personalized medicine in asthma: Current approach and future perspectives. J. Pers. Med. 2023, 13, 1459. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Guo, T.; Yang, H.; Deng, P.; Gu, Q.; Liu, C.; Wu, M.; Lizaso, A.; Li, B.; Zhang, S.; et al. The utility of sputum as an alternative liquid biopsy specimen for next-generation sequencing-based somatic variation profiling. Ann. Transl. Med. 2022, 10, 462. [Google Scholar] [CrossRef] [PubMed]
- Lone, S.N.; Nisar, S.; Masoodi, T.; Singh, M.; Rizwan, A.; Hashem, S.; El-Rifai, W.; Bedognetti, D.; Batra, S.K.; Haris, M.; et al. Liquid biopsy: A step closer to transform diagnosis, prognosis and future of cancer treatments. Mol. Cancer 2022, 21, 79. [Google Scholar] [CrossRef]
- Wang, N.; Wang, Q.; Du, T.; Gabriel, A.N.A.; Wang, X.; Sun, L.; Li, X.; Xu, K.; Jiang, X.; Zhang, Y. The potential roles of exosomes in chronic obstructive pulmonary disease. Front. Med. 2020, 7, 618506. [Google Scholar] [CrossRef]
- Hortin, G.L.; Carr, S.A.; Anderson, N.L. Introduction: Advances in protein analysis for the clinical laboratory. Clin. Chem. 2010, 56, 149–151. [Google Scholar] [CrossRef]
- Harti, J.; Kurth, F.; Kappert, K.; Mülleder, M.; Hartman, G.; Raiser, M. Quantitative protein biomarker panel: A path to improved clinical practice through proteomics. EMBO Mol. Med. 2023, 15, 4. [Google Scholar]
- Captur, G.; Moon, J.C.; Topriceanu, C.-C.; Joy, G.; Swading, L.; Hallqvist, J.; Doykov, I.; Patel, N.; Spiewak, J.; Balswin, T.; et al. Plasma proteomics signature predicts who will get persistent symptoms following SARS-CoV-2 infection. eBiomedicine 2022, 85, 104293. [Google Scholar] [CrossRef] [PubMed]
- Wallace, L.E.; Liu, M.; van Kuppeveld, F.J.M.; de Vries, E.; de Haan, C.A.M. Respiratory mucus as a virus-host range determinant. Trends Microbiol. 2021, 29, 983–992. [Google Scholar] [CrossRef] [PubMed]
- Farzan, S. Cough and sputum production. In Clinical Methods: The History, Physical, and Laboratory Examinations, 3rd ed.; Walker, H.K., Hall, W.D., Hurst, J.W., Eds.; Butterworths: Boston, MA, USA, 1990; Chapter 38. [Google Scholar]
- Charriots, J.; Volpato, M.; Petit, A.; Vachier, I.; Bourdin, A. Methods of sputum and mucus assessment for muco-obstructive lung diseases in 2022: Time to “unplug” from our daily routine! Cells 2022, 11, 812. [Google Scholar] [CrossRef] [PubMed]
- Dao, T.L.; Hoang, V.T.; Ly, T.D.A.; Lagier, J.C.; Baron, S.A.; Raoult, D.; Parola, C.; Courjon, J.; Marty, P.; Chaudet, H.; et al. Sputum proteomic analysis for distinguishing between pulmonary tuberculosis and non-tuberculosis using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS): Preliminary results. Clin. Microbiol. Infect. 2021, 27, e1–e1694. [Google Scholar] [CrossRef] [PubMed]
- Radicioni, G.; Ceppe, A.; Ford, A.A.; Alexis, N.E.; Barr, R.G.; Bleecker, E.R.; Christenson, S.A.; Cooper, C.B.; Han, M.K.; Hansel, N.N.; et al. Airway mucin MUC5AC and MUC5B concentrations and the initiation and progression of chronic obstructive pulmonary disease: An analysis of the SPIROMICS cohort. Lancet Respir. Med 2021, 9, 1241–1254. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, M.; Yu, Y.; Han, T.; Zhou, J.; Bi, L. Sputum characteristics and airway clearance methods in patients with severe COVID-19. Medicine 2020, 99, e23257. [Google Scholar] [CrossRef]
- Panchabhai, T.S.; Methta, A.C. Historical perspectives of bronchoscopy. Connecting the dots. Ann. Am. Thorac. Soc. 2015, 12, 631–641. [Google Scholar] [CrossRef]
- Aravena, C.; Metha, A.C.; Almeida, F.A.; Lamb, C.; Maldonado, F.; Gildea, T.R. Innovation in rigid broncoscopy—Past, present, and future. J. Thorac. Dis. 2023, 15, 2836–2847. [Google Scholar] [CrossRef]
- Elsevier Health. Specimen Collection: Sputum (Pediatrics). January 2024. Available online: https://elsevier.health/en-US/preview/specimen-sputum-peds (accessed on 25 January 2024).
- Guzman, N.A.; Guzman, D.E. A COVID-19 infection with positive sputum and negative nasopharyngeal/oropharyngeal rapid antigen-based testing: A case report and pilot study. Med. Clin. Case Rep. J. 2023, 1, 77–84. [Google Scholar] [CrossRef]
- Dragonieri, S.; Bilov, A.; Capuano, A.; Scarlata, S.; Carpagnano, G.E. Methodological aspects of induced sputum. Adv. Respir. Med. 2023, 91, 397–406. [Google Scholar] [CrossRef]
- Paggiaro, P.L.; Chanez, P.; Holz, O.; Ind, P.W.; Djukanović, T.; Maestrelli, P.; Sterk, P.J. Sputum induction. Eur. Respir. J. 2020, 20, 3s–8s. [Google Scholar]
- Taube, C.; Holz, O.; Mücke, M.; Jörres, R.A.; Magnussen, H. Airway response to inhaled saline in patients with moderate to severe chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2001, 164, 1810–1815. [Google Scholar] [CrossRef]
- Bart, I.Y.; Mourits, M.; van Gent, R.; van Leuken, M.H.; Hilbink, M.; Warris, A.; Wever, P.C.; de Vries, E. Sputum induction in children is feasible and useful in a bustling general hospital practice. Glob. Pediatr. Health 2016, 3, 2333794X16636504. [Google Scholar] [CrossRef] [PubMed]
- Rueda, Z.V.; Bermúdez, M.; Restrepo, A.; Garcés, C.; Morales, O.; Roya-Pabón, C.; Carmona, L.F.; Arango, C.; Albarracín, J.L.; López, L.; et al. Induced sputum as an adequate clinical specimen for the etiological diagnosis of community-acquired pneumonia (CAP) in children and adolescents. Int. J. Infect. Dis. 2022, 116, 348–354. [Google Scholar] [CrossRef]
- Gibson, P.G.; Grootendorst, D.C.; Henry, R.; Pin, I.; Rytila, P.L.; Wark, P.; Wilson, N.; Gjukanović, R. Sputum induction in children. Eur. Respir. J. 2002, 20, 44s–46s. [Google Scholar]
- Weiszhar, Z.; Horvath, I. Induced sputum ana lysis: Step by step. Breathe 2013, 9, 301–306. [Google Scholar] [CrossRef]
- Goncalves, B.; Eze, U.A. Sputum induction and its diagnostic applications in inflammatory airway disorders: A review. Front. Allergy 2023, 4, 1282782. [Google Scholar] [CrossRef] [PubMed]
- Suri, R.; Marshall, L.J.; Wallis, C.; Metcalfe, C.; Shute, J.K.; Bush, A. Safety and use of sputum induction in children with cystic fibrosis. Pediatr. Pulmonol. 2003, 35, 309–313. [Google Scholar] [CrossRef] [PubMed]
- Kang, A.; Yeom, M.; Kim, H.; Yoon, S.-W.; Jeong, D.-G.; Moon, H.-J.; Lyoo, K.-S.; Na, W.; Song, D. Sputum processing method for lateral flow immunochromatography assays to detect coronaviruses. Immune Netw. 2021, 21, e11. [Google Scholar] [CrossRef]
- Barber, C.; Lau, K.; Ward, J.A.; Daniels, T.; Watson, A.; Staples, K.J.; Wilkinson, T.M.A.; Howarth, P.H. Sputum processing by mechanical dissociation: A rapid alternative to traditional sputum assessment approaches. Clin. Respir. J. 2021, 15, 800–807. [Google Scholar] [CrossRef]
- Clemente, A.; Alba-Patiño, A.; Santopolo, G.; Rojo-Molinero, E.; Oliver, A.; Borges, M.; Aranda, M.; del Castillo, A.; de la Rica, R. Immunodetection of lung IgG and IgM antibodies against SARS-CoV-2 via enzymatic liquefaction of respiratory samples from COVID-19 patients. Anal. Chem. 2021, 93, 5259–5266. [Google Scholar] [CrossRef] [PubMed]
- Hiraiwa, M.; Kim, J.-H.; Lee, H.-B.; Inoue, S.; Becker, A.L.; Weigel, K.W.; Cangelosi, G.A.; Lee, K.-H.; Chung, J.-H. Amperometric immunosensor for rapid detection of Mycobacterium tuberculosis. J. Micromech. Microeng. 2015, 25, 055013. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, T.M.; Weigel, K.M.; Becker, A.L.; Ontengco, D.; Narita, M.; Tolstorukov, I.; Doebler, R.; Cangelosi, G.A.; Niemz, A. Pilot study of a rapid and minimally instrumented sputum sample preparation for molecular diagnosis of tuberculosis. Sci. Rep. 2016, 6, 19541. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.R.; Guan, S.Z.; Zhang, S.Y.; Li, M. Proteomic analysis of sputum in patients with active pulmonary fibrosis. Clin. Microbiol. Infect. 2012, 18, 1241–1247. [Google Scholar] [CrossRef]
- Johnson-Arbor, K.; Reid, N.; Smolinske, S. Human toxicity from COVID-19 rapid home test kits. Am. J. Emerg. Med. 2022, 57, 215–216. [Google Scholar] [CrossRef]
- Richardson, S.; Kohn, M.A.; Bollyky, J.; Parsonnet, J. Validity of at-home rapid antigen lateral flow assay and artificial intelligence read to detect SARS-CoV-2. Diagn. Microbiol. Infect. Dis. 2022, 104, 115763. [Google Scholar] [CrossRef] [PubMed]
- Filchakova, O.; Dossym, D.; Ilyas, A.; Kuanysheva, T.; Abdizhamil, A.; Bukasov, R. Review of COVID-19 testing and diagnostic methods. Talanta 2022, 244, 123409. [Google Scholar] [CrossRef] [PubMed]
- Lee, H. At Home COVID-19 Antigen Test Kits: Where to Buy and What You Should Know. 2023. Available online: https://www.nytimes.com/wirecutter/reviews/at-home-covid-test-kits/ (accessed on 21 November 2023).
- Omidfar, K.; Riahi, F.; Kashanian, S. Lateral flow assay: A summary of recent progress for improving assay performance. Biosensors 2023, 13, 837. [Google Scholar] [CrossRef] [PubMed]
- Chiu, W.-H.; Kong, W.-Y.; Chueh, Y.-H.; Wen, J.-W.; Tsai, C.-M.; Hong, C.; Chen, P.-Y.; Ko, C.-H. Using an ultra-compact optical system to improve lateral flow immunoassay results quantitatively. Heliyon 2022, 8, e12116. [Google Scholar] [CrossRef]
- Panferov, V.G.; Ivanov, N.A.; Brinc, D.; Fabros, A.; Krylov, S.N. Electrophoretic assembly of antibody-antigen complexes facilitates 1000 times improvement in the limit of detection of serological paper-based assay. ACS Sens. 2023, 8, 1792–1798. [Google Scholar] [CrossRef]
- Panferov, V.G.; Zherdev, A.V.; Dzantiev, B.B. Post-assay chemical enhancement for highly sensitive lateral flow immunoassay: A critical review. Biosensors 2023, 13, 866. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Chen, Y.; Zhu, Y.; Zhao, M.; Xu, Y. Enhancing the sensitivity of lateral flow immunoassay by centrifugation-assisted flow control. Anal. Chem. 2019, 91, 4814–4820. [Google Scholar] [CrossRef] [PubMed]
- Le, T.S.; He, S.; Takahashi, M.; Enomoto, Y.; Matsumura, Y.; Maenosono, S. Enhancing the sensitivity of lateral flow immunoassay by magnetic enrichment using multifunctional nanocomposite probes. Langmuir 2021, 37, 6566–6577. [Google Scholar] [CrossRef]
- Cho, W.C.S. Proteomics technologies and challenges. Genom. Proteom. Bioinform. 2007, 5, 77–85. [Google Scholar] [CrossRef]
- Al-Amrani, S.; Al-Jabri, Z.; Al-Zaabi, A.; Alshekaili, J.; Al-Khabori, M. Proteomics: Concepts and applications in human medicine. World J. Biol. Chem. 2021, 12, 57–69. [Google Scholar] [CrossRef]
- Smith, L.M.; Agar, J.N.; Chamot-Rooke, J.; Danis, P.O.; Ge, Y.; Loo, J.A.; Paša-Tolić, L.; Tsybin, Y.O.; Kelleher, N.L. The Consortium for Top-Down Proteomics. Sci. Adv. 2021, 7, eabk0734. [Google Scholar] [CrossRef]
- Guzman, N.A.; Guzman, D.E. Immunoaffinity capillary electrophoresis in the era of proteoforms, liquid biopsy and preventive medicine. A potential impact in the diagnosis and monitoring of disease progression. Biomolecules 2021, 11, 1443. [Google Scholar] [CrossRef] [PubMed]
- Seydel, C. Diving deeper into the proteome. Nat. Methods 2022, 19, 1036–1040. [Google Scholar] [CrossRef]
- Huang, L.; Shao, D.; Wang, Y.; Cui, X.; Li, Y.; Chen, Q.; Cui, J. Human body-fluid proteome: Quantitative profiling and computational prediction. Brief. Bioinform. 2021, 22, 315–333. [Google Scholar] [CrossRef]
- Nicholas, B.; Skipp, P.; Mould, R.; Rennard, S.; Davies, D.E.; O’Connor, C.D.; Djukanović, R. Shotgun proteomic analysis of human-induced sputum. Proteomics 2006, 6, 4390–4401. [Google Scholar] [CrossRef]
- Terracciano, R.; Preianò, M.; Palladino, G.P.; Carpagnano, G.E.; Barbaro, M.P.F.; Pelaia, G.; Savino, R.; Maselli, R. Peptidome profiling of induced sputum by mesoporous silica beads and MALDI-TOF MS for non-invasive biomarker discovery of chronic inflammatory lung diseases. Proteomics 2011, 11, 3402–3414. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, T.; Riedel, S.; Kapcia, J., III; Caverly, L.J.; Carmody, L.; Kalikin, L.M.; Lu, J.; Phan, J.; Gargus, M.; Kagawa, M.; et al. Liquid chromatography mass spectrometry detection of antibiotic agents in sputum from persons with cystic fibrosis. Antimicrob. Agents Chemother. 2021, 65, e00927. [Google Scholar] [CrossRef] [PubMed]
- Mateos, J.; Estévez, O.; González-Fernández, A.; Anibarro, L.; Pallerés, A.; Reljic, R.; Gallardo, J.M.; Medina, I.; Carrera, M. High-resolution quantitative proteomics applied to the study of the specific protein signature in the sputum and saliva of active tuberculosis patients and their infected and uninfected contacts. J. Proteom. 2019, 195, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Casado, B.; Iadarola, P.; Pannell, L.K.; Luisetti, M.; Corsico, A.; Ansaldo, E.; Ferrarotti, I.; Boschetto, P.; Baraniuk, J.N. Protein expression in sputum of smokers and chronic obstructive pulmonary disease patients: A pilot study by CapLC-ESI-Q-TOF. J. Proteome Res. 2007, 6, 46. [Google Scholar] [CrossRef] [PubMed]
- Pattison, S.H.; Gibson, D.S.; Johnston, E.; Peacock, S.; Rivera, K.; Tunney, M.M.; Pappin, D.J.; Elborn, J.S. Proteomic profile of cystic fibrosis sputum cells in adults chronically infected with Pseudomonas aeruginosa. Eur. Respir. J. 2017, 50, 1601569. [Google Scholar] [CrossRef] [PubMed]
- Asamoah, K.; Chung, K.F.; Kermani, N.Z.; Bodinier, B.; Dahlen, S.-E.; Djukanovic, R.; Bhavsar, P.K.; Adcock, I.M.; Vuckovic, D.; Chadeau-Hyam, M. Proteomic signatures of eosinophilic and neutrophilic asthma from serum and sputum. eBioMedicine 2024, 19, 104936. [Google Scholar] [CrossRef] [PubMed]
- HaileMariam, M.; Yu, Y.; Singh, H.; Teklu, T.; Wondale, B.; Worku, A.; Zewude, A.; Mounaud, S.; Tsitrin, T.; Legesse, M.; et al. Protein and microbial biomarkers in sputum discern acute and latent tuberculosis in investigation of pastoral Ethiopian cohort. Front. Cell. Infect. Microbiol. 2021, 11, 595554. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Santos, D.; Yang, Z.; Yang, S.; Kirkhus, N.E. Sputum and salivary protein biomarkers and point-of-care biosensors for the management of COPD. Analyst 2020, 145, 1583–1604. [Google Scholar] [CrossRef] [PubMed]
- Dennis, E.G.; Hornbrook, M.M.; Ishizaka, K. Serum proteins in sputum of patients with asthma. J. Allergy Clin. Immunol. 1964, 35, 464–471. [Google Scholar] [CrossRef]
- Houston, C.J.; Alkhatib, A.; Einarsson, G.G.; Tunney, M.M.; Taggart, C.C.; Downey, D.G. Diminished airway host innate response in people with cystic fibrosis who experience frequent pulmonary exacerbations. Eur. Respir. J. 2013, in press. [Google Scholar] [CrossRef]
- Ali, M.M.; Mukherjee, M.; Radford, K.; Patel, Z.; Capretta, A.; Nair, P.; Brennan, J.D. A rapid sputum-based lateral flow assay for airway eosinophilia using RNA-cleaving DNAzyme selected for eosinophil peroxidase. Angew. Chem. Int. Ed. 2023, 62, e202307451. [Google Scholar] [CrossRef] [PubMed]
- Gharib, S.A.; Nguyen, E.V.; Lai, Y.; Plampin, J.D.; Goodlett, D.R.; Hallstrand, T.S. Induced sputum proteome in healthy subjects and asthmatic patients. J. Allergy Clin. Immunol. 2011, 128, 1176–1184.e6. [Google Scholar] [CrossRef] [PubMed]
- Rezaeeyan, H.; Arabfard, M.; Rasouli, H.R.; Shahriary, A.; Gh, B.F.N.M. Evaluation of common protein biomarkers involved in the pathogenesis of respiratory diseases with proteomic methods: A systematic approach. Immun. Inflamm. Dis. 2023, 11, e1090. [Google Scholar] [CrossRef] [PubMed]
- Polachek, A.; Vree Egberts, W.; Fireman, E.; Druckman, I.; Stark, M.; Paran, D.; Kaufman, I.; Wigler, I.; Levartovsky, D.; Caspi, D.; et al. Sputum anticitrullinated protein antibodies in patients with long-standing rheumatoid arthritis. J. Clin. Rheumatol. 2018, 24, 122–126. [Google Scholar] [CrossRef]
- Devlin, R.B.; Alexis, N.E.; Carter, J.; Ghio, A.J.; Richards, J.H. Iron-binding and storage proteins in sputum. Inhal. Toxicol. 2004, 14, 387–400. [Google Scholar]
- Bodaghi, A.; Fattahi, N.; Ramazani, A. Biomarkers: Promising and valuable tools towards diagnosis, prognosis and treatment of Covid-19 and other diseases. Heliyon 2023, 9, e13323. [Google Scholar] [CrossRef] [PubMed]
- McNerney, M.P.; Zhang, Y.; Silverman, A.D.; Jewett, M.C.; Styczynski, M.P. Point-of-care biomarker quantification enabled by sample-specific calibration. Sci. Adv. 2019, 5, eaax4473. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.G.; Gerszten, R.E. Emerging affinity-based proteomic technologies for large-scale plasma profiling in cardiovascular disease. Circulation 2017, 135, 1651–1664. [Google Scholar] [CrossRef] [PubMed]
- Ling, X.; Li, G. Accelerating sample preparation for the analysis of complex samples. LC-GC 2022, 40, 374–376. [Google Scholar] [CrossRef]
- Huang, P.-H.; Ren, L.; Nama, N.; Li, S.; Li, P.; Yao, X.; Cuento, R.A.; Wei, C.-H.; Chen, Y.; Xie, Y.; et al. An acoustofluidic sputum liquefier. Lab Chip 2015, 15, 3125–3131. [Google Scholar] [CrossRef]
- Kim, S.-H.; Weiß, C.; Hoffman, U.; Borggrefe, M.; Akin, I.; Behnes, M. Advantages and limitations of current biomarker research: From experimental research to clinical application. Curr. Pharm. Biotechnol. 2017, 18, 445–455. [Google Scholar] [CrossRef] [PubMed]
- St. John, A.; Price, C.P. Existing technologies for point-of-care testing. Clin. Biochem. Rev. 2014, 35, 155–167. [Google Scholar]
- Verrills, N.M. Clinical proteomics: Present and future prospects. Clin. Biochem. Rev. 2006, 27, 99–116. [Google Scholar] [PubMed]
- Papatheocharidou, C.; Samanidou, V. Two-dimensional high-performance liquid chromatography as a powerful tool for bioanalysis: The paradigm of antibiotics. Molecules 2003, 28, 5056. [Google Scholar] [CrossRef] [PubMed]
- Kitteringham, N.R.; Jenkins, R.E.; Lane, C.S.; Elliott, V.L.; Park, B.K. Multiple reaction for quantitative biomarker analysis in proteomics and metabolomics. J. Chromatogr. B 2009, 877, 1229–1239. [Google Scholar] [CrossRef]
- Banerjee, S. Empowering clinical diagnostics with mass spectrometry. ACS Omega 2020, 5, 2041–2048. [Google Scholar] [CrossRef]
- Di Nardo, F.; Chiarello, M.; Cavalera, S.; Baggiani, C.; Anfossi, L. Ten years of lateral flow immunoassay technique applications: Trends, challenges and future perspectives. Sensors 2021, 21, 5185. [Google Scholar] [CrossRef] [PubMed]
- Sena-Torralba, A.; Álvarez-Diduk, R.; Parolo, C.; Mercoçi, A. Toward next generation lateral flow assays: Integration of nanomaterials. Chem. Rev. 2022, 122, 14881–14910. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Jiang, H.; Li, X.; Lv, X. Recent advances in sensitivity enhancement for lateral flow assay. Microchim. Acta 2021, 188, 379. [Google Scholar] [CrossRef]
- Guzman, N.A.; Guzman, D.E.; Blanc, T. Advancements in portable instruments based on affinity- capture-migration and affinity-capture-separation for use in clinical testing and life applications. J. Chromatogr. A 2023, 1704, 464109. [Google Scholar] [CrossRef]
- Guzman, N.A. Determination of sputum constituents by capillary electrophoresis. in preparation.
- Guzman, N.A.; Phillips, T.M. Immunoaffinity capillary electrophoresis: A new versatile tool for determining protein biomarkers in inflammatory processes. Electrophoresis 2011, 32, 1565–1578. [Google Scholar] [CrossRef] [PubMed]
- Esene, J.E.; Burningham, A.J.; Tahir, A.; Nordin, G.P.; Wooley, A.T. 3D printed microfluidic devices for integrated solid-phase extraction microchip electrophoresis of preterm birth biomarkers. Anal. Chim. Acta 2024, 1296, 342338. [Google Scholar] [CrossRef] [PubMed]
- Pont, L.; Benavente, F.; Barbosa, J.; Sanz-Nebot, V. On-line immunoaffinity solid-phase extraction capillary electrophoresis mass spectrometry using Fab’ antibody fragments for the analysis of serum transthyretin. Talanta 2017, 170, 224–232. [Google Scholar] [CrossRef] [PubMed]
- Vergara-Barberán, M.; Pont, L.; Salim, H.; Giménez, E.; Benavente, F. On-line aptamer affinity solid-phase extraction capillary electrophoresis-mass spectrometry for the analysis of protein biomarkers in biological fluids and food: A tutorial. Adv. Sample Prep. 2023, 7, 100082. [Google Scholar] [CrossRef]
- Effenhauser, C.S.; Manz, A.; Widmer, H.M. Glass chips for high-speed capillary electrophoresis separations with submicrometer plate heights. Anal. Chem. 1993, 65, 2637–2642. [Google Scholar] [CrossRef]
- Garcia, C.D.; Henry, C.S. Direct detection of renal function markers using microchip CE with pulsed electrochemical detection. Analyst 2004, 129, 579–584. [Google Scholar] [CrossRef]
- Ávila, M.; Floris, A.; Staal, S.; Ríos, Á.; Eijkel, J.; van den Berg, A. Point of care creatinine measurement for diagnosis of renal disease using disposable microchip. Electrophoresis 2013, 34, 2956–2961. [Google Scholar] [CrossRef] [PubMed]
- Eijkel, J. Chip-based capillary electrophoresis platforms: Toward point-of-care applications. Bioanalysis 2015, 7, 1385–1387. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.-Z.; Fang, P.; Fang, X.-X.; Hu, T.-T.; Fang, J.; Fang, Q. A low-cost palmtop high-speed capillary electrophoresis bioanalyzer with laser induced fluorescence detection. Sci. Rep. 2018, 8, 1791. [Google Scholar] [CrossRef]
- Nguyen, V.D.; Nguyen, H.Q.; Bui, K.H.; Ko, Y.S.; Park, B.J.; Seo, T.S. A handheld-type total integrated capillary electrophoresis system for SARS-CoV-2 diagnostics: Power, fluorescence detection, and data analysis by smartphone. Biosen. Bioelectron. 2022, 195, 113632. [Google Scholar] [CrossRef]
- Harrison, D.J.; Fluri, K.; Seiler, K.; Fan, Z.; Effenhauser, C.S.; Manz, A. Micromachining a miniaturized capillary electrophoresis-based chemical analysis system on a chip. Science 1993, 261, 895–897. [Google Scholar] [CrossRef] [PubMed]
- Kubáň, P.; Kubáň, P. Novel developments in capillary electrophoresis miniaturization, sampling, detection and portability: An overview of the last decade. Trends Anal. Chem. 2023, 151, 116941. [Google Scholar] [CrossRef]
- He, M.; Xue, Z.; Zhang, Y.; Huang, Z.; Fang, X.; Qu, F.; Ouyang, Z.; Xu, W. Development and characterizations of a miniature capillary electrophoresis mass spectrometry system. Anal. Chem. 2015, 87, 2236–2241. [Google Scholar] [CrossRef]
- Annese, V.F.; Hu, C. Integrating microfluidics and electronics in point-of-care diagnostics: Current and future challenges. Micromachines 2022, 13, 1923. [Google Scholar] [CrossRef] [PubMed]
- Behera, P.P.; Kumar, N.; Kumari, M.; Kumar, S.; Mondal, P.K.; Arun, R.K. Integrated microfluidic devices for point-of-care detection of bio-analytes and disease. Sens. Diagn. 2023, 2, 1437–1459. [Google Scholar] [CrossRef]
- Choi, B.C.K.; Morrison, H.; Wong, T.; Wu, J.; Yan, Y.-P. Bringing chronic disease and infectious disease epidemiology back together. J. Epidemiol. Community Health 2007, 61, 832. [Google Scholar] [CrossRef] [PubMed]
- Peeling, R.W.; Mabey, D. Point-of-care tests for diagnosing infections in the developing world. Clin. Microbiol. Infect. 2010, 16, 1062–1069. [Google Scholar] [CrossRef]
- Goldstein, S. Chronic, noncommunicable diseases (NCDs): A silent scourge threatening to overwhelm global health. Fogarty Int. Cent. NIH 2011, 10, 5. Available online: https://www.fic.nih.gov/News/GlobalHealthMatters/Sept-Oct-2011/Pages/chronic-disease.aspx (accessed on 4 March 2024).
- Priyadarshani, W.V.D.; de Namor, A.F.D.; Silva, S.R.P. Rising of a global silent killer: Critical analysis of chronic kidney disease if uncertain aetiology (CKDu) worldwide and mitigation steps. Environ. Geochem. Health 2023, 45, 2647–2662. [Google Scholar] [CrossRef]
- Marrif, H.I.; Al-Sunousi, S.I. Pancreatic ß cell mass death. Front. Pharmacol. 2016, 7, 83. [Google Scholar] [CrossRef]
- Guzman, N.A.; Guzman, D.E. An emerging microscale immuno-analytical diagnostic tool to see the unseen. Holding promise for precision medicine and P4 medicine. J. Chromatogr. B 2016, 1021, 14–29. [Google Scholar] [CrossRef] [PubMed]
- Guzman, N.A.; Guzman, D.E. From a central laboratory to the bedside: A point-of-care instrument for monitoring wellness and disease using two-dimensional immunoaffinity capillary electrophoresis technology. Arch. Biomed. Res. 2018, 1, 1. [Google Scholar]
- Guzman, N.A. Disease Detection System and Method. U.S. Patent 10,408,789, 10 September 2019. [Google Scholar]
- Guzman, N.A. Method and System for Simultaneous Determination of Multiple Measurable Biomarkers during the Development of a Communicable Disease. U.S. Patent 11,740,204, 29 August 2023. [Google Scholar]
- Thiha, A.; Ibrahim, F. A colorimetric enzyme-linked immunosorbent assay (ELISA) detection platform for a point-of-care dengue detection system on a lab-on-compact-disc. Sensors 2015, 15, 11431–11441. [Google Scholar] [CrossRef] [PubMed]
- Alhajj, M.; Zubair, M.; Farhana, A. Enzyme linked immunosorbent assay. StatPearls 2023. Available online: https://www.ncbi.nlm.nih.gov/books/NBK555922 (accessed on 23 April 2023).
- Bar-Haim, E.; Rotem, S.; Elia, U.; Bercovich-Kinori, A.; Israeli, M.; Chen-Gihon, I.; Israeli, O.; Erez, N.; Achdout, H.; Zauberman, A.; et al. Early diagnosis of pathogen infection by cell-based activation immunoassay. Cells 2019, 8, 952. [Google Scholar] [CrossRef] [PubMed]
- Leng, S.X.; McElhaney, J.E.; Walston, J.D.; Xie, D.; Fedarko, N.S.; Kuchel, G.A. ELISA and multiplex technologies for cytokine measurement in inflammation and aging research. J. Gerontol. 2008, 63A, 879–884. [Google Scholar] [CrossRef] [PubMed]
- Ungaro, C.T.; Wolfe, A.S.; Brown, S.D. Comparison of serum cytokine measurement techniques between ELISA vs Multiplex. FASEB J. 2020, 34, 1. [Google Scholar] [CrossRef]
- Kim, C.H.; Tworoger, S.S.; Stampfer, M.; Dillon, S.T.; Gu, X.; Sawyer, S.J.; Chan, A.T.; Libermann, T.A.; Eliassen, A.H. Stability and reproducibility of proteomic profiles measured with an aptamer-based platform. Sci. Rep. 2018, 8, 8382. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tam, W.W.; Yu, Y.; Zhuo, Z.; Xue, Z.; Tsang, C.; Qiao, X.; Wang, X.; Li, Y.; Tu, Y.; et al. The application of aptamer in biomarker discovery. Biomark. Res. 2023, 11, 70. [Google Scholar] [CrossRef]
- Huang, J.; Chen, X.; Fu, X.; Li, Z.; Huang, Y.; Liang, C. Advances in aptamer-based biomarker discovery. Front. Cell. Dev. Biol. 2021, 9, 659760. [Google Scholar] [CrossRef]
- Petrera, A.; von Toerne, C.; Behler, J.; Huth, C.; Thorand, B.; Hilgendorff, A.; Hauck, S.M. Multiplatform approach for plasma proteomics: Complementary of Olink proximity extension assay technology to mass spectrometry-based protein profiling. J. Proteome Res. 2021, 20, 751–762. [Google Scholar] [CrossRef] [PubMed]
- Lind, L.; Ärnlöv, J.; Lindahl, B.; Siegbahn, A.; Sundström, J.; Ingelsson, R. Use of a proximity extension assay proteomic chip to discover new biomarkers for human atherosclerosis. Atherosclerosis 2015, 242, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Carlyle, B.; Kitchen, R.R.; Mattingly, Z.; Celia, A.M.; Trombetta, B.A.; Das, S.; Hyman, B.T.; Kivisäkk, P.; Arnold, S.A. Technical performance evaluation of Olink proximity extension assay for blood-based biomarker discovery in longitudinal studies of Alzheimer’s disease. Front. Neurol. 2022, 13, 889647. [Google Scholar] [CrossRef] [PubMed]
- Wik, L.; Nordberg, N.; Broberg, J.; Björkesten, J.; Assarsson, E.; Henriksson, S.; Grundberg, I.; Petterson, E.; Westerberg, C.; Liljeroth, E.; et al. Proximity extension assay in combination with next-generation sequencing for high-throughput proteome-wide analysis. Mol. Cell. Proteom. 2021, 20, 100168. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.bioxpedia.com/olink-proteomics/ (accessed on 4 March 2024).
- Al-Aly, Z.; Topol, E. Solving the puzzle of Long COVID. Science 2024, 383, 830–832. [Google Scholar] [CrossRef] [PubMed]
- Palstrom, N.B.; Matthlesen, R.; Rasmussen, L.M.; Beck, H.C. Recent developments in clinical plasma proteomics—Applied to cardiovascular research. Biomedicines 2022, 10, 162. [Google Scholar] [CrossRef] [PubMed]
- Raffield, L.M.; Pratte, K.A.; Jacobson, S.; Gillenwater, L.A.; Ampleford, E.; Barjaktaveric, I.; Basta, P.; Clish, C.B.; Comellas, A.P.; Cornell, E.; et al. Comparison of proteomic assessment methods in multiple cohort studies. Proteomics 2020, 20, e1900278. [Google Scholar] [CrossRef] [PubMed]
- Phillips, T.M.; Wellner, E.F. Analysis of inflammatory biomarkers from tissue biopsies by chip-based immunoaffinity CE. Electrophoresis 2007, 28, 3041–3048. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Liu, J.; Yang, J.; Yang, L. Aptamer affinity-based microextraction in-line coupled to capillary electrophoresis mass spectrometry using porous layer/nanoparticle-modified open tubular column. Anal. Chim. Acta 2023, 1239, 340750. [Google Scholar] [CrossRef]
- Nge, P.N.; Pagaduan, J.V.; Yu, M.; Woolley, A.T. Microfluidic chips with reversed-phase monoliths for solid phase extraction and on-chip labeling. J. Chromatogr. A 2012, 1261, 129–135. [Google Scholar] [CrossRef]
- Almughamsi, H.M.; Howell, M.K.; Parry, S.R.; Esene, J.E.; Nielsen, J.B.; Nordin, G.P.; Woolley, A.T. Immunoaffinity monoliths for multiplexed extraction of preterm birth biomarkers from human blood serum in 3D printed microfluidic devices. Analyst 2022, 147, 734. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Yu, X.; Sun, X.; Woolley, A.T. Microdevices integrating affinity columns and capillary electrophoresis for multi-biomarker analysis in human serum. Lab Chip 2010, 10, 2527–2533. [Google Scholar] [CrossRef] [PubMed]
- Pero-Gascon, R.; Pont, L.; Sanz-Nebot, V.; Benavente, F. On-line immunoaffinity solid-phase extraction capillary electrophoresis-mass spectrometry for the analysis of serum transthyretin. Methods Mol. Biol. 2019, 1972, 57–76. [Google Scholar] [PubMed]
- Tůma, P. Progress in on-line, at-line, and in-line coupling of sample pretreatment with capillary and microchip electrophoresis over the past 10 years: A review. Anal. Chim. Acta 2023, 1261, 341249. [Google Scholar] [CrossRef] [PubMed]
- Kašička, V. Recent developments in capillary and microchip electroseparations of peptides (2021- mid-2023). Electrophoresis 2024, 45, 165–198. [Google Scholar] [CrossRef] [PubMed]
- Guzman, N.A. Determination of immunoreactive gonadotropin-releasing hormone in serum and in urine by on-line immunoaffinity capillary coupled to mass spectrometry. J. Chromatogr. B Biomed. Sci. Appl. 2000, 749, 197–213. [Google Scholar] [CrossRef] [PubMed]
- Kammeijer, G.S.M.; Nouta, J.; de la Rosette, J.J.M.C.H.; de Reijke, T.M.; Wuhrer, M. An in-depth glycosylation assay for urinary prostate-specific antigen. Anal. Chem. 2018, 90, 4414–4421. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Zhao, Q.; Zhang, D.; Yuan, Z.; Wang, H. Affinity interactions by capillary electrophoresis: Binding, separation, and detection. Anal. Chem. 2019, 91, 372–387. [Google Scholar] [CrossRef] [PubMed]
- Puerta, A.; Martin-Alvarez, P.J.; Ongay, S.; Diez-Masa, J.C.; de Frutos, M. Immunoaffinity, capillary electrophoresis, and statistics for studying intact alpha 1-acid glycoprotein isoforms as atherothrombosis biomarker. Methods Mol. Biol. 2013, 919, 215–230. [Google Scholar]
- Okanda, F.M.; El Rassi, Z. Biospecific interaction (affinity) CEC and affinity nano-LC. Electrophoresis 2007, 28, 89–98. [Google Scholar] [CrossRef]
- Guzman, N.A.; Trebilcock, M.A.; Advis, J.P. The use of a concentration step to collect urinary components separated by capillary electrophoresis and further characterization of collected analytes by mass spectrometry. J. Liq. Chromatogr. 1991, 14, 997–1015. [Google Scholar] [CrossRef]
- Guzman, N.A. Biomedical applications of on-line preconcentration-capillary electrophoresis using an analyte concentrator: Investigation of design options. J. Liq. Chromatogr. 1995, 18, 3751–3768. [Google Scholar] [CrossRef]
- Nevídalová, H.; Michalcová, L.; Glatz, Z. Capillary electrophoresis-based immunoassay and aptamer assay: A review. Electrophoresis 2020, 41, 414–433. [Google Scholar] [CrossRef] [PubMed]
- Guzman, N.A. Method and System for Simultaneous Determination of Multiple Measurable Biomarkers during the Development of a Communicable Disease. U.S. Patent 11,287,396, 29 March 2022. [Google Scholar]
- Guzman, N.A.; Stubbs, R.J.; Phillips, T.M. Determination of inflammatory biomarkers by immunoaffinity capillary electrophoresis. Drug Discov. Today Technol. 2006, 3, 29–37. [Google Scholar]
- Guzman, N.A.; Guzman, D.E. A two-dimensional affinity capture and separation mini-platform for the isolation, enrichment, and quantification of biomarkers and its potential use for liquid biopsy. Biomedicines 2020, 8, 255. [Google Scholar] [CrossRef] [PubMed]
- Guzman, N.A.; Guzman, D.E. Improving diagnostic testing and disease analysis with IACE. Res. Features 2022. [Google Scholar] [CrossRef]
- Shimura, K.; Nagai, T. Capillary isoelectric focusing after enrichment with immunochromatography in a single capillary. Sci. Rep. 2016, 6, 39221. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Miao, S.; Tan, J.; Zhang, Q.; Chen, D.D.Y. Capillary Electrophoresis: A three-year literature review. Anal. Chem. 2024, in press. [Google Scholar] [CrossRef]
- Guzman, N.A. Consecutive protein digestion and peptide derivatization employing an on-line analyte concentrator to map proteins using capillary electrophoresis. In Capillary Electrophoresis in Analytical Biotechnology; Righetti, P.G., Ed.; CRC Press: Boca Raton, FL, USA; New York, NY, USA, 1996; Chapter 4; pp. 101–121. [Google Scholar]
- Nashabeth, W.; El Rassi, Z. Enzymophoresis of nucleic acids by tandem capillary enzyme reactor-capillary zone electrophoresis. J. Chromatogr. 1992, 596, 251–264. [Google Scholar] [CrossRef]
- Balaram, V. Current advances in the miniaturization of analytical instruments—Applications in cosmochemistry, geochemistry, exploration, and environmental sciences. Spectroscopy 2016, 31, 40–44. [Google Scholar]
- Thomas, S.; Ahmadi, M.; Nguyen, T.A.; Afkhami, A.; Madrakian, T. (Eds.) Micro- and Nano Technology Enabled Applications for Portable Miniaturized Analytical Systems; Elsevier: Amsterdam, The Netherlands, 2022; Chapters 1–17; pp. 1–424. [Google Scholar]
- Hansen, G.T. Point-of-care testing in microbiology: A mechanism for improving patient outcomes. Clin. Chem. 2020, 66, 124–137. [Google Scholar] [CrossRef] [PubMed]
- Drain, P.K.; Heichman, K.A.; Wilson, D. A new point-of-care test to diagnose tuberculosis. Lancet Infect. Dis. 2019, 19, 794–796. [Google Scholar] [CrossRef] [PubMed]
- Drain, P.K. Rapid diagnostic testing for SARS-CoV-2. N. Engl. J. Med. 2022, 386, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Schito, M.; Peter, T.F.; Cavanaugh, S.; Piatek, A.S.; Young, G.J.; Alexander, H.; Coggin, W.; Domingo, G.J.; Ellenberger, D.; Ermantraut, E.; et al. Opportunities and challenges for cost-efficient implementation of new point-of-care diagnostics for HIV and tuberculosis. J. Infect. Dis. 2022, 205 (Suppl. S2), S169–S180. [Google Scholar] [CrossRef] [PubMed]
- Johnson, E.M.; Ellis, W.R., Jr.; Powers, L.S.; Wysocki, V.H. Affinity capture mass spectrometry of biomarker proteins using peptide ligands from biopanning. Anal. Chem. 2009, 81, 5999–6005. [Google Scholar] [CrossRef] [PubMed]
- Panagides, N.; Zacchi, L.F.; De Souza, M.J.; Morales, R.A.V.; Karnowsi, A.; Liddament, M.T.; Owczarek, C.M.; Mahler, S.M.; Panoulis, C.; Jones, M.L.; et al. Evaluation of phage display biopanning for the selection of anti-cell surface receptor antibodies. Int. J. Mol. Sci. 2022, 23, 8470. [Google Scholar] [CrossRef] [PubMed]
- Castro, G.; Schrandt, S. Improving diagnosis through technology. HealthManagent.org-J. 2020, 20, 472–473. [Google Scholar]
- Tockman, M.S. Advances in sputum analysis for screening and early detection of lung cancer. Cancer Control 2000, 7, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Willis, V.C.; Demourelle, M.K.; Derber, L.A.; Chartier-Logan, C.J.; Parish, M.C.; Pedraza, I.F.; Weisman, M.H.; Norris, J.M.; Holers, V.M.; Deane, K.D. Sputum autoantibodies in patients with established rheumatoid arthritis and subjects at risk of future clinically apparent disease. Arthritis Rheum. 2013, 65, 2545–2554. [Google Scholar] [CrossRef]
- Ren, C.; Gao, Y.; Zhang, C.; Zhou, C.; Hong, Y.; Qu, M.; Zhao, Z.; Du, Y.; Yang, L.; Liu, B.; et al. Respiratory mucosal immunity: Kinetics of secretory immunoglobulin A in sputum and throat swabs from COVID-19 patients and vaccine recipients. Front. Microbiol. 2022, 13, 782421. [Google Scholar] [CrossRef]
- Kelemen, K.; Cslszér, E.; Barta, I.; Antus, B. Detection of antibodies against Pseudomonas in the sputum of cystic fibrosis patients: A pilot study. Eur. Respir. J. 2012, 40, P1449. [Google Scholar]
- Chan, M.; Linn, M.M.N.; O’Hagan, T.; Guerra-Assunção, J.A.; Lackenby, A.; Workman, S.; Dacre, A.; Burns, S.O.; Breuer, J.; Hart, J.; et al. Persistent SARS-CoV-2 PCR positivity despite anti-viral treatment in immunodeficient patients. J. Clin. Immunol. 2023, 43, 1083–1092. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Chu, D.; Kalantar-Zadeh, K.; George, J.; Young, H.A.; Liu, G. Cytokines: From clinical significance to quantification. Adv. Sci. 2021, 8, 2004433. [Google Scholar] [CrossRef] [PubMed]
- Tholey, A.; Schlüter, H. Top-down proteomics and proteoforms—Special issue (Editorial). Proteomics 2024, 24, 2200375. [Google Scholar] [CrossRef] [PubMed]
- Nedelkov, D. Human proteoforms as new targets for clinical mass spectrometry protein tests. Exp. Rev. Proteom. 2017, 14, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Forgrave, L.M.; Wang, M.; Yang, D.; DeMarco, M.L. Proteoforms and their expanding role in laboratory medicine. Pract. Lab. Med. 2022, 28, e00260. [Google Scholar] [CrossRef]
- Su, J.; Yang, L.; Sun, Z.; Zhan, X. Personalized drug therapy: Innovative concept guided with proteoformics. Mol. Cell. Proteom. 2024, 12, 100737. [Google Scholar] [CrossRef]
- Cramer, D.A.T.; Yin, V.; Caval, T.; Franc, V.; Yu, D.; Wu, G.; Lloyd, G.; Langendorf, C.; Chisstock, J.C.; Law, R.H.P.; et al. Proteoform-resolved profiling of plasminogen activation reveals novel abundant phosphorylation site and primary N-terminal cleavage site. Mol. Cell. Proteom. 2024, 23, 100696. [Google Scholar] [CrossRef]
- Tamara, S.; den Boer, M.A.; Heck, A.J.R. High-resolution mass spectrometry. Chem. Rev. 2022, 122, 7269–7326. [Google Scholar] [CrossRef]
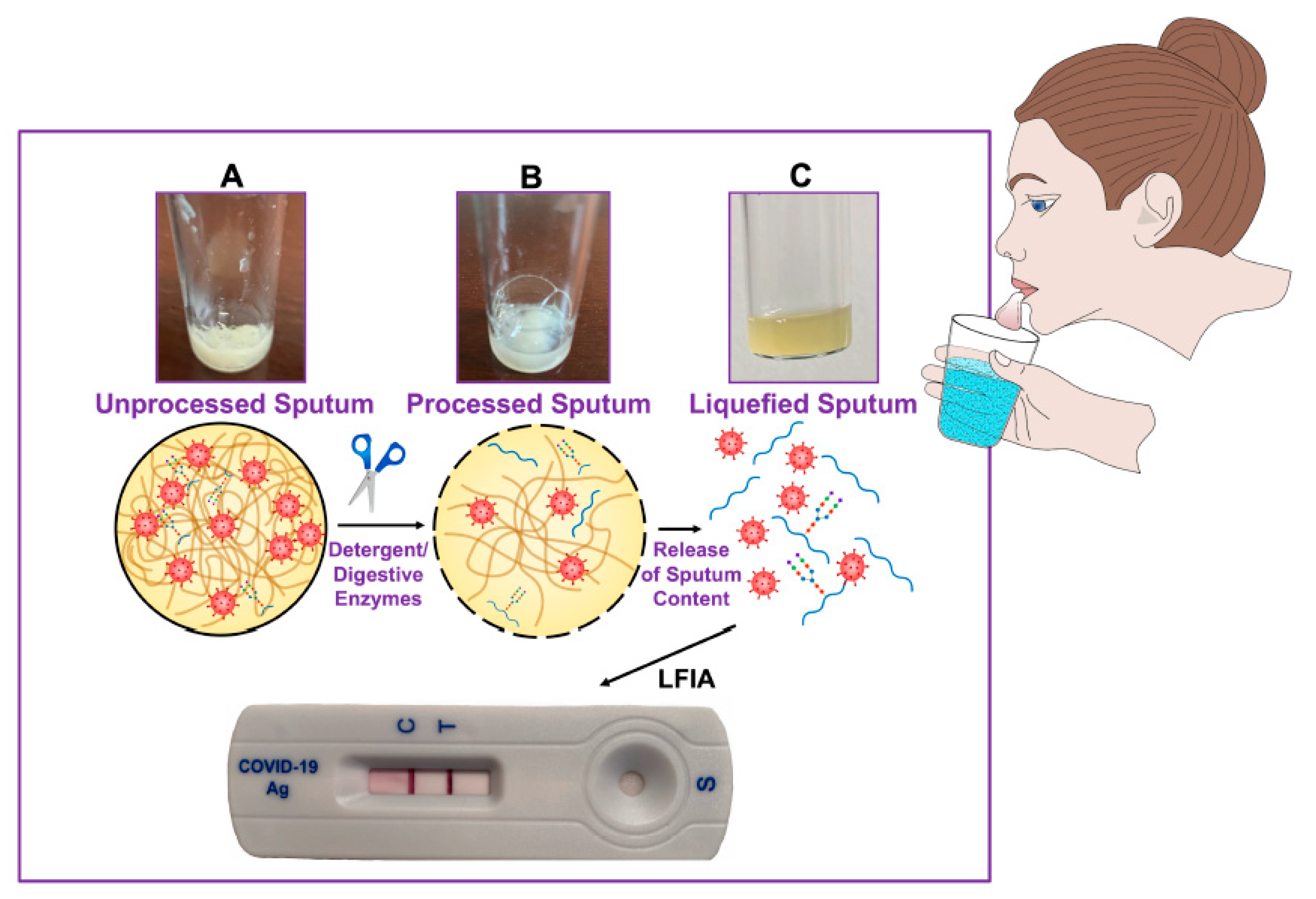

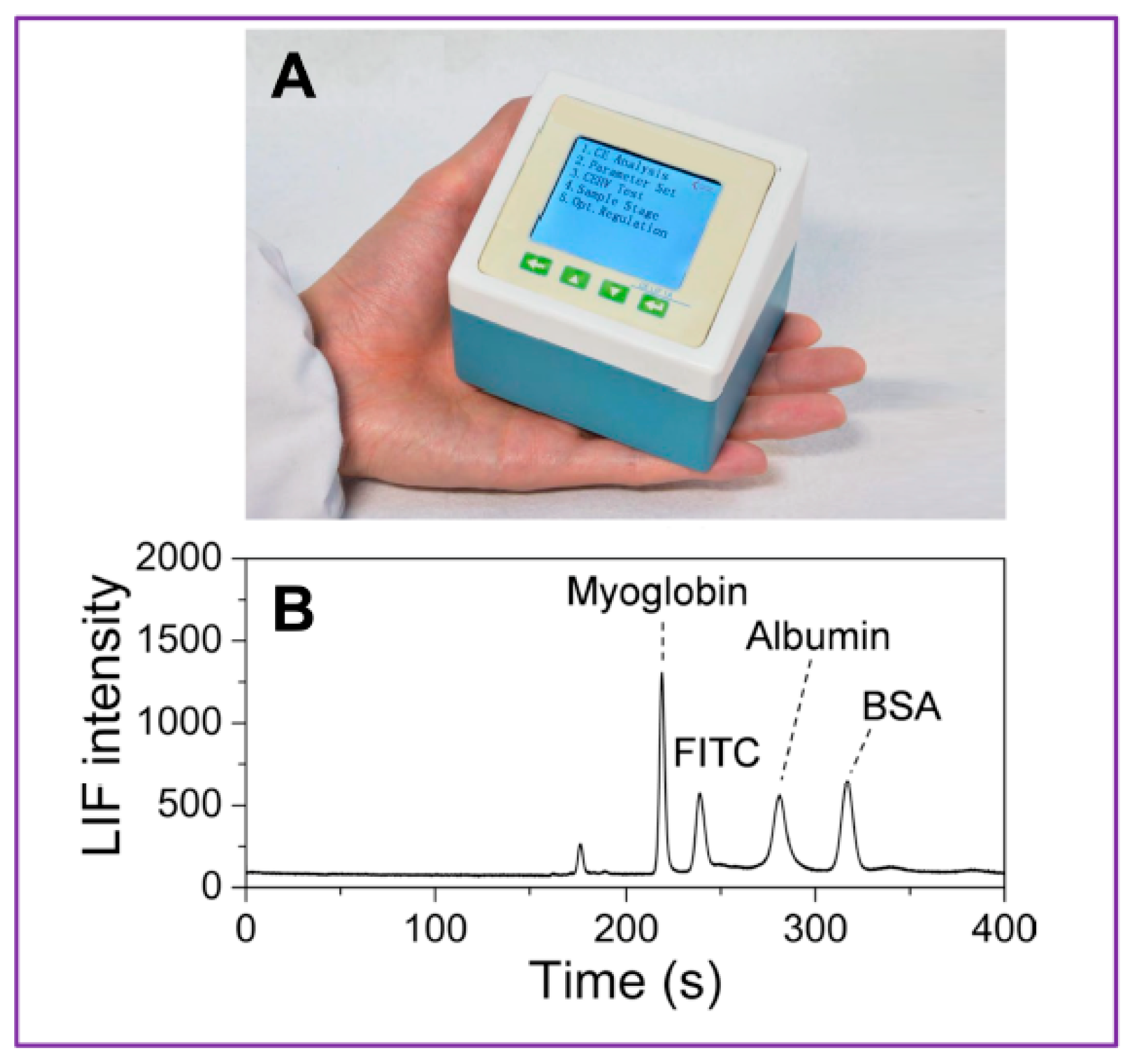
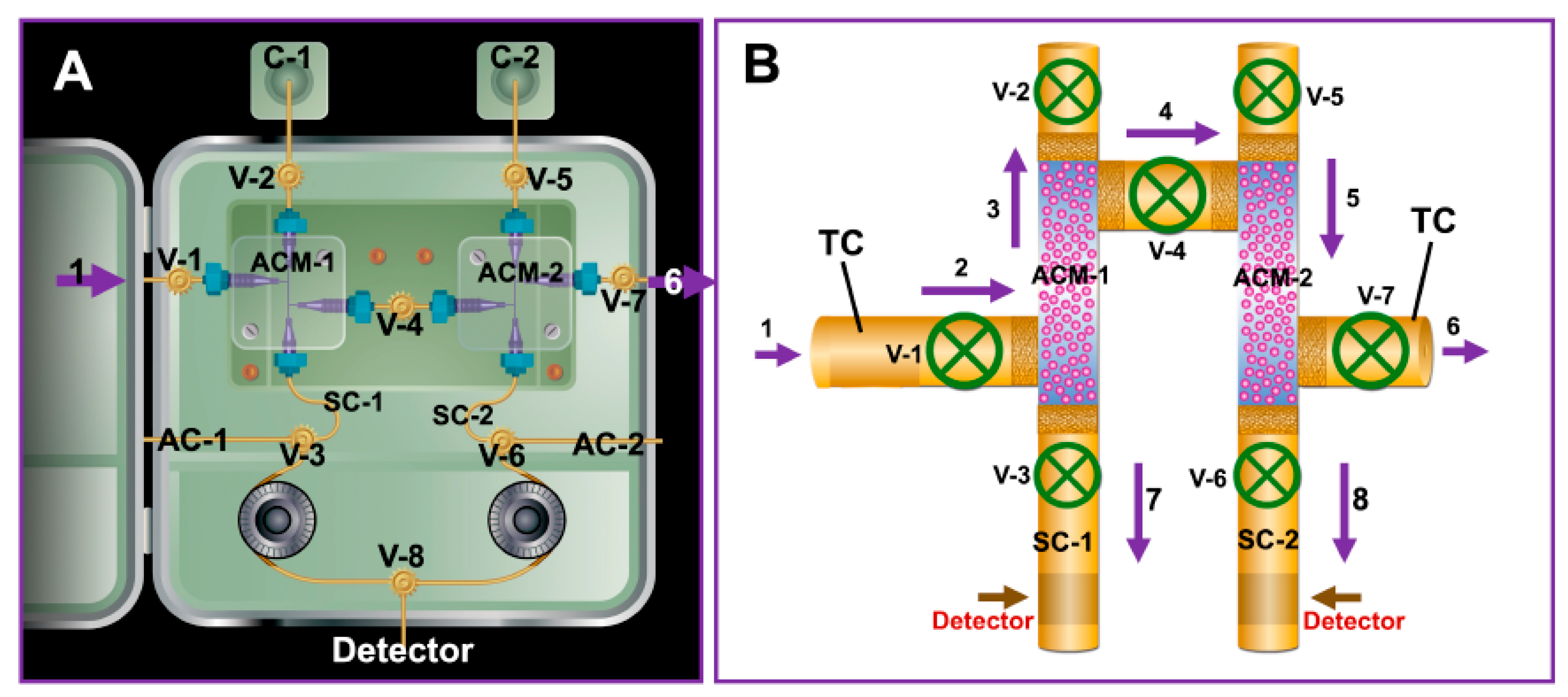
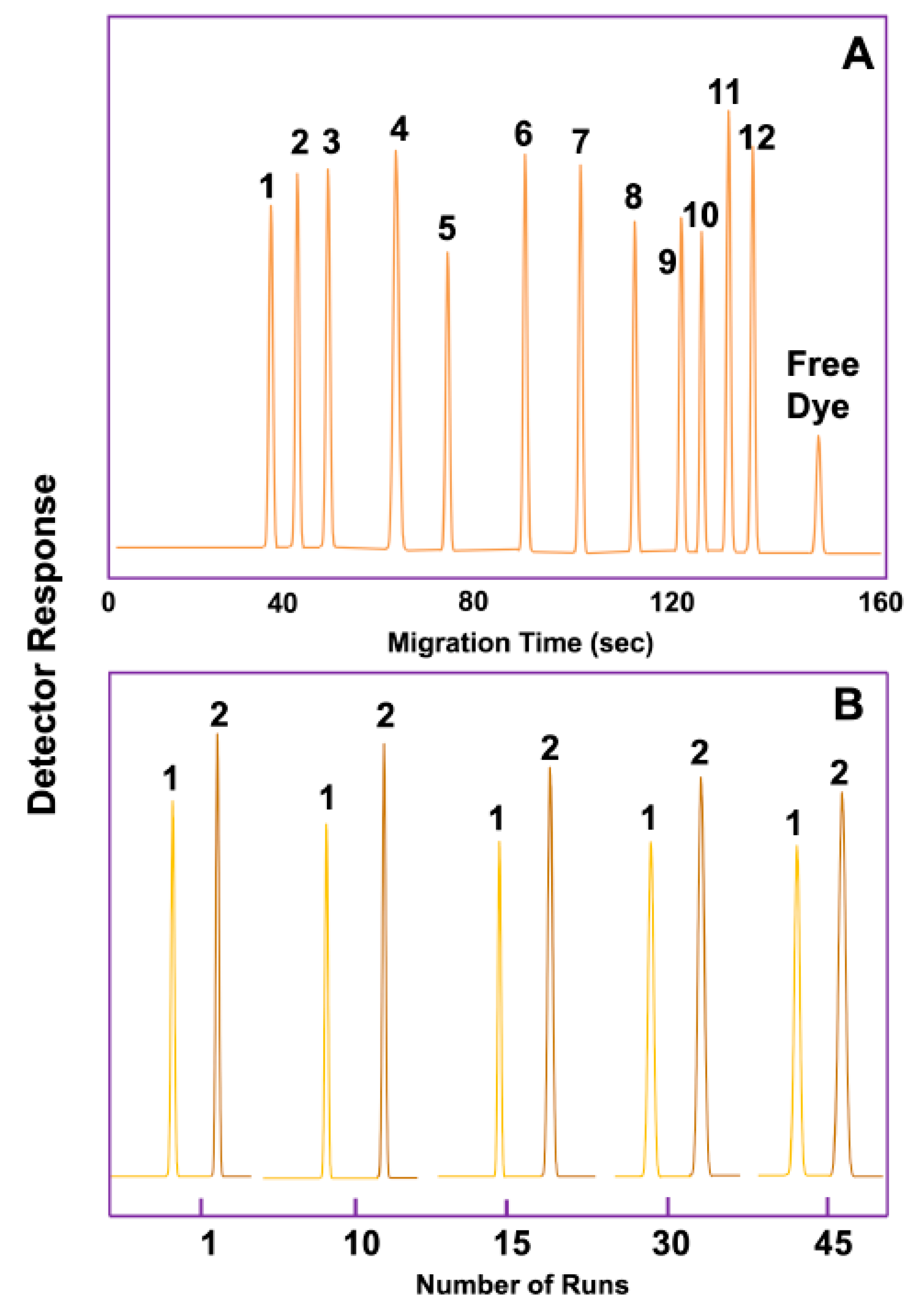
| Procedure Performed or Substances Added to Sputum or Bronchial Aspirate Samples | Rationale | Reference |
|---|---|---|
| Mechanical homogenization. | To macerate or crush the sputum in the presence of a suitable buffer, to obtain viable single-cell suspensions, or to disperse fragments of the sputum gathered evenly throughout the mixture. | [34,52] |
| Acetone sedimentation. | Acetone-precipitated samples are dissolved in 8M urea, reduced with Tris-(2-carboxyethyl)phosphine, and alkylated with iodoacetamide. | [17] |
| Tris(2-carboxyethyl)phosphine (TCEP), N-acetyl-L-cysteine (NALC), bovine serum albumin (BSA), and protease inhibitor cocktail. | TCEP and NALC (reducing agents) are used to break the disulfide bonds of sputum. BSA is used as a blocking agent. | [51] |
| Hydrogen peroxide (H2O2). | Enzymatic liquefaction. The peroxide solution, in the presence of endogenous catalase, is used to trigger the formation of an oxygen bubble. | [53] |
| NaOH, sodium dodecyl sulfate (SDS), glass beads, temperature (60 °C). | Breakdown of sample mixture. | [54] |
| PureLyse® bead blender, solid phase extraction, which does not require chaotropic salts or organic solvents. | A miniaturized bead beating system for mechanical pathogen lysis (effectively disrupts tough-walled microorganisms). This is a disinfection and liquefaction method. | [55] |
| Dithiothreitol (DTT), homogenization, centrifugation, and supernatant desalting. | Release of proteins from sputum. | [56] |
| Procedure Performed to Analyze Proteins/Peptides in Sputum | Number of Proteins/Peptides Analyzed | Reference |
|---|---|---|
| Two-dimensional electrophoresis gel (2-DE), and one-dimensional gel electrophoresis followed by liquid chromatography–tandem mass spectrometry (GeLC-MS/MS) | 191 | [73] |
| Mesoporous silica beads and matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry (MALDI-TOF-MS) | >400 | [74] |
| Liquid chromatography–mass spectrometry (LC-MS) | 192–1666 | [23,24,75,76] |
| Capillary liquid chromatography (capLC-MS) | 203 | [77] |
| Multi-dimensional protein identification technology (MudPIT) | 2210 | [78] |
| Aptamer-based assay (SOMAscan) | 1129 | [79] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guzman, N.A.; Guzman, A. Human Sputum Proteomics: Advancing Non-Invasive Diagnosis of Respiratory Diseases with Enhanced Biomarker Analysis Methods. Int. J. Transl. Med. 2024, 4, 309-333. https://doi.org/10.3390/ijtm4020020
Guzman NA, Guzman A. Human Sputum Proteomics: Advancing Non-Invasive Diagnosis of Respiratory Diseases with Enhanced Biomarker Analysis Methods. International Journal of Translational Medicine. 2024; 4(2):309-333. https://doi.org/10.3390/ijtm4020020
Chicago/Turabian StyleGuzman, Norberto A., and Andrea Guzman. 2024. "Human Sputum Proteomics: Advancing Non-Invasive Diagnosis of Respiratory Diseases with Enhanced Biomarker Analysis Methods" International Journal of Translational Medicine 4, no. 2: 309-333. https://doi.org/10.3390/ijtm4020020
APA StyleGuzman, N. A., & Guzman, A. (2024). Human Sputum Proteomics: Advancing Non-Invasive Diagnosis of Respiratory Diseases with Enhanced Biomarker Analysis Methods. International Journal of Translational Medicine, 4(2), 309-333. https://doi.org/10.3390/ijtm4020020







