The Co-Localization of NLRP3 and ASC Specks Does Not Automatically Entail NLRP3 Inflammasome Functionality in PDAC Cell Lines
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culturing
2.2. RNA Extraction, cDNA Synthesis and Real-Time PCR
2.3. The Cytokines IL-1β and IL-18 in the Cell Media
2.4. Immunofluorescence and Co-Localization
2.5. Protein Extraction and WB
2.6. R2 Gene Expression Analysis
2.7. Statistical Analysis
3. Results
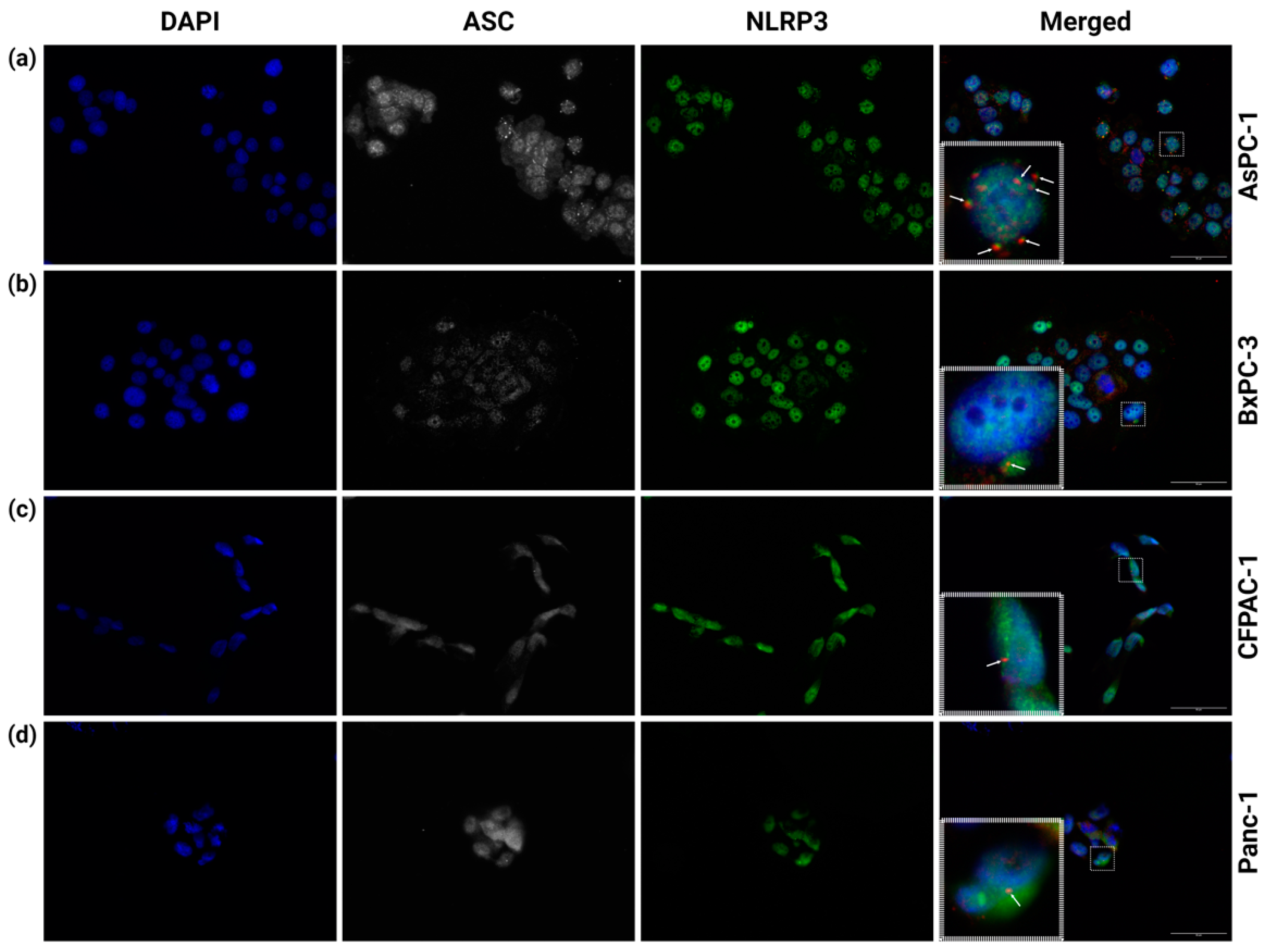
3.1. Multiple NLRP3 Inflammasomes Form Spontaneously in AsPC-1 Cells
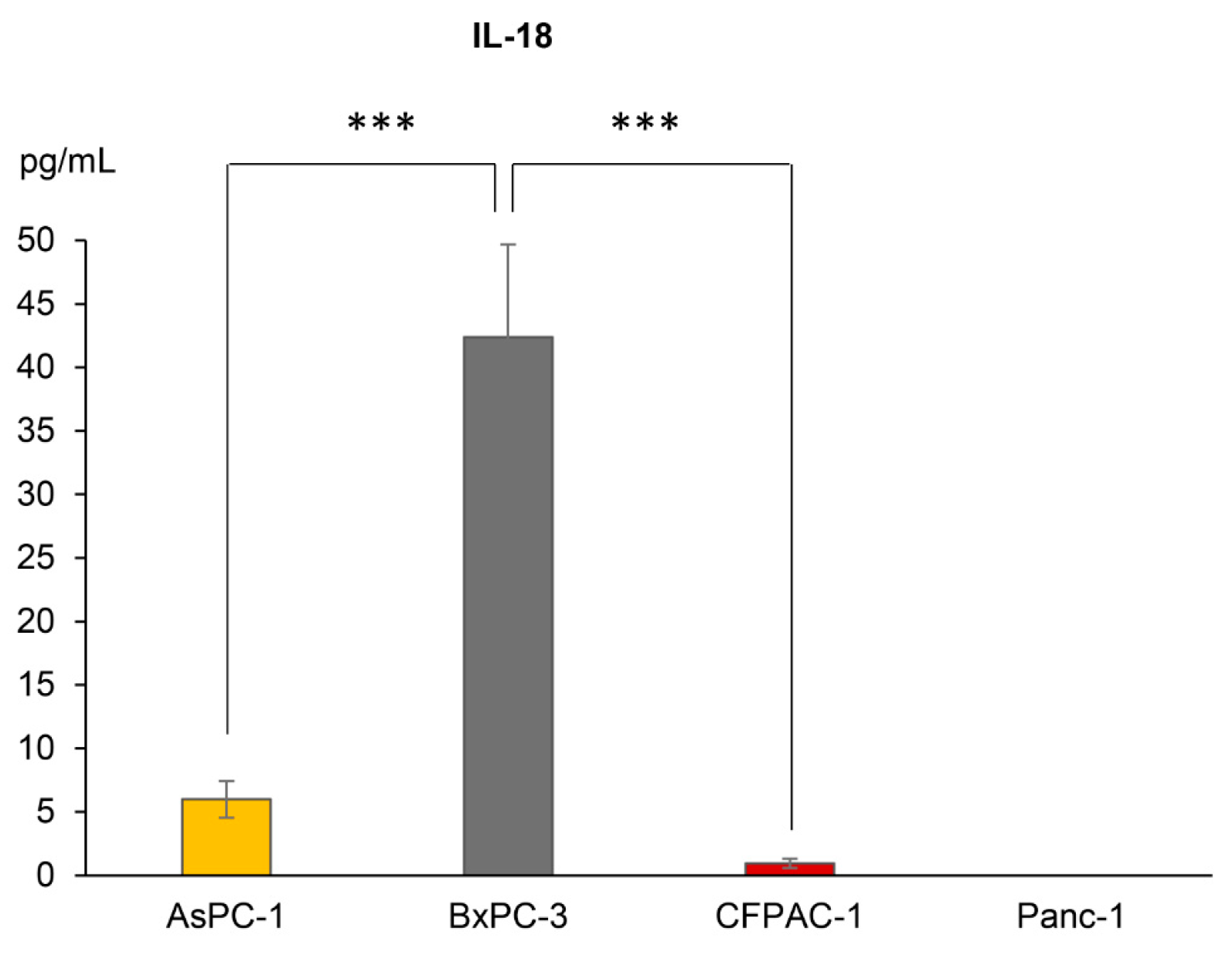
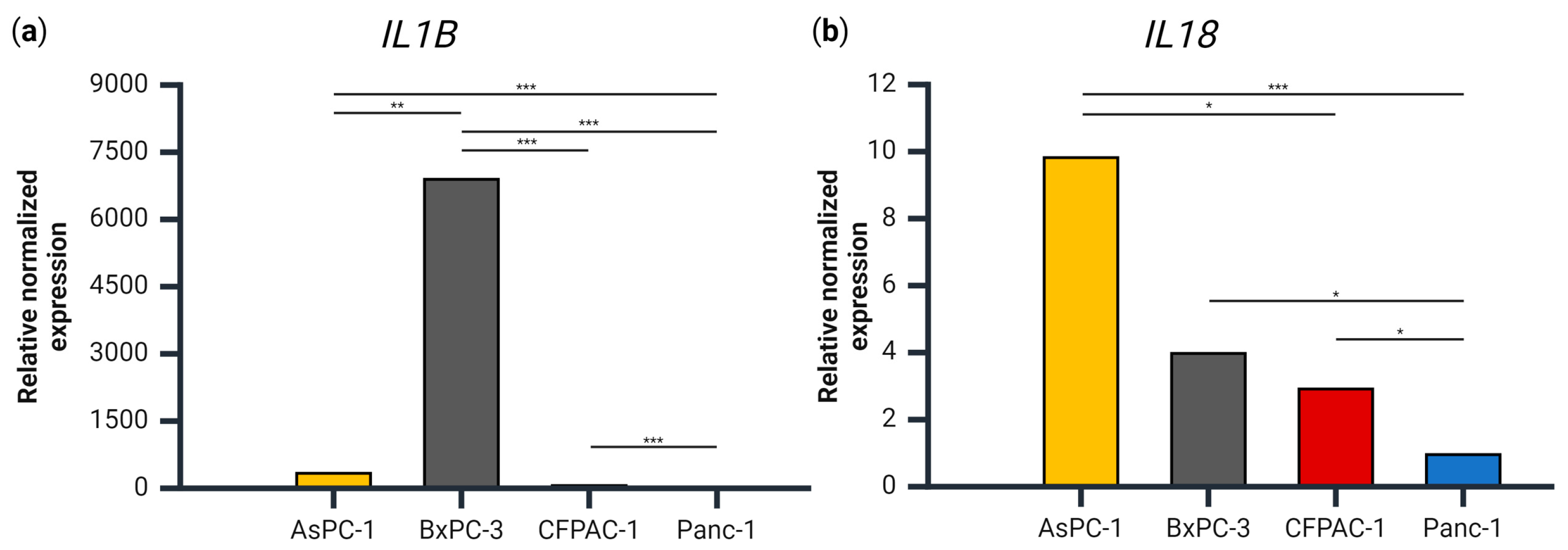
3.2. IL1B, but Not IL18, Gene Expression Levels Coincide with Secretion Levels
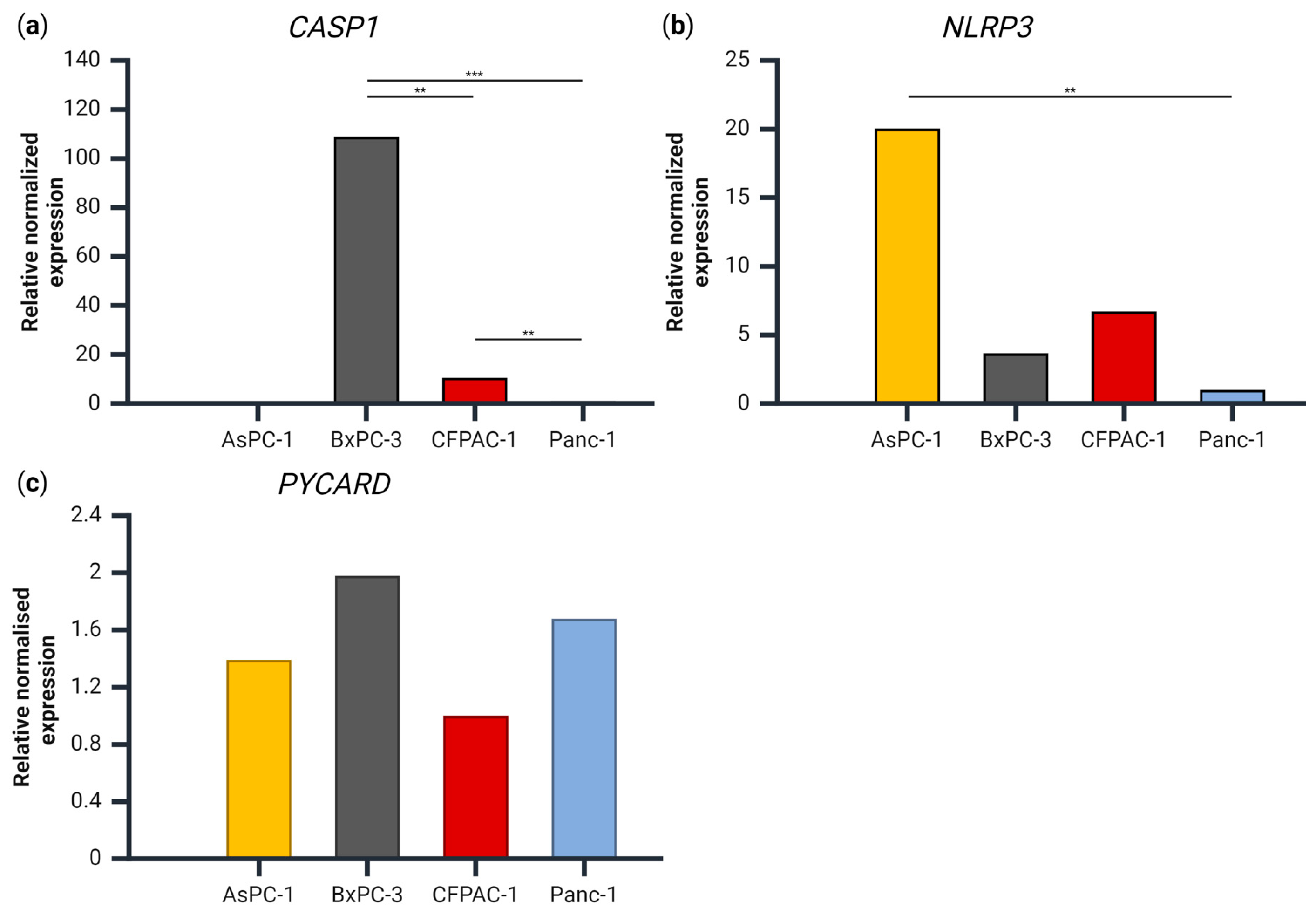
3.3. Gene Expression of CASP1, PYCARD and NLRP3 in the PDAC Cell Lines
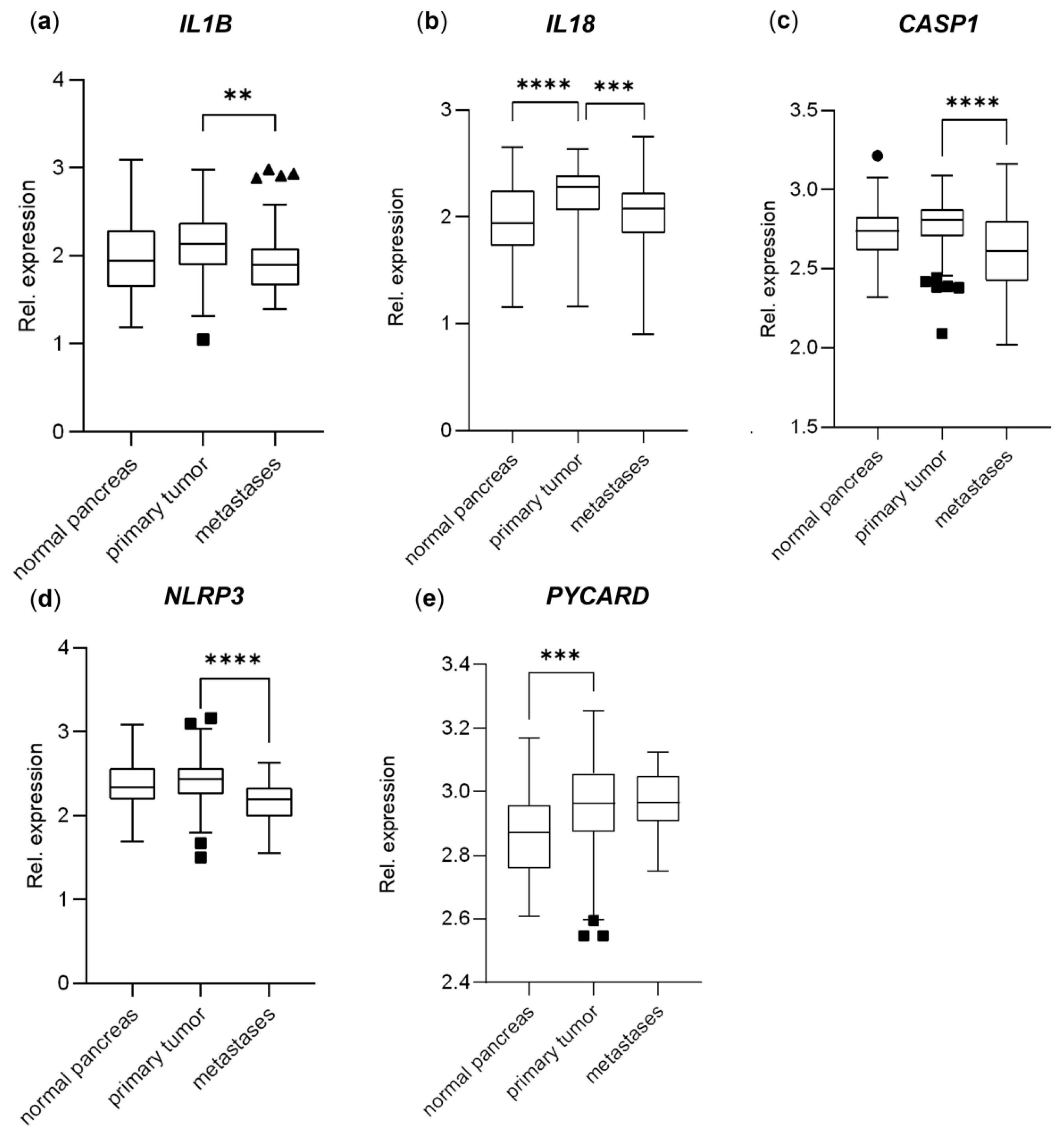
3.4. Genes Important to Inflammasome Response Are Affected in PDAC Primary Tumors and Metastases and Affect Overall Survival
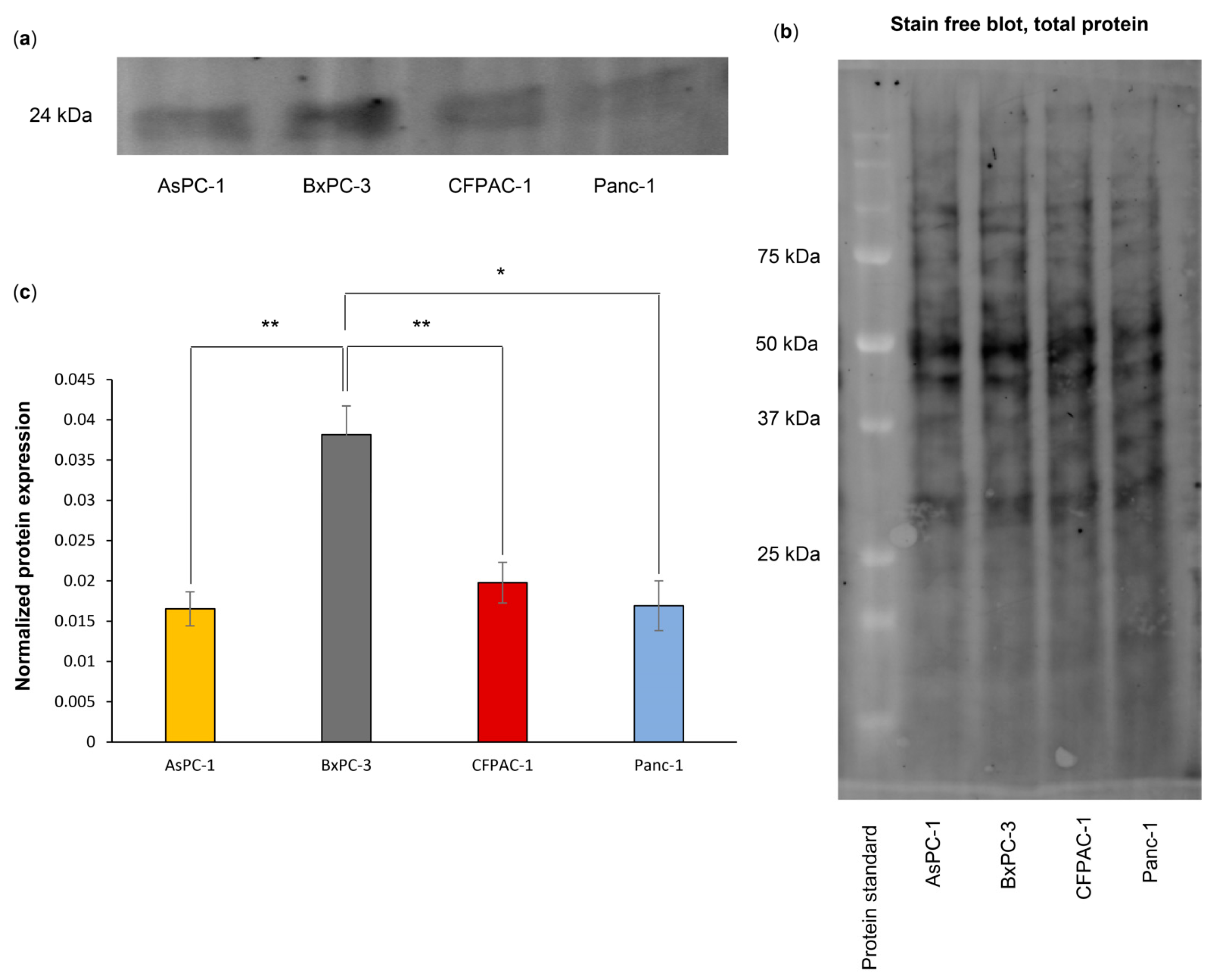
3.5. BxPC-3 Cells Have a High Total ASC Level
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lesina, M.; Kurkowski, M.U.; Ludes, K.; Rose-John, S.; Treiber, M.; Klöppel, G.; Yoshimura, A.; Reindl, W.; Sipos, B.; Akira, S.; et al. Stat3/Socs3 Activation by IL-6 Transsignaling Promotes Progression of Pancreatic Intraepithelial Neoplasia and Development of Pancreatic Cancer. Cancer Cell 2011, 19, 456–469. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, A.; Wang Sam, C.; Morris John, P.I.V.; Folias Alexandra, E.; Liou, A.; Kim Grace, E.; Akira, S.; Boucher Kenneth, M.; Firpo Matthew, A.; Mulvihill Sean, J.; et al. Stat3 and MMP7 Contribute to Pancreatic Ductal Adenocarcinoma Initiation and Progression. Cancer Cell 2011, 19, 441–455. [Google Scholar] [CrossRef]
- Farrow B, Evers BM: Inflammation and the development of pancreatic cancer. Surg. Oncol. 2002, 10, 153–169. [CrossRef] [PubMed]
- Kanak, M.A.; Shahbazov, R.; Yoshimatsu, G.; Levy, M.F.; Lawrence, M.C.; Naziruddin, B. A small molecule inhibitor of NFκB blocks ER stress and the NLRP3 inflammasome and prevents progression of pancreatitis. J. Gastroenterol. 2017, 52, 352–365. [Google Scholar] [CrossRef]
- Liu, T.; Wang, Q.; Du, Z.; Yin, L.; Li, J.; Meng, X.; Xue, D. The trigger for pancreatic disease: NLRP3 inflammasome. Cell Death Discov. 2023, 9, 246. [Google Scholar] [CrossRef] [PubMed]
- Latz, E.; Xiao, T.S.; Stutz, A. Activation and regulation of the inflammasomes. Nat. Rev. Immunol. 2013, 13, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Zhiyu, W.; Wang, N.; Wang, Q.; Peng, C.; Zhang, J.; Liu, P.; Ou, A.; Zhong, S.; Cordero, M.D.; Lin, Y. The inflammasome: An emerging therapeutic oncotarget for cancer prevention. Oncotarget 2016, 7, 50766–50780. [Google Scholar] [CrossRef] [PubMed]
- Evavold, C.L.; Ruan, J.; Tan, Y.; Xia, S.; Wu, H.; Kagan, J.C. The Pore-Forming Protein Gasdermin D Regulates Interleukin-1 Secretion from Living Macrophages. Immunity 2018, 48, 35–44.e36. [Google Scholar] [CrossRef]
- He, W.T.; Wan, H.; Hu, L.; Chen, P.; Wang, X.; Huang, Z.; Yang, Z.-H.; Zhong, C.-Q.; Han, J. Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell Res. 2015, 25, 1285–1298. [Google Scholar] [CrossRef]
- Heilig, R.; Dick, M.S.; Sborgi, L.; Meunier, E.; Hiller, S.; Broz, P. The Gasdermin-D pore acts as a conduit for IL-1β secretion in mice. Eur. J. Immunol. 2018, 48, 584–592. [Google Scholar] [CrossRef]
- Bryan, N.B.; Dorfleutner, A.; Rojanasakul, Y.; Stehlik, C. Activation of inflammasomes requires intracellular redistribution of the apoptotic speck-like protein containing a caspase recruitment domain. J. Immunol. 2009, 182, 3173–3182. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zeng, M.Y.; Yang, D.; Motro, B.; Núñez, G. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature 2016, 530, 354–357. [Google Scholar] [CrossRef] [PubMed]
- Amo-Aparicio, J.; Dominguez, A.; Atif, S.M.; Dinarello, A.; Azam, T.; Alula, K.M.; Piper, M.; Lieu, C.H.; Lentz, R.W.; Leal, A.D.; et al. Pancreatic Ductal Adenocarcinoma Cells Regulate NLRP3 Activation to Generate a Tolerogenic Microenvironment. Cancer Res. Commun. 2023, 3, 1899–1911. [Google Scholar] [CrossRef] [PubMed]
- Turner, M.D.; Nedjai, B.; Hurst, T.; Pennington, D.J. Cytokines and chemokines: At the crossroads of cell signalling and inflammatory disease. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2014, 1843, 2563–2582. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Barajon, I.; Garlanda, C. IL-1 and IL-1 regulatory pathways in cancer progression and therapy. Immunol. Rev. 2018, 281, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Bent, R.; Moll, L.; Grabbe, S.; Bros, M. Interleukin-1 Beta—A Friend or Foe in Malignancies? Int. J. Mol. Sci. 2018, 19, 2155. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Shapiro, B.; Vucic, E.A.; Vogt, S.; Bar-Sagi, D. Tumor Cell-Derived IL1β Promotes Desmoplasia and Immune Suppression in Pancreatic Cancer. Cancer Res. 2020, 80, 1088–1101. [Google Scholar] [CrossRef] [PubMed]
- Qiang, R.; Li, Y.; Dai, X.; Lv, W. NLRP3 inflammasome in digestive diseases: From mechanism to therapy. Front. Immunol. 2022, 13, 978190. [Google Scholar] [CrossRef] [PubMed]
- Tjomsland, V.; Spångeus, A.; Välilä, J.; Sandström, P.; Borch, K.; Druid, H.; Falkmer, S.; Falkmer, U.; Messmer, D.; Larsson, M. Interleukin 1α sustains the expression of inflammatory factors in human pancreatic cancer microenvironment by targeting cancer-associated fibroblasts. Neoplasia 2011, 13, 664–675. [Google Scholar] [CrossRef]
- Brunetto, E.; De Monte, L.; Balzano, G.; Camisa, B.; Laino, V.; Riba, M.; Heltai, S.; Bianchi, M.; Bordignon, C.; Falconi, M.; et al. The IL-1/IL-1 receptor axis and tumor cell released inflammasome adaptor ASC are key regulators of TSLP secretion by cancer associated fibroblasts in pancreatic cancer. J. Immunother. Cancer 2019, 7, 45. [Google Scholar] [CrossRef]
- Zheng, L.; Liu, H. Prognostic association between NLRP3 inflammasome expression level and operable pancreatic adenocarcinoma. Int. J. Biol. Markers 2022, 37, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, M.; Watanabe, T.; Masumoto, J.; Sunago, K.; Imamura, Y.; Kanemitsu, K.; Kumagi, T.; Hiasa, Y. Apoptosis-associated speck-like protein containing a CARD regulates the growth of pancreatic ductal adenocarcinoma. Sci. Rep. 2021, 11, 22351. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Yang, G.; Huang, H.; Fu, Z.; Cao, Z.; Zheng, L.; You, L.; Zhang, T. Preclinical models of pancreatic ductal adenocarcinoma: Challenges and opportunities in the era of precision medicine. J. Exp. Clin. Cancer Res. 2021, 40, 8. [Google Scholar] [CrossRef] [PubMed]
- Daussy, C.F.; Monard, S.C.; Guy, C.; Muñoz-González, S.; Chazal, M.; Anthonsen, M.W.; Jouvenet, N.; Henry, T.; Dreux, M.; Meurs, E.F.; et al. The Inflammasome Components NLRP3 and ASC Act in Concert with IRGM To Rearrange the Golgi Apparatus during Hepatitis C Virus Infection. J. Virol. 2021, 95, 10–1128. [Google Scholar] [CrossRef]
- Bodnar-Wachtel, M.; Huber, A.L.; Gorry, J.; Hacot, S.; Burlet, D.; Gérossier, L.; Guey, B.; Goutagny, N.; Bartosch, B.; Ballot, E.; et al. Inflammasome-independent NLRP3 function enforces ATM activity in response to genotoxic stress. Life Sci. Alliance 2023, 6. [Google Scholar] [CrossRef] [PubMed]
- Moffitt, R.A.; Marayati, R.; Flate, E.L.; Volmar, K.E.; Loeza, S.G.H.; Hoadley, K.A.; Rashid, N.U.; Williams, L.A.; Eaton, S.C.; Chung, A.H.; et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 2015, 47, 1168–1178. [Google Scholar] [CrossRef]
- Janky Rs Binda, M.M.; Allemeersch, J.; Van den Broeck, A.; Govaere, O.; Swinnen, J.V.; Roskams, T.; Aerts, S.; Topal, B. Prognostic relevance of molecular subtypes and master regulators in pancreatic ductal adenocarcinoma. BMC Cancer 2016, 16, 632. [Google Scholar] [CrossRef]
- Zhang, G.; He, P.; Tan, H.; Budhu, A.; Gaedcke, J.; Ghadimi, B.M.; Ried, T.; Yfantis, H.G.; Lee, D.H.; Maitra, A.; et al. Integration of metabolomics and transcriptomics revealed a fatty acid network exerting growth inhibitory effects in human pancreatic cancer. Clin. Cancer Res. 2013, 19, 4983–4993. [Google Scholar] [CrossRef]
- Yang, S.; He, P.; Wang, J.; Schetter, A.; Tang, W.; Funamizu, N.; Yanaga, K.; Uwagawa, T.; Satoskar, A.R.; Gaedcke, J.; et al. A Novel MIF Signaling Pathway Drives the Malignant Character of Pancreatic Cancer by Targeting NR3C2. Cancer Res. 2016, 76, 3838–3850. [Google Scholar] [CrossRef]
- Bolte, S.; Cordelières, F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006, 224, 213–232. [Google Scholar] [CrossRef]
- Hamarsheh, S.; Zeiser, R. NLRP3 Inflammasome Activation in Cancer: A Double-Edged Sword. Front. Immunol. 2020, 11, 538030. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Lu, L.; Li, B.; Shi, X.; Jin, H.; Hu, W. The roles of inflammasomes in cancer. Front. Immunol. 2023, 14, 1195572. [Google Scholar] [CrossRef] [PubMed]
- Moossavi, M.; Parsamanesh, N.; Bahrami, A.; Atkin, S.L.; Sahebkar, A. Role of the NLRP3 inflammasome in cancer. Mol. Cancer 2018, 17, 158. [Google Scholar] [CrossRef] [PubMed]
- Saponaro, C.; Scarpi, E.; Sonnessa, M.; Cioffi, A.; Buccino, F.; Giotta, F.; Pastena, M.I.; Zito, F.A.; Mangia, A. Prognostic Value of NLRP3 Inflammasome and TLR4 Expression in Breast Cancer Patients. Front. Oncol. 2021, 11, 705331. [Google Scholar] [CrossRef] [PubMed]
- Missiroli, S.; Perrone, M.; Boncompagni, C.; Borghi, C.; Campagnaro, A.; Marchetti, F.; Anania, G.; Greco, P.; Fiorica, F.; Pinton, P.; et al. Targeting the NLRP3 Inflammasome as a New Therapeutic Option for Overcoming Cancer. Cancers 2021, 13, 2297. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xu, Y.; Liang, K.; Liu, R. Immune Cells Combined With NLRP3 Inflammasome Inhibitor Exert Better Antitumor Effect on Pancreatic Ductal Adenocarcinoma. Front. Oncol. 2020, 10, 1378. [Google Scholar] [CrossRef] [PubMed]
- Greco, E.; Basso, D.; Fogar, P.; Mazza, S.; Navaglia, F.; Zambon, C.-F.; Falda, A.; Pedrazzoli, S.; Ancona, E.; Plebani, M. Pancreatic Cancer Cells Invasiveness is Mainly Affected by Interleukin-1β not by Transforming Growth Factor-β1. Int. J. Biol. Markers 2005, 20, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Herremans, K.M.; Szymkiewicz, D.D.; Riner, A.N.; Bohan, R.P.; Tushoski, G.W.; Davidson, A.M.; Lou, X.; Leong, M.C.; Dean, B.D.; Gerber, M.; et al. The interleukin-1 axis and the tumor immune microenvironment in pancreatic ductal adenocarcinoma. Neoplasia 2022, 28, 100789. [Google Scholar] [CrossRef] [PubMed]
- Carbone, A.; Vizio, B.; Novarino, A.; Mauri, F.A.; Geuna, M.; Robino, C.; Brondino, G.; Prati, A.; Giacobino, A.; Campra, D.; et al. IL-18 Paradox in Pancreatic Carcinoma: Elevated Serum Levels of Free IL-18 are Correlated with Poor Survival. J. Immunother. 2009, 32, 920–931. [Google Scholar] [CrossRef]
- Xu, S.; Li, X.; Liu, Y.; Xia, Y.; Chang, R.; Zhang, C. Inflammasome inhibitors: Promising therapeutic approaches against cancer. J. Hematol. Oncol. 2019, 12, 64. [Google Scholar] [CrossRef]
- Li, X.; Thome, S.; Ma, X.; Amrute-Nayak, M.; Finigan, A.; Kitt, L.; Masters, L.; James, J.R.; Shi, Y.; Meng, G.; et al. MARK4 regulates NLRP3 positioning and inflammasome activation through a microtubule-dependent mechanism. Nat. Commun. 2017, 8, 15986. [Google Scholar] [CrossRef] [PubMed]
- Fernandes-Alnemri, T.; Wu, J.; Yu, J.W.; Datta, P.; Miller, B.; Jankowski, W.; Rosenberg, S.; Zhang, J.; Alnemri, E.S. The pyroptosome: A supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ 2007, 14, 1590–1604. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Wang, P.; Zhang, B.; Dong, X. Expression of the NEK7/NLRP3 inflammasome pathway in patients with diabetic lower extremity arterial disease. BMJ Open Diabetes Res. Care 2020, 8, e001808. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.F.; Chen, L.; Li, Y.C.; Wu, L.; Yu, G.T.; Zhang, W.F.; Sun, Z.J. NLRP3 inflammasome activation promotes inflammation-induced carcinogenesis in head and neck squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2017, 36, 116. [Google Scholar] [CrossRef] [PubMed]
- Mao, K.; Chen, S.; Chen, M.; Ma, Y.; Wang, Y.; Huang, B.; He, Z.; Zeng, Y.; Hu, Y.; Sun, S.; et al. Nitric oxide suppresses NLRP3 inflammasome activation and protects against LPS-induced septic shock. Cell Res. 2013, 23, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Mishra, N.; Schwerdtner, L.; Sams, K.; Mondal, S.; Ahmad, F.; Schmidt, R.E.; Coonrod, S.A.; Thompson, P.R.; Lerch, M.M.; Bossaller, L. Cutting Edge: Protein Arginine Deiminase 2 and 4 Regulate NLRP3 Inflammasome-Dependent IL-1β Maturation and ASC Speck Formation in Macrophages. J. Immunol. 2019, 203, 795–800. [Google Scholar] [CrossRef]
- Saponaro, C.; Fanizzi, A.; Sonnessa, M.; Mondelli, P.; Vergara, D.; Loisi, D.; Massafra, R.; Latorre, A.; Zito, F.A.; Schirosi, L. Downstream Signaling of Inflammasome Pathway Affects Patients’ Outcome in the Context of Distinct Molecular Breast Cancer Subtypes. Pharmaceuticals 2022, 15, 651. [Google Scholar] [CrossRef]
- Xue, Y.; Zhang, Y.; Chen, L.; Wang, Y.; Lv, Z.; Yang, L.-Q.; Li, S. Citrulline protects against LPS-induced acute lung injury by inhibiting ROS/NLRP3-dependent pyroptosis and apoptosis via the Nrf2 signaling pathway. Exp. Ther. Med. 2022, 24, 632. [Google Scholar] [CrossRef]
- Xue, Y.; Du, H.D.; Tang, D.; Zhang, D.; Zhou, J.; Zhai, C.W.; Yuan, C.C.; Hsueh, C.Y.; Li, S.J.; Heng, Y.; et al. Correlation Between the NLRP3 Inflammasome and the Prognosis of Patients With LSCC. Front. Oncol. 2019, 9, 588. [Google Scholar] [CrossRef]
- Liang, A.; Zhong, S.; Xi, B.; Zhou, C.; Jiang, X.; Zhu, R.; Yang, Y.; Zhong, L.; Wan, D. High expression of PYCARD is an independent predictor of unfavorable prognosis and chemotherapy resistance in glioma. Ann. Transl. Med. 2021, 9, 986. [Google Scholar] [CrossRef]
- Nagar, A.; Rahman, T.; Harton, J.A. The ASC Speck and NLRP3 Inflammasome Function Are Spatially and Temporally Distinct. Front. Immunol. 2021, 12, 752482. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lindholm, H.; Herring, M.; Faresjö, M.; Haux, J.; Szekeres, F.; Ejeskär, K. The Co-Localization of NLRP3 and ASC Specks Does Not Automatically Entail NLRP3 Inflammasome Functionality in PDAC Cell Lines. Int. J. Transl. Med. 2024, 4, 224-237. https://doi.org/10.3390/ijtm4020013
Lindholm H, Herring M, Faresjö M, Haux J, Szekeres F, Ejeskär K. The Co-Localization of NLRP3 and ASC Specks Does Not Automatically Entail NLRP3 Inflammasome Functionality in PDAC Cell Lines. International Journal of Translational Medicine. 2024; 4(2):224-237. https://doi.org/10.3390/ijtm4020013
Chicago/Turabian StyleLindholm, Heléne, Matthew Herring, Maria Faresjö, Johan Haux, Ferenc Szekeres, and Katarina Ejeskär. 2024. "The Co-Localization of NLRP3 and ASC Specks Does Not Automatically Entail NLRP3 Inflammasome Functionality in PDAC Cell Lines" International Journal of Translational Medicine 4, no. 2: 224-237. https://doi.org/10.3390/ijtm4020013
APA StyleLindholm, H., Herring, M., Faresjö, M., Haux, J., Szekeres, F., & Ejeskär, K. (2024). The Co-Localization of NLRP3 and ASC Specks Does Not Automatically Entail NLRP3 Inflammasome Functionality in PDAC Cell Lines. International Journal of Translational Medicine, 4(2), 224-237. https://doi.org/10.3390/ijtm4020013






