Abyssal DNA: Eukaryotic Diversity in Atlantic Equatorial Deep-Sea Sediments Assessed Through DNA Metabarcoding
Abstract
1. Introduction
2. Materials and Methods
2.1. Core Sampling
2.2. Sedimentology and Chronological Analysis of the Cores
2.3. Selection of Sediment Core Material for Environmental DNA Analysis
2.4. DNA Sampling, Extraction and Sequencing
2.5. Data Analyses and Taxonomic Assignment
2.6. Eukaryotic Diversity
3. Results
3.1. Sedimentological and Chronological Analysis of the Cores
3.2. Eukaryotic Taxonomy
3.3. Eukaryotic Diversity and Distribution
4. Discussion
4.1. Eukaryotic Taxonomy and Diversity
4.2. Possible Relationships Between the Eukaryotic Diversity Detected and Core Sedimentology and Chronology
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Ramirez-Llodra, E.; Brandt, R.; Danovaro, R.; De Mol, B.; Escovar, E.; German, C.R.; Levin, L.A.; Arbizu, P.M.; Menot, L.; Buhl-Mortensen, P.; et al. Deep, diverse and definitely different: Unique attributes of the world’s largest ecosystem. Biogeosciences 2010, 7, 2851–2899. [Google Scholar] [CrossRef]
- Watling, L.; Guinotte, J.; Clark, M.R.; Smith, C.R. A proposed biogeography of the deep ocean floor. Prog. Oceanogr. 2013, 111, 91–112. [Google Scholar] [CrossRef]
- Riehl, T.; Wolf, A.-C.; Augustin, N.; Brandt, A. Discovery of widely available abyssal rock patches reveals overlooked habitat type and prompts rethinking deep-sea biodiversity. Proc. Natl. Acad. Sci. USA 2020, 117, 15450–15459. [Google Scholar] [CrossRef] [PubMed]
- Voelker, D. Abyssal Plains. In Encyclopedia of Marine Geosciences; Harff, J., Meschede, M., Petersen, S., Thiede, J., Eds.; Springer Reference: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Lyle, M. Deep-Sea Sediments. In Encyclopedia of Marine Geosciences; Harff, J., Meschede, M., Petersen, S., Thiede, J., Eds.; Springer: Dordrecht, The Netherlands, 2015. [Google Scholar] [CrossRef]
- Hurtado, C.; Roddaz, M.; Santos, R.V.; Baby, P.; Antoine, P.-O.; Dantas, E.L. Cretaceous-early paleocene drainage shift of amazonian rivers driven by Equatorial Atlantic Ocean opening and andean uplift as deduced from the provenance of northern Peruvian sedimentary rocks (Huallaga basin). Gondwana Res. 2018, 63, 152–168. [Google Scholar] [CrossRef]
- Jones, E.J.; Cande, S.C.; Spathopoulos, F. Evolution of a major oceanographic pathways: The equatorial atlantic, the tectonics, sedimentation and paleoceanography of the North Atlantic Region. Geol. Soc. Spec. Publ. 1995, 90, 199–213. [Google Scholar] [CrossRef]
- Munsell, C. Munsell Soil Color Charts, New Revised Edition; Macbeth Division of Kollmorgen Instruments; New Windsor: New York, NY, USA, 2009. [Google Scholar]
- Walker, R.G.; James, N.P. Facies Models and Modern Stratigrahic Concepts. In Facies Models—Response to Sea Level Change; Walker, R.G., James, N.P., Eds.; Geological Association of Canada: St. John’s, ON, Canada, 1992. [Google Scholar]
- Tauhata, L.; Salati, I.; Di Prinzio, R.; Di Prinzio, A.R. Radioproteção e Dosimetria: Fundamentos; CBPF: Dampier Peninsula, Australia, 2003; Volume 254. [Google Scholar]
- Ferretti, M. Princípios e aplicações de espectroscopia de fluorescência de Raios X (FRX) com instrumentação portátil para estudo de bens culturais. Rev. CPC 2009, 7, 74–98. [Google Scholar] [CrossRef][Green Version]
- Azevedo, A.Q.; Gonçalves, T.R.; Soares, V.G.; Quina, D.F.; Ayres, A.; Rosa, L.H.; Jovane, L. Tropical Atlantic: Upper ocean structure from the last 80 kyr. In preparation.
- Blaauw, M.; Christeny, J.A. Flexible paleoclimate age-depth models using an autoregressive gamma process. Bayesian Anal. 2011, 6, 457–474. [Google Scholar] [CrossRef]
- Stuiver, M.; Reimer, P.J.; Reimer, R.W. CALIB 8.2 [WWW Program]. 2021. Available online: http://calib.org (accessed on 1 April 2021).
- Chen, S.; Yao, H.; Han, J.; Liu, C.; Song, J.; Shi, L.; Zhu, Y.; Pang, X.X.; Luo, K.; Li, Y.; et al. Validation of the ITS2 region as a novel DNA barcode of identifying medicinal plant species. PLoS ONE 2010, 5, e8613. [Google Scholar] [CrossRef]
- Richardson, R.T.; Lin, C.-H.; Sponsler, D.B.; Quijia, J.O.; Goodell, K.; Johnson, R.M. Application of ITS2 metabarcoding to determine the provenance of pollen collected by honey bees in an agroecosystem. Appl. Plant Sci. 2015, 3, 1400066. [Google Scholar] [CrossRef]
- Ruppert, K.M.; Kline, R.J.; Rahman, M.S. Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: A systematic review in methods, monitoring, and applications of global eDNA. Glob. Ecol. Conserv. 2019, 17, e00547. [Google Scholar] [CrossRef]
- da Silva, M.K.; De Souza, L.M.D.; Vieira, R.; Neto, A.A.; Lopes, F.A.C.; De Oliveira, F.S.; Convey, P.; Carvalho-Silva, M.; Duarte, A.W.F.; Câmara, P.E.A.S.; et al. Fungal and fungal-like diversity in marine sediments from the maritime Antarctic assessed using DNA Metabarcoding. Sci. Rep. 2022, 12, 21044. [Google Scholar] [CrossRef]
- Bushnell, B. BBMap: A Fast, Accurate, Splice-Aware Aligner; Lawrence Berkeley National Lab (LBNL): Berkeley, CA, USA, 2014. [Google Scholar]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, B.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution sample inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.G.; Caporaso, J.G. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2′s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Banchi, E.; Ametrano, C.G.; Greco, S.; Stankovic, D.; Muggia, L.; Pallavicini, A. PLANiTS: A curated sequence reference dataset for plant ITS DNA metabarcoding. Database 2020, 2020, baz155. [Google Scholar] [CrossRef] [PubMed]
- Abarenkov, K.; Zirk, A.; Piirmann, T.; Pohonen, R.; Ivanov, F.; Nilsson, R.H.; Kõljalg, U. UNITE QIIME Release for Eukaryotes; Version 04.02.2020; UNITE Community: London, UK, 2020. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Hudson, D.H.; Beier, S.; Flade, I.; Górska, A.; El-Hadidi, M.S.; Ruscheweyh, H.-J.; Tappu, R. MEGAN community edition-interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput. Biol. 2016, 12, e1004957. [Google Scholar] [CrossRef]
- Giner, C.R.; Forn, I.; Romac, S.; Logares, R.; Vargas, C.; Massana, R. Environmental sequencing provides reasonable estimates of the relative abundance of specific picoeukaryotes. Appl. Environ. Microbiol. 2016, 82, 4757–4766. [Google Scholar] [CrossRef]
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive metagenomic visualization in a web browser. BMC Bioinform. 2011, 12, 385. [Google Scholar] [CrossRef]
- Rosa, L.H.; Pinto, O.H.B.; Convey, P.; Carvalho-Silva, M.; Rosa, C.A.; Câmara, P.E.A.S. DNA metabarcoding to assess the diversity of airborne fungi present over Keller Peninsula, King George Island, Antarctica. Microb. Ecol. 2021, 82, 165–172. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1. [Google Scholar]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. Jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef]
- Thompson, S.B.; Craveling, J.R. A global database of marine isotope substage 5a and 5c marine terraces and paleoshoreline indicators. Earth Syst. Sci. Data 2021, 13, 3467–3490. [Google Scholar] [CrossRef]
- Breyer, E.; Baltar, F. The largely neglected ecological role of oceanic pelagic fungi. Trends Ecol. Evol. 2023, 38, 870–888. [Google Scholar] [CrossRef] [PubMed]
- Musa, H.; Kasim, F.H.; Gunny, A.A.N.; Gopinath, S.C.B. Salt-adapted moulds and yeasts: Potentials in industrial and environmental biotechnology. Process Biochem. 2018, 69, 33–44. [Google Scholar] [CrossRef]
- Chen, Q.; Hou, L.W.; Duan, W.J.; Crous, P.W.; Cai, L. Didymellaceae revisited. Stud. Mycol. 2017, 87, 105–159. [Google Scholar] [CrossRef]
- Swathi, J.; Sowjanya, K.M.; Narendra, K.; Reddy, K.V.N.R.; Satya, A.K. Isolation, identification and production of bioactive metabolites from marine fungi collected from coastal area of Andhra Pradesh, India. J. Pharm. Res. 2013, 6, 663–666. [Google Scholar] [CrossRef]
- Godinho, V.M.; Gonçalves, V.N.; Santiago, I.F.; Figueredo, H.M.; Vitoreli, G.A.; Schaefer, C.E.G.R.; Barbosa, E.C.; Oliveira, J.G.; Alves, T.M.A.; Zani, C.L.; et al. Diversity and bioprospection of fungal community present in oligotrophic soil of continental Antarctica. Extremophiles 2015, 19, 585–596. [Google Scholar] [CrossRef]
- Bensch, K.; Groenewald, J.Z.; Dijksterhuis, J.; Starink-Willemse, M.; Andersen, B.; Summerell, B.A.; Shin, H.-D.; Dugan, F.M.; Schroes, H.-J.; Braun, U.; et al. Species and ecological diversity within the Cladosporium cladosporioides complex (Davidiellaceae, Capnodiales). Stud. Mycol. 2010, 67, 1–94. [Google Scholar] [CrossRef]
- Apangu, G.P.; Frisk, C.A.; Petch, G.M.; Muggia, L.; Pallavicini, A.; Hanson, M.; Skjoth, C.A. Environmental DNA reveals diversity and abundance of Alternaria species in neighbouring heterogeneous landscapes in Worcester, UK. Aerobiologia 2022, 38, 457–481. [Google Scholar] [CrossRef]
- Gunde-Cimerman, N.; Zalarb, P.; de Hoogs, S.; Plemenitasd, A. Hypersaline water in salterns—Natural ecological niches for halophilic black yeasts. FEMS Microbiol. Ecol. 2000, 32, 235–240. [Google Scholar]
- Gonçalves, V.N.; Soares, F.O.; Corrêa, G.R.; Senra, E.O.; Lopes, F.A.C.; Silva, M.C.; Convey, P.; Câmara, P.E.A.S.; Duarte, A.W.F.; Rosa, L.H. Fungal diversity present in ornithogenic soils of extreme equatorial Atlantic São Pedro and São Paulo archipelago using DNA metabarcoding. Braz. J. Microbiol. 2025, 56, 1619–1629. [Google Scholar] [CrossRef]
- Rotem, J. The genus Alternaria: Biology, Epidemiology and Pathogenicity; American Phytopathological Society: Saint Paul, MN, USA, 1994. [Google Scholar]
- Ding, H.; Zhang, D.; Zhou, B.; Ma, Z. Inhibitors of BRD4 Protein from a marine-derived fungus Alternaria sp. NH-F6. Mar. Drugs. 2017, 15, 76. [Google Scholar] [CrossRef]
- Nag Raj, T.R.; Ponnappa, K.M. Blight of water-hyacinth caused by Alternaria eichhorniae sp. nov. Trans. Br. Mycol. Soc. 1970, 55, 123–130. [Google Scholar] [CrossRef]
- Charudattanl, R.; Rao, R.V. Bostrycin and 4-deoxybostrycin: Two nonspecific phytotoxins produced by Alternaria eichhorniae. Appl. Environ. Microbiol. 1982, 43, 846–849. [Google Scholar] [CrossRef] [PubMed]
- Mehta, T.; Meena, M.; Nagda, A. Bioactive compounds of Curvularia species as a source of various biological activities and biotechnological applications. Front. Microbiol. 2022, 8, 1069095. [Google Scholar] [CrossRef] [PubMed]
- Yurchenko, A.N.; Smetanina, O.F.S.; Khudyakova, Y.V.; Kirichuk, N.N.; Yurchenko, E.A.; Afiyatullov, S.S. Metabolites of the marine isolate of the fungus Curvularia inaequalis. Chem. Nat. Compd. 2013, 49, 163–164. [Google Scholar] [CrossRef]
- Morton, F.J.; Smith, G. The genera Scopulariopsis Bainier, Microascus Zukal, and Doratomyces Corda. Mycol. Pap. 1963, 86, 1–96. [Google Scholar]
- Tosti, A.; Piraccini, B.M.; Stinchi, C.; Lorenzi, S. Onychomycosis due to Scopulariopsis brevicaulis: Clinical features and response to systemic antifungals. Br. J. Dermatol. 1996, 135, 799–802. [Google Scholar] [CrossRef]
- Vaupotič, T.; Plemenitaš, A. Differential gene expression and Hog1 interaction with osmoresponsive genes in the extremely halotolerant black yeast Hortaea werneckii. BMC Genom. 2007, 8, 280. [Google Scholar] [CrossRef]
- Gostinčar, C.; Stajich, J.E.; Gunde-Cimerman, N. Extremophilic and extremotolerant fungi. Curr. Biol. 2023, 33, R743–R759. [Google Scholar] [CrossRef]
- Ianiri, G.; LeibundGut-Landmann, S.; Dawson, T.L., Jr. Malassezia: A commensal, pathogen, and mutualist of human and animal skin. Annu. Rev. Microbiol. 2022, 8, 757–782. [Google Scholar] [CrossRef] [PubMed]
- Rosa, L.H.; Da Silva, T.H.; Ogaki, M.B.; Pinto, O.H.B.; Stech, M.; Convey, P.; Carvalho-Silva, M.; Rosa, C.A.; Câmara, P.E.A.S. DNA metabarcoding uncovers fungal diversity in soils of protected and non-protected areas on Deception Island, Antarctica. Sci. Rep. 2020, 10, 21986. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Cao, X.; Wang, Z.; Dai, Y.; Xing, B. Mechanistic understanding toward the toxicity of graphene-family materials to freshwater algae. Water Res. 2017, 111, 18–27. [Google Scholar] [CrossRef]
- Mayén-Estrada, R.; Dávila, S.; Dias, R.J.P. Ciliate symbionts of bivalves with notes on their worldwide geographic distribution. Zootaxa 2024, 14, 451–481. [Google Scholar] [CrossRef]
- Mabberley, D.J. Mabberley’s Plant-Book: A Portable Dictionary of Plants, Their Classification and Uses, 4th ed.; Cambridge University Press: Cambridge, UK, 2017. [Google Scholar]
- Armbrecht, L.; Weber, M.E.; Raymo, M.E.; Peck, V.L.; Williams, T.; Warnock, J.; Kato, Y.; Hernández-Almeida, I.; Hoem, F.; Reilly, B.; et al. Ancient marine sediment DNA reveals diatom transition in Antarctica. Nat. Commun. 2022, 13, 5787. [Google Scholar] [CrossRef]
- Pickering, J.L. Terrace formation in the upper Bengal basin since the Middle Pleistocene: Brahmaputra fan delta construction during multiple highstands. Basin Res. 2017, 30, 550–567. [Google Scholar] [CrossRef]


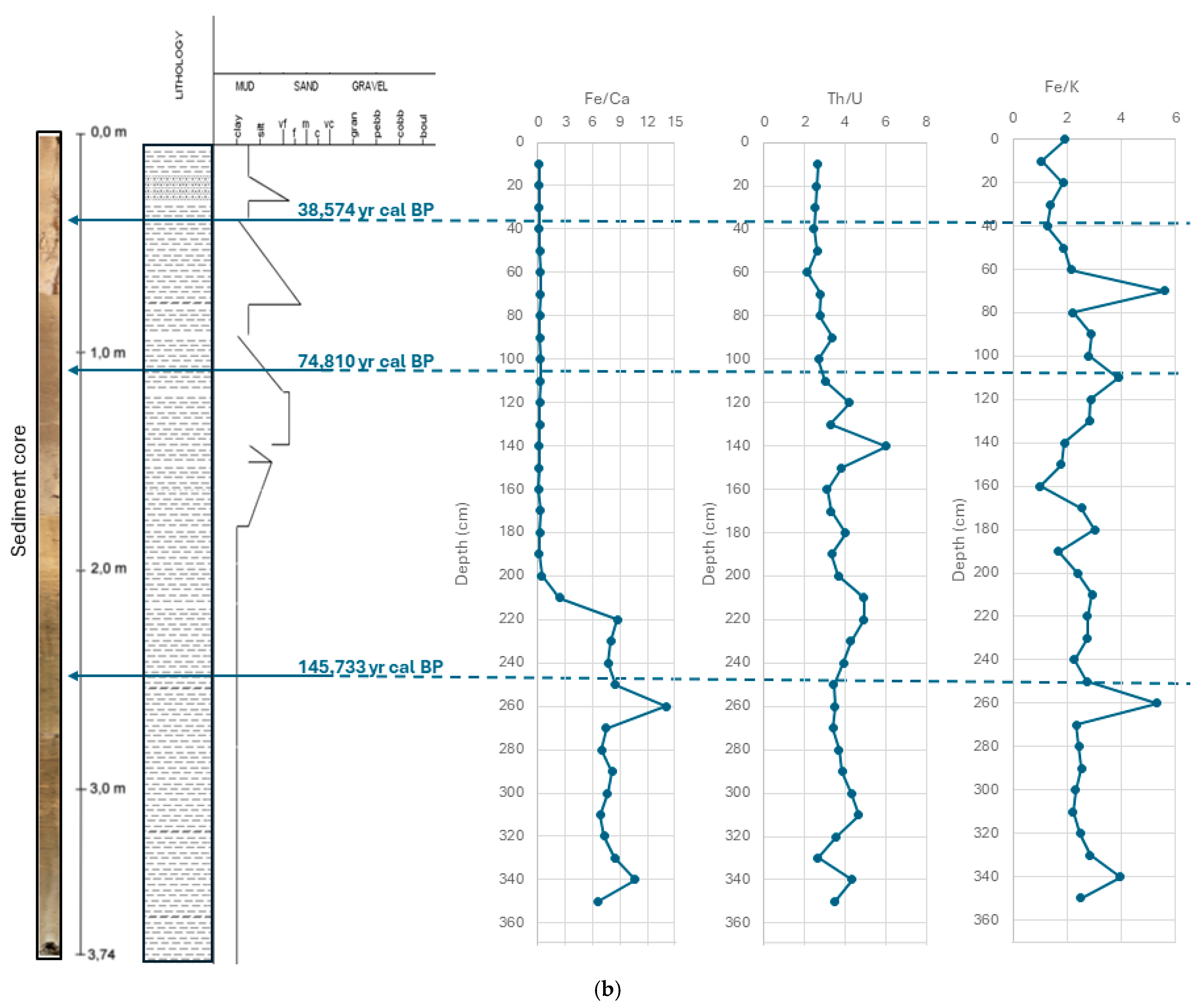
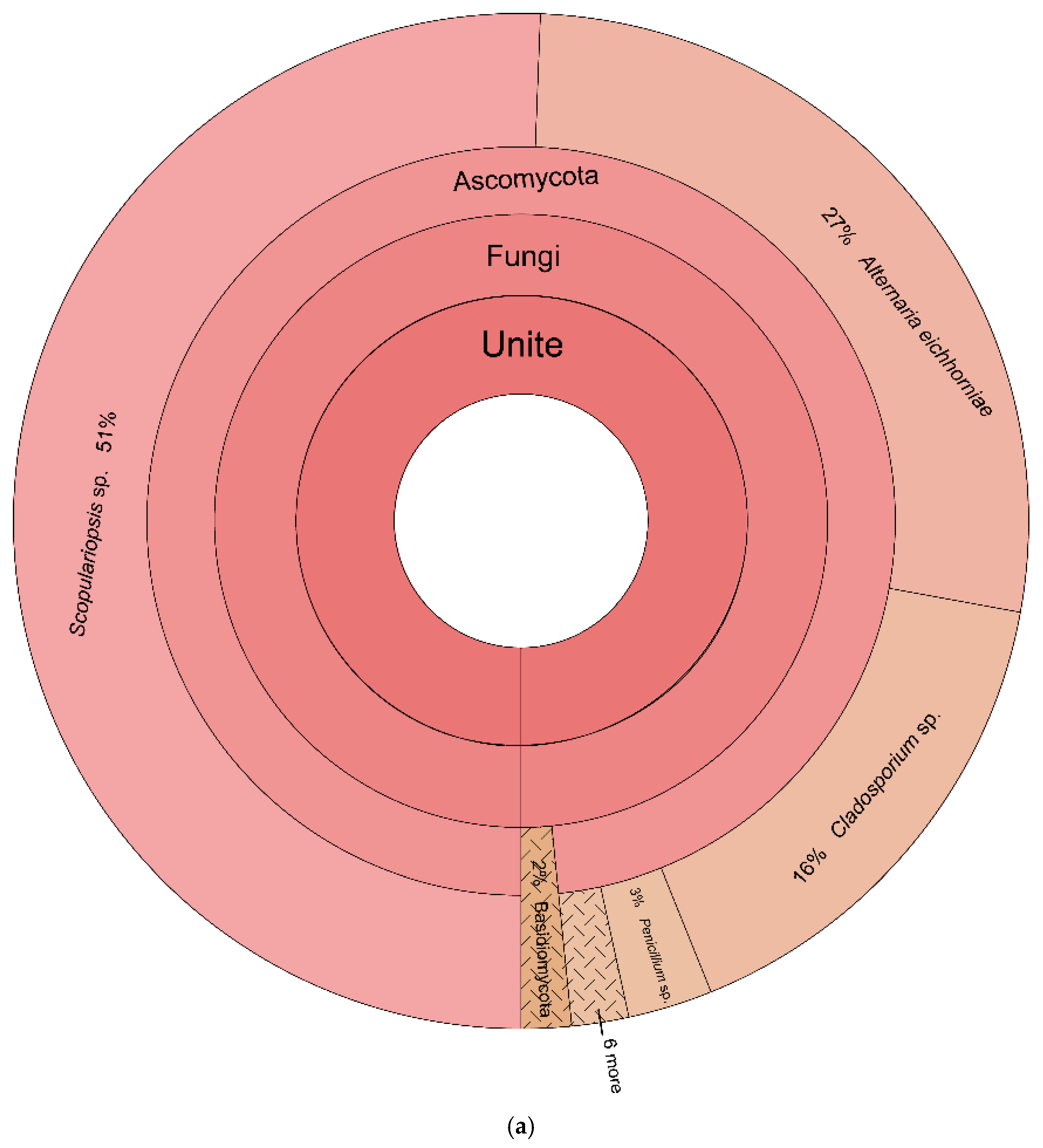
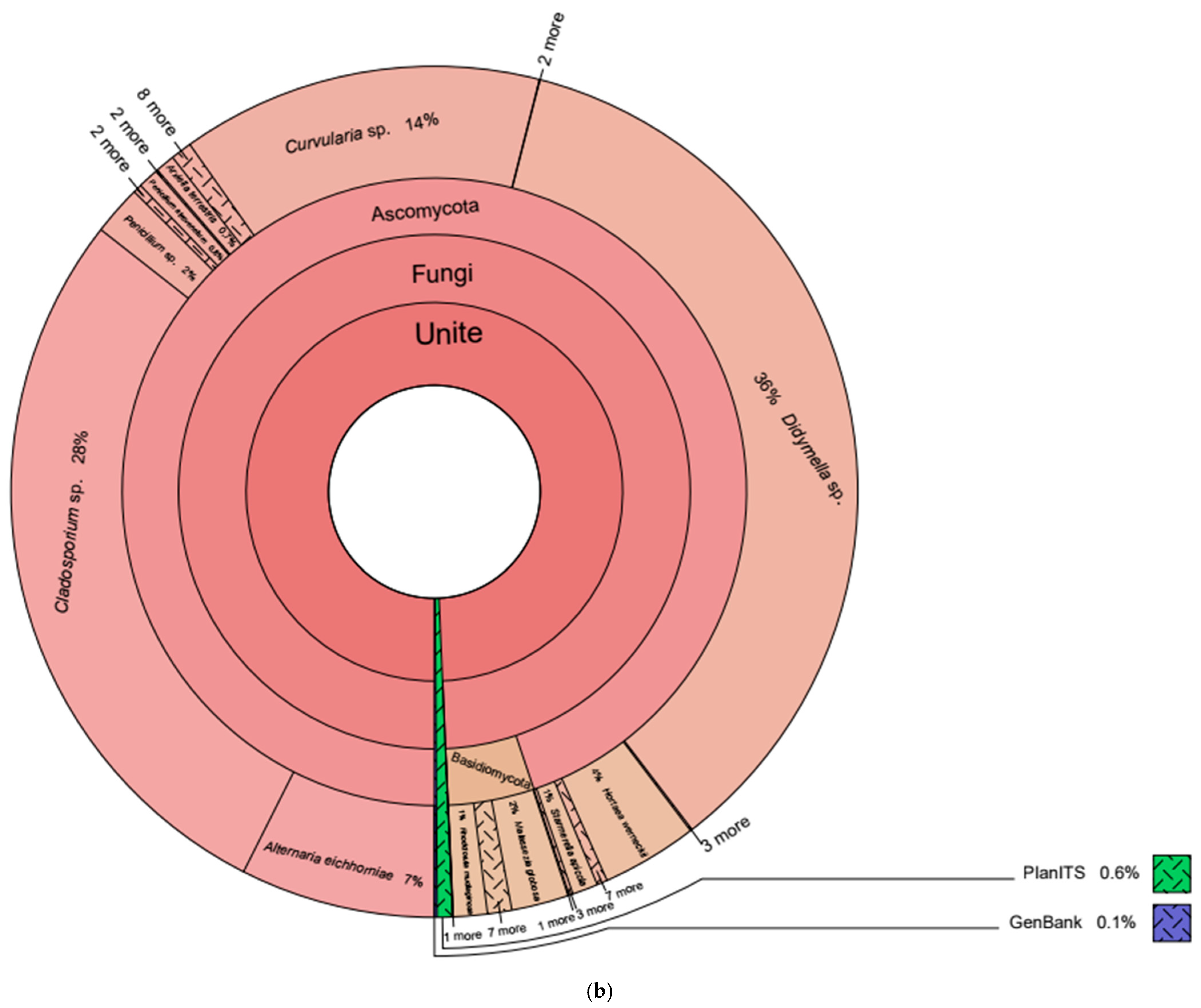


| Database | Kingdom | Phylum | Amplicon Sequence Variant | Core Depth and Relative Abundance (%) | |
|---|---|---|---|---|---|
| 4280 | 4444 | ||||
| Unite | Fungi | Ascomycota | Didymella sp. | 0.0000 | 25.1674 |
| Cladosporium sp. | 4.6853 | 19.8931 | |||
| Scopulariopsis sp. | 14.8085 | 0.0000 | |||
| Alternaria eichhorniae | 7.9806 | 5.2631 | |||
| Curvularia sp. | 0.0000 | 9.7575 | |||
| Hortaea werneckii | 0.0000 | 2.5857 | |||
| Penicillium sp. | 0.7895 | 1.3387 | |||
| Starmerella apicola | 0.0000 | 0.6914 | |||
| Penicillium atrovenetum | 0.0599 | 0.5548 | |||
| Arxiella terrestris | 0.0000 | 0.5231 | |||
| Aspergillus versicolor | 0.1081 | 0.2210 | |||
| Cryomyces funiculosus | 0.0000 | 0.2121 | |||
| Pseudogymnoascus pannorum | 0.0000 | 0.2109 | |||
| Chaetothyriales sp. | 0.0000 | 0.1860 | |||
| Colletotrichum sp. | 0.1008 | 0.0165 | |||
| Acremonium charticola | 0.1109 | 0.0000 | |||
| Pichia terricola | 0.0775 | 0.0000 | |||
| Fusarium sp. | 0.0731 | 0.0000 | |||
| Trapeliopsis sp. | 0.0000 | 0.0635 | |||
| Chaetothyriales sp. | 0.0000 | 0.0579 | |||
| Agyriales sp. | 0.0000 | 0.0362 | |||
| Catenulostroma sp. | 0.0000 | 0.0297 | |||
| Sugiyamaella sp. | 0.0000 | 0.0297 | |||
| Candida albicans | 0.0000 | 0.0293 | |||
| Dothideomycetes sp. | 0.0000 | 0.0273 | |||
| Aspergillus sp. | 0.0000 | 0.0233 | |||
| Physciaceae sp. | 0.0000 | 0.0221 | |||
| Phaeosphaeria sp. | 0.0000 | 0.0177 | |||
| Acarospora sp. | 0.0000 | 0.0169 | |||
| Elasticomyces elasticus | 0.0000 | 0.0149 | |||
| Buellia sp. | 0.0000 | 0.0129 | |||
| Capnodiales sp. | 0.0000 | 0.0129 | |||
| Phylliscum sp. | 0.0000 | 0.0117 | |||
| Paraphaeosphaeria sp. | 0.0000 | 0.0112 | |||
| Debaryomyces sp. | 0.0000 | 0.0104 | |||
| Candida orthopsilosis | 0.0000 | 0.0088 | |||
| Exophiala cancerae | 0.0000 | 0.0080 | |||
| Starmerella etchellsii | 0.0000 | 0.0076 | |||
| Lachancea thermotolerans | 0.0000 | 0.0052 | |||
| Lecanora sp. | 0.0000 | 0.0048 | |||
| Diaporthaceae sp. | 0.0000 | 0.0028 | |||
| Basidiomycota | Malassezia globosa | 0.1149 | 1.5403 | ||
| Rhodotorula mucilaginosa | 0.0000 | 0.9144 | |||
| Cutaneotrichosporon debeurmannianum | 0.0000 | 0.4962 | |||
| Malassezia restricta | 0.1611 | 0.0378 | |||
| Papiliotrema terrestris | 0.1245 | 0.0000 | |||
| Radulomyces sp. | 0.0663 | 0.0000 | |||
| Naganishia diffluens | 0.0000 | 0.0558 | |||
| Exidiaceae sp. | 0.0000 | 0.0333 | |||
| Malassezia arunalokei | 0.0000 | 0.0325 | |||
| Malasseziaceae sp. | 0.0000 | 0.0145 | |||
| Phenoliferia psychrophenolica | 0.0000 | 0.0129 | |||
| Gjaerumia minor | 0.0000 | 0.0117 | |||
| Robbauera albescens | 0.0000 | 0.0096 | |||
| Chromista | Ciliophora | Spirotrachelostyla tani | 0.0000 | 0.0072 | |
| GenBank | Fungi | Ascomycota | Rhizocarpon sp. | 0.0000 | 0.0088 |
| Fungal sp. | 0.0000 | 0.0679 | |||
| PlanITS | Viridiplantae | Chlorophyta | Pseudochlorella pyrenoidosa | 0.0000 | 0.2828 |
| Bracteacoccus bullatus | 0.0000 | 0.0325 | |||
| Stichococcus mirabilis | 0.0000 | 0.0233 | |||
| Diplosphaera sp. | 0.0000 | 0.0217 | |||
| Trebouxia flava | 0.0000 | 0.0108 | |||
| Streptophyta | Laportea aestuans | 0.0000 | 0.0229 | ||
| Tortula mucronifolia | 0.0000 | 0.0100 | |||
| Oxyria digyna | 0.0000 | 0.0056 | |||
| Diversity Indices | Core Water Depth (m) | |
|---|---|---|
| 4280 | 4444 | |
| Number of fungal ASVs | 14 | 59 |
| Number of DNA reads | 72,832 | 176,073 |
| Fisher’s α | 1.30 | 5.71 |
| Margalef | 1.16 | 4.80 |
| Simpson | 0.64 | 0.77 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gontijo, N.R.; Gonçalves, V.N.; Neto, A.A.; Vieira, R.; Caram, T.N.; Malheiros, M.M.; Lopes, F.A.C.; Silva, M.C.; Azevedo, A.Q.; Gonçalves, T.R.; et al. Abyssal DNA: Eukaryotic Diversity in Atlantic Equatorial Deep-Sea Sediments Assessed Through DNA Metabarcoding. DNA 2025, 5, 45. https://doi.org/10.3390/dna5030045
Gontijo NR, Gonçalves VN, Neto AA, Vieira R, Caram TN, Malheiros MM, Lopes FAC, Silva MC, Azevedo AQ, Gonçalves TR, et al. Abyssal DNA: Eukaryotic Diversity in Atlantic Equatorial Deep-Sea Sediments Assessed Through DNA Metabarcoding. DNA. 2025; 5(3):45. https://doi.org/10.3390/dna5030045
Chicago/Turabian StyleGontijo, Natana Rabelo, Vívian Nicolau Gonçalves, Arthur Ayres Neto, Rosemary Vieira, Tainá Napoleão Caram, Marina Martins Malheiros, Fabyano A. C. Lopes, Micheline C. Silva, Allana Queiroz Azevedo, Thauana Rodrigues Gonçalves, and et al. 2025. "Abyssal DNA: Eukaryotic Diversity in Atlantic Equatorial Deep-Sea Sediments Assessed Through DNA Metabarcoding" DNA 5, no. 3: 45. https://doi.org/10.3390/dna5030045
APA StyleGontijo, N. R., Gonçalves, V. N., Neto, A. A., Vieira, R., Caram, T. N., Malheiros, M. M., Lopes, F. A. C., Silva, M. C., Azevedo, A. Q., Gonçalves, T. R., Jovane, L., Convey, P., Câmara, P. E. A. S., & Rosa, L. H. (2025). Abyssal DNA: Eukaryotic Diversity in Atlantic Equatorial Deep-Sea Sediments Assessed Through DNA Metabarcoding. DNA, 5(3), 45. https://doi.org/10.3390/dna5030045









