Abstract
Background/Objectives: Male infertility is a major health concern with a complex etiopathology, yet a substantial proportion of cases remain idiopathic. Mitochondrial dysfunction and non-coding RNA (ncRNA) deregulation have both been implicated in impaired spermatogenesis, but their interplay remains poorly understood. This study aimed to identify infertility-specific variants in ncRNAs that affect mitochondrial dynamics and homeostasis and to explore their roles. Methods: Whole-genome sequencing (WGS) was performed on genomic DNA samples from teratozoospermic, asthenozoospermic, oligozoospermic, and normozoospermic men. Variants uniquely present in infertile individuals and mapped to ncRNAs that affect mitochondrial dynamics were selected and prioritized using bioinformatics tools. An independent transcriptomic validation was conducted using RNA-sequencing data from testicular biopsies of men with non-obstructive azoospermia (NOA) to determine whether the ncRNAs harboring WGS-derived variants were transcriptionally altered. Results: We identified several infertility-specific variants located in lncRNAs known to interact with mitochondrial regulators, including GAS5, HOTAIR, PVT1, MEG3, and CDKN2B-AS1. Transcriptomic analysis confirmed significant deregulation of these lncRNAs in azoospermic testicular samples. Bioinformatic analysis also implicated the disruption of lncRNA–miRNA–mitochondria networks, potentially contributing to mitochondrial membrane potential loss, elevated reactive oxygen species (ROS) production, impaired mitophagy, and germ cell apoptosis. Conclusions: Our integrative genomic and transcriptomic analysis highlights lncRNA–mitochondrial gene interactions as a novel regulatory layer in male infertility, while the identified lncRNAs hold promise as biomarkers and therapeutic targets. However, future functional studies are warranted to elucidate their mechanistic roles and potential for clinical translation in reproductive medicine.
1. Introduction
Infertility is a significant global health concern, affecting approximately one in six couples of reproductive age, according to the World Health Organization (WHO). It is defined as the inability to achieve pregnancy after 12 months of regular, unprotected sexual intercourse and can result from various factors affecting either partner or both [1]. Male infertility contributes to nearly 50% of all infertility cases, emphasizing its crucial role in reproductive health [2]. It is commonly classified based on semen analysis parameters into azoospermia (absence of sperm in the ejaculate), oligozoospermia (low sperm count), asthenozoospermia (reduced sperm motility), and teratozoospermia (abnormal sperm morphology) [3]. These sperm abnormalities often arise from a complex interplay of genetic, hormonal, environmental, and lifestyle factors [3,4]. More importantly, a large proportion of male infertility cases remain unexplained and are classified as idiopathic [5]. As semen quality has declined globally over recent decades [6,7], the burden of male infertility is expected to increase, further amplifying its psychosocial, economic, and public health implications [8,9]. Given its profound impact on affected individuals and couples, there is an urgent need for more advanced diagnostic approaches, targeted therapeutic interventions, and broader research efforts to improve male reproductive health outcomes.
Mitochondria play a crucial role in male fertility, particularly in sperm function, as they are responsible for ATP production, oxidative stress regulation, and apoptosis [10,11]. Spermatozoa have a high energy demand, relying on mitochondrial oxidative phosphorylation to support motility, capacitation, and fertilization potential [11]. Numerous studies have demonstrated a strong association between mitochondrial dysfunction and male infertility, providing evidence that mitochondrial DNA (mtDNA) mutations are linked to impaired sperm parameters, such as reduced motility and increased oxidative stress [12,13,14,15]. Furthermore, excessive production of reactive oxygen species (ROS) due to mitochondrial dysfunction has been shown to induce lipid peroxidation, DNA damage, and apoptosis in sperm cells [16,17]. These findings highlight the importance of mitochondrial integrity in sperm physiology and underscore the need for further research, as the upstream regulatory mechanisms affecting mitochondrial behavior in the testis and the context of male infertility remain largely undefined.
Emerging evidence suggests that non-coding RNAs (ncRNAs) are key regulators of gene expression in spermatogenesis and play a significant role in male infertility [18,19]. ncRNAs, including microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and PIWI-interacting RNAs (piRNAs), have been implicated in the post-transcriptional regulation of genes essential for sperm development, maturation, and function [20]. Studies have identified differential expression patterns of ncRNAs in infertile men, indicating their involvement in conditions such as azoospermia, asthenozoospermia, oligozoospermia, and teratozoospermia [21,22,23]. miRNAs, for instance, have been found to modulate pathways related to apoptosis, oxidative stress, and mitochondrial function [23,24,25], while lncRNAs and piRNAs contribute to epigenetic regulation and transposon silencing during spermatogenesis [18,26]. Considering their regulatory potential, ncRNAs represent promising biomarkers and therapeutic targets for diagnosing and managing male infertility. Furthermore, they are promising research candidates for uncovering regulatory networks that contribute to male infertility.
While mitochondrial dysfunction and ncRNA-mediated regulation have each been independently recognized as critical contributors to spermatogenesis, their interplay in the context of male infertility remains largely unexplored. Distinct expression profiles of ncRNAs between fertile and infertile individuals suggest their diagnostic and regulatory potential, yet our understanding of how genetic variants within ncRNA loci influence sperm function is still limited. Similarly, mitochondria—key regulators of energy metabolism, apoptosis, and germ cell maturation—are increasingly implicated in various forms of sperm dysfunction, including oligozoospermia, asthenozoospermia, and azoospermia. Despite this, the connection between mitochondrial dynamics and ncRNA regulation has not been adequately addressed in male reproductive genetics.
To bridge this gap, the present study investigates the interaction between ncRNAs, mitochondrial function, and genetic variation by leveraging whole-genome sequencing (WGS) to identify novel variants located within ncRNAs known to regulate mitochondrial dynamics. These variants are further prioritized using bioinformatics tools to assess their potential functional impact. Through this integrative approach, we aim to uncover novel ncRNA–mitochondrial gene regulatory networks that may underlie male infertility, with the ultimate goal of identifying new biomarkers, therapeutic targets, and contributing to the advancement of personalized reproductive medicine.
2. Materials and Methods
2.1. Biological Material
The study was approved by the Ethics Committee of the University of Thessaly in Volos, Greece, and all participants provided their informed written consent. Subsequently, blood and sperm samples were collected from male volunteers. The samples were obtained by masturbation after a minimum period of abstaining from sexual intercourse for two to five days. The volunteers filled out a questionnaire regarding their medical history, health and lifestyle habits, and other relevant information. To focus exclusively on idiopathic male infertility, the exclusion criteria had to be very strict. Specifically, patients with varicocele; reproductive tract infections; testicular injuries or pathologies; or a history of cryptorchidism, orchitis, or epididymitis, as well as those with specific systemic diseases, including endocrine disorders (diabetes mellitus, thyroid disorders, hypogonadism), autoimmune disorders (systemic lupus erythematosus, rheumatoid arthritis), metabolic syndromes, and active or past cancer, were excluded. Furthermore, extensive genetic testing was conducted to exclude males with Y microdeletions, chromosomal abnormalities, or any other genetic causes of infertility.
Every volunteer underwent an andrological examination, and his semen sample was used for subsequent analysis. The World Health Organization (WHO) manual for the examination and processing of human semen, fifth edition (https://apps.who.int/iris/handle/10665/44261, accessed on 9 June 2025), was followed for the semen analyses. More specifically, the analyses were conducted at an accredited andrology laboratory that adheres to standardized procedures for evaluating sperm count, motility, morphology, and other related parameters. The results of the analysis were used to diagnose abnormal sperm parameters and subsequently classify the individuals as normozoospermic, asthenozoospermic, oligozoospermic, or teratozoospermic, according to the reference values reported in the WHO manual. As for the abnormal findings, it is of great importance to repeat the analysis after a period of time to account for any variability in sperm parameters. Thus, repeated semen analyses were performed after three to six months to confirm the consistency of the initial findings.
2.2. DNA Extraction and Sample Preparation
Tubes containing ethylenediaminetetraacetic acid (EDTA) were used to store blood samples from volunteers. DNA extraction was performed using 200 μL of blood samples and the PureLink Genomic DNA Mini Kit (Invitrogen, Waltham, MA, USA—Catalog number: K182002), according to the manufacturer’s guidelines. Quantitative DNA analysis was conducted using the Qubit 2.0 fluorometer and the Qubit dsDNA BR Assay Kit (Invitrogen, Waltham, MA, USA—Catalog number: Q32850). Additionally, agarose gel electrophoresis was used to evaluate the integrity of the DNA. Only high-quality samples were used for further analysis.
Subsequently, five separate sequencing pools were established for whole-genome sequencing (WGS). The first two pools contained DNA from five men each with normozoospermia. The third and fourth pools consisted of DNA from five individuals with asthenozoospermia and teratozoospermia, respectively. The final pool contained DNA from five men with oligozoospermia. The DNA in each pool was mixed to achieve equimolar concentrations, resulting in a uniform final concentration of 100 ng/uL and a total quantity of 2 mg.
2.3. Whole-Genome Sequencing (WGS) and Data Analysis
After preparation, the DNA samples were sent to Novogene Co. (Cambridge, UK) where 100 bp paired-end libraries were constructed and sequenced using an Illumina HiSeq 3000 with an average sequencing coverage of 30×.
Following WGS, the quality of the generated FASTQ files was evaluated using FASTQC [27]. Low-quality reads (minimum PHRED score of 30) and adapter sequences were trimmed using Trimmomatic (v0.39) [28]. After quality control, the processed reads were aligned to the human reference genome (GRCh37/hg19) and retrieved from the Ensembl database [29] using the Burrows-Wheeler aligner (BWA) (version 0.7.17) [30]. Duplicate reads produced by polymerase chain reaction (PCR) were removed using Picard tools (http://broadinstitute.github.io/picard/, accessed on 9 June 2025), and SAM files were converted to BAM format with SAMtools (v1.19.2) [31]. SAMtools (v1.19.2) [31] were also used to merge the BAM files from normozoospermic samples into a single file for further analysis. FreeBayes (v1.3.7) [32] was used for variant calling, and the results were stored in variant call format (VCF). A comparative analysis of the VCF files was then performed using BCFtools (v1.17) [31]. Specifically, the VCF files for asthenozoospermic, oligozoospermic, and teratozoospermic men were compared to those of normozoospermic men, resulting in three novel VCF files containing genetic variants unique to each infertile group. These variants were used for subsequent analyses to investigate their potential involvement in pathogenic phenotypes and to provide insights into the molecular mechanisms underlying male infertility. Finally, the variants were annotated using the Variant Effect Predictor (VEP) tool (https://www.ensembl.org/Tools/VEP, accessed on 9 June 2025) [33] provided by the Ensembl database.
2.4. Bioinformatics Analysis
To investigate the role of non-coding RNAs in mitochondrial regulation and male infertility, this study focused on analyzing the unique variants in asthenozoospermic, oligozoospermic, and teratozoospermic men by prioritizing and investigating variants in these genomic regions. For this reason, initially, the analysis involved compiling a list of all ncRNAs (miRNAs and lncRNAs) associated with mitochondrial function, based on their roles in targeting related genes, as described by Gusic and Prokisch (2020) [34] and Sun et al. (2022) [35]. Specifically, this study provided a basis for identifying 91 miRNAs and 22 lncRNAs involved in mitochondrial dynamics. The ncRNAs were then evaluated using the Ensembl Genome Browser [29]. For each ncRNA, the corresponding gene name and Ensembl ID were recorded to facilitate the identification of variants from the annotation files generated during comparisons between normozoospermic and non-normozoospermic men. Additionally, searches were conducted in GeneCards [36] for lncRNAs with multiple aliases to ensure accurate matching with Ensembl GRCh37. The final ncRNA list was then used to identify unique variants mapped to these genes in infertile men (asthenozoospermic, teratozoospermic, oligozoospermic).
After identifying unique variants found only in infertile men, in ncRNAs associated with mitochondrial dynamics, a filtering process was applied to prioritize those with a potential role in male infertility via mitochondrial regulation. This prioritization was conducted using multiple bioinformatics tools and stringent selection criteria. At first, common variants with an allele frequency >0.05 in the European population, based on data from the 1000 Genomes Project [37], were excluded to focus on rare variants. Rare variants are more likely to be associated with pathogenic phenotypes, as they are less frequent in the population and can provide insights into common diseases [38]. Additionally, SNPs lacking population genetic data in Ensembl were also chosen for analysis due to their potential pathogenic significance. It should also be noted that allele frequency data were restricted to the specified geographical regions, as this study specifically focuses on the Greek population. Furthermore, a CADD Score > 10 was applied as an additional filter to prioritize variants, as scores above this threshold place a variant within the top 10% of likely functional and deleterious variants in the human genome [39]. Variants were further analyzed using SNPnexus [40] to explore their potential associations with male infertility, reproductive system diseases, reproductive cancers, or other diseases. Specifically, batch queries were performed with GRCh37 genome annotations to extract functional and phenotypic data for the filtered variants. Finally, SNPs in ncRNAs with potential roles in mitochondrial dynamics were assessed using the lncRNASNP2 database [41] to evaluate their impact on lncRNA-miRNA binding interactions. This analysis identified variants that may disrupt or create miRNA binding sites, potentially affecting gene expression regulation and mitochondrial homeostasis and thereby contributing to male infertility phenotypes in the studied population.
2.5. Validation of lncRNA Expression in Azoospermic Samples
To further evaluate the functional relevance of the lncRNAs harboring infertility-associated variants identified via whole-genome sequencing (WGS), we examined their expression patterns in an independent RNA-sequencing (RNA-seq) dataset. This dataset originated from our previously published transcriptomic study of testicular biopsies from azoospermic men [42]. In brief, total RNA was extracted from 19 testicular biopsy samples from patients with idiopathic non-obstructive azoospermia (iNOA). Samples were categorized based on the presence of testicular spermatozoa into three groups: “No presence” (n = 9), “High presence” (n = 5), and “Rare presence” (n = 5). The term “No presence” was used for samples in which no spermatozoa were found after at least 30 min of searching under an inverted microscope by three different experienced operators. The term “High presence” referred to samples where the first spermatozoon was identified within a maximum of 2 min of searching, while “Rare presence” described samples where the first spermatozoon was found after more than 10 min of searching. Samples from patients with obstructive azoospermia served as controls (n = 7). RNA sequencing (RNA-seq) and comparative transcriptomics analysis were performed, followed by differential gene expression analysis, applying standard quality control, normalization, and filtering procedures. We specifically assessed whether the lncRNAs identified through WGS were differentially expressed between different comparisons (“High presence” vs. “No presence”, “High presence” vs. “Rare presence”, and “Rare presence” vs. “No presence”). Expression changes were considered significant at |log2 fold change| > 1 and a false discovery rate (FDR) < 0.05. This analysis provided an additional layer of validation, supporting a potential link between genetic variation, transcriptional deregulation, and the involvement of lncRNA–target gene axes in mitochondrial dynamics and male infertility.
2.6. Statistical Analysis
All statistical analyses were performed using SPSS software (version 28) and R (version 4.3.3). Quantitative data are presented as mean ± standard deviation (SD). The normality of the data was assessed using the Shapiro–Wilk test. Comparisons between groups were made using one-way ANOVA, followed by Tukey’s post hoc test. Categorical variables were compared using the chi-square test. A p-value of <0.05 was considered statistically significant. Where applicable, adjustments for multiple comparisons were made, and corrected p-values were reported. Furthermore, for RNA-seq data analysis, differential expression was assessed using DESeq2, and volcano plots of differentially expressed genes were generated using the ggplot2 package in R.
3. Results
3.1. Sample Characteristics
A total of 38 volunteers were initially recruited for the study. After applying all inclusion and exclusion criteria, 25 samples were retained and used for subsequent analyses (normozoospermic, n = 10; asthenozoospermic, n = 5; oligozoospermic, n = 5; teratozoospermic, n = 5). Table 1 summarizes the demographic characteristics of the study participants across the analyzed variables, including age, Body Mass Index (BMI), smoking status, and alcohol consumption. Statistical analysis revealed no significant differences between groups regarding BMI, smoking, or alcohol intake (p-value > 0.05). However, a significant age difference was observed (ANOVA, p-value = 0.049717), with post hoc analysis indicating a specific difference between the asthenozoospermic and oligozoospermic groups (Tukey’s test, p-value = 0.037). Since all subsequent comparisons were performed between fertile and infertile men, this age difference was not considered relevant.

Table 1.
Demographic characteristics of study participants.
3.2. Whole-Genome Sequencing and Variant Annotation
Following whole-genome sequencing (WGS), data analysis was performed. Table 2 summarizes the number of unique variants identified in each group of infertile men compared to normozoospermic individuals. The analysis revealed a higher number of exclusive variants in normozoospermic men across all comparisons. These variants were mapped to thousands of genes and non-coding regions, highlighting potential genetic differences between the groups (Table 2). For subsequent analyses, we focused exclusively on unique variants identified in infertile males (asthenozoospermic, oligozoospermic, teratozoospermic), as these may contribute to the male infertility phenotype and serve as potential biomarkers.

Table 2.
Summary of unique variants identified in each group compared to normozoospermic individuals, along with the genes to which these variants are mapped.
3.3. Identification and Filtering of Unique Variants Affecting Mitochondrial Dynamics
This study aimed to investigate ncRNAs involved in mitochondrial homeostasis in the context of male infertility. To achieve this, a list of relevant lncRNAs and miRNAs was compiled based on the publications by Gusic and Prokisch (2020) [34] and Sun et al. (2022) [35]. Using this curated list and the corresponding Ensembl IDs, a custom script was developed to search the annotated files, including unique variants between normozoospermic and non-normozoospermic individuals. The goal was to identify unique variants in the selected ncRNA genes of interest (Table S1a,b). Notably, no variants were identified in miRNA genes across the three male infertility subtypes (asthenozoospermia, oligozoospermia, teratozoospermia), likely due to the small size of these genes. Consequently, this study focused on variants identified in lncRNA genes associated with mitochondrial dynamics, which were exclusively found in infertile men from these three subgroups.
The identified variants were then filtered to pinpoint those most likely to contribute to male infertility, potentially having a functional role, and serve as potential biomarkers. The filtering criteria included an MAF (Minor Allele Frequency) <5% (1000 Genome Project—Europe), as rarer mutations are more likely to be implicated in pathogenic phenotypes. Additionally, a second filtering criterion was applied, using a CADD score >10, which indicates potentially damaging mutations. This filtering process identified a total of 121 variants in nine lncRNAs, found exclusively in infertile men, which may potentially affect mitochondrial function. Specifically, for asthenozoospermia, 35 variants were detected in six lncRNAs (Table S2), while for oligozoospermia, 56 variants were found in five lncRNAs (Table S3). For teratozoospermia, after filtering, 51 variants in six lncRNAs were identified (Table S4). Notably, 11 variants were shared across at least two subcategories of infertility, with 5 of these found in all three subcategories (Table 3).

Table 3.
List of unique variants shared across the three male infertility subcategories.
3.4. Investigation of the Role of Variants—Association with Diseases and Impact on lncRNA-miRNA Interactions
In the next step of the analysis, we investigated the role of the identified variants on lncRNAs affecting mitochondrial dynamics according to Gusic and Prokisch (2020) [34] and Sun et al. (2022) [35]. At first, the identified variants were analyzed using the SNPnexus database [40] to assess their potential role and determine whether they had been previously associated with male infertility, other diseases, or reproductive system cancer. As shown in Table 4, SNPnexus [40] provided results for three variants.

Table 4.
Unique variants associated with male infertility or other diseases according to SNPnexus [40].
As shown in Table 4, the identified variants are associated with rs1011970 and are located in the CDKN2B-AS1 gene, which encodes the lncRNA ANRIL. Two of these variants (rs61743293, rs4977754) have previously been associated with breast cancer and were, interestingly, detected in all three subcategories of infertility. The third variant (rs16935753) is also linked to rs17694493 and is associated with prostate cancer, too. This variant was identified exclusively in the teratozoospermia subtype.
The filtered variants were subsequently analyzed using the lncRNASNP2 database [41] to assess their potential impact on lncRNA-miRNA interactions. This analysis identified 23 SNPs leading to either the gain or loss of miRNA binding sites on lncRNAs. These alterations are crucial for the dysregulation of gene expression, potentially affecting mitochondrial function and dynamics. Given their potential influence, these variants are likely contributors to the development of male infertility.
As illustrated in Table 5, a total of 18 variants found in seven lncRNAs resulted in the creation of 36 new miRNA binding sites. The formation of these sites facilitates molecular interactions that typically do not occur under physiological conditions, potentially leading to changes in gene expression that affect mitochondrial function and are likely associated with male infertility.

Table 5.
Variants leading to the gain of miRNA binding sites on lncRNAs according to lncRNASNP2 [41].
Furthermore, Table 6 indicates that a total of 15 variants found in six lncRNAs lead to the loss of 26 miRNA binding sites. This loss is significant because it eliminates interactions that may be crucial for mitochondrial function and dynamics, both of which are essential for male fertility.

Table 6.
Variants leading to the loss of miRNA binding sites on lncRNAs according to lncRNASNP2 [41].
3.5. Expression-Based Validation of lncRNAs Harboring Infertility-Associated Variants
To determine whether the lncRNAs harboring unique variants identified through WGS are transcriptionally dysregulated in testicular tissue of infertile men, we analyzed their expression profiles using an independent RNA-seq dataset derived from testicular biopsies of patients with idiopathic non-obstructive azoospermia (iNOA) [42]. This dataset included samples categorized by the presence of testicular spermatozoa (“No presence”, “Rare presence”, “High presence”) and a control group of men with obstructive azoospermia.
Differential expression analysis between the “No presence” and “High presence” groups revealed that several lncRNAs were significantly dysregulated. In total, 16,866 genes were differentially expressed in the “High Presence” group, with 10,997 upregulated and 5869 downregulated genes (|log2 fold change| > 1, FDR < 0.05), as shown in the volcano plot (Figure 1). Among these, 2497 lncRNAs were significantly upregulated and 409 lncRNAs were significantly downregulated in samples with detectable spermatozoa, suggesting widespread transcriptional changes associated with spermatogenic activity.
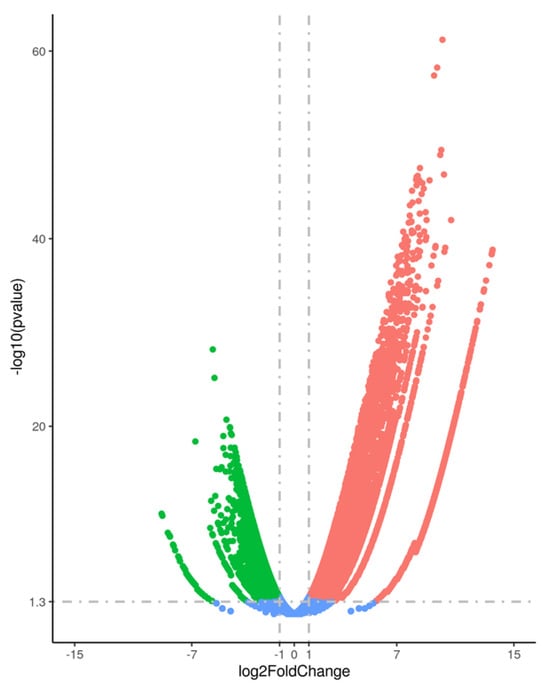
Figure 1.
Volcano plot illustrating the differential gene expression between testicular samples with “High Presence” and “No Presence” of spermatozoa. Significantly upregulated genes are highlighted in red, while downregulated genes are shown in green. Genes with no significant differential expression are depicted in blue. The x-axis represents the log2 fold change, and the y-axis represents the −log10 of the adjusted p-value (FDR).
Regarding the lncRNAs identified with infertility-specific variants through WGS, several exhibited significant transcriptional deregulation in the RNA-seq dataset (Table 7). For example, MEG3, GAS5, and PVT1 were significantly downregulated in the “High Presence” group, with FDR values of 0.00035, 0.00053, and 0.00013, respectively. Conversely, HOTAIR and CDKN2B-AS1 were found to be upregulated, with FDRs ranging from 0.00015 to 0.052. These expression patterns are consistent with the presence of WGS-identified variants and support the hypothesis that dysregulation of these lncRNAs may contribute to impaired spermatogenesis, potentially through pathways involving mitochondrial homeostasis and function.

Table 7.
Differentially expressed lncRNAs harboring infertility-specific variants identified through whole-genome sequencing (WGS). The table presents lncRNAs significantly deregulated between testicular samples with “High Presence” and “No Presence” of spermatozoa (|log2 fold change| > 1, FDR < 0.05), along with the type(s) of spermatogenic defect in which each variant was detected.
In the comparison between the “High Presence” and “Rare Presence” groups, a total of 4091 genes were significantly downregulated, and 9674 were upregulated in the “High Presence” group (|log2 fold change| > 1, FDR < 0.05), as shown in Figure 2. Among these, 347 downregulated and 2331 upregulated transcripts were identified as long non-coding RNAs (lncRNAs).
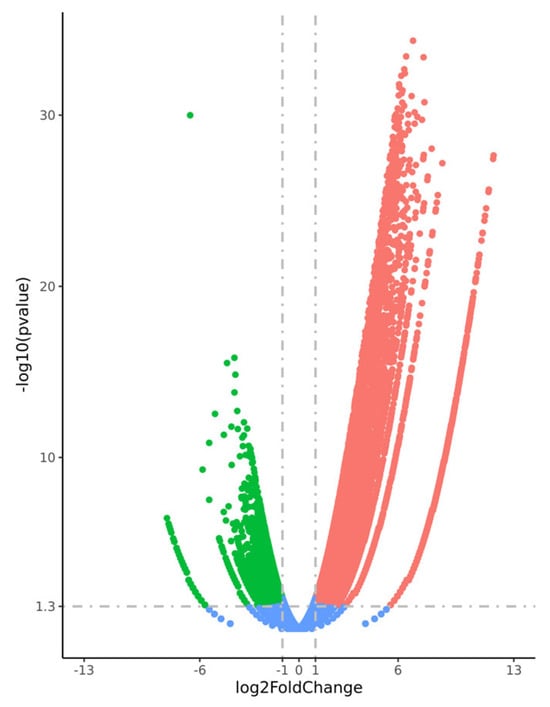
Figure 2.
Volcano plot illustrating the differential gene expression between testicular samples with “High Presence” and “Rare Presence” of spermatozoa. Significantly upregulated genes are highlighted in red, while downregulated genes are shown in green. Genes with no significant differential expression are depicted in blue. The x-axis represents the log2 fold change, and the y-axis represents the –log10 of the adjusted p-value (FDR).
Among the lncRNAs identified with infertility-specific variants through WGS, several showed significant transcriptional deregulation in this comparison (Table 8). For example, MEG3 and GAS5 were significantly downregulated in the “High Presence” group, whereas HOTAIR was found to be upregulated.

Table 8.
Differentially expressed lncRNAs harboring infertility-specific variants identified through whole-genome sequencing (WGS). The table presents lncRNAs significantly deregulated between testicular samples with “High Presence” and “Rare Presence” of spermatozoa (|log2 fold change| > 1, FDR < 0.05), along with the type(s) of spermatogenic defect in which each variant was detected.
Finally, in the comparison between the “Rare Presence” and “No Presence” groups, 6524 genes were significantly upregulated, and 2721 were downregulated (Figure 3). Among these, 878 upregulated and 344 downregulated transcripts were identified as long non-coding RNAs (lncRNAs).
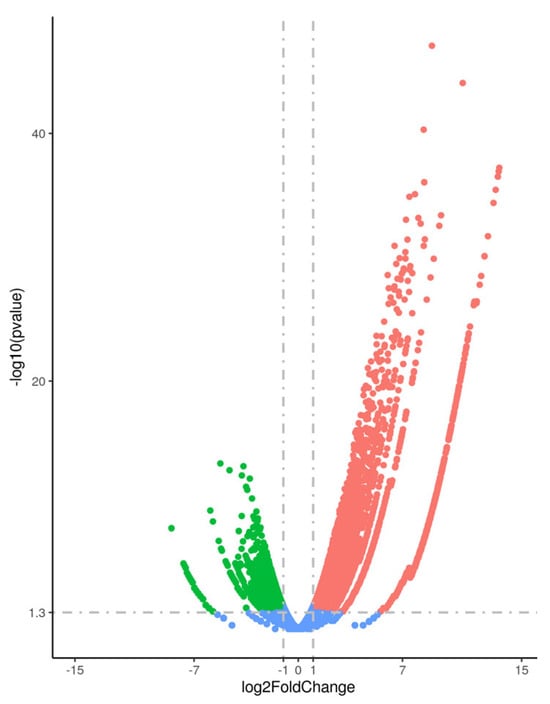
Figure 3.
Volcano plot illustrating the differential gene expression between testicular samples with “Rare Presence” and “No Presence” of spermatozoa. Significantly upregulated genes are highlighted in red, while downregulated genes are shown in green. Genes with no significant differential expression are depicted in blue. The x-axis represents the log2 fold change, and the y-axis represents the –log10 of the adjusted p-value (FDR).
In this comparison, several lncRNAs identified with infertility-specific variants through WGS also exhibited differential expression in the RNA-seq dataset. As shown in Table 9, PVT1 was downregulated, while HOTAIR and CDKN2B-AS1 were upregulated.

Table 9.
Differentially expressed lncRNAs harboring infertility-specific variants identified through whole-genome sequencing (WGS). The table presents lncRNAs significantly deregulated between testicular samples with “Rare Presence” and “No Presence” of spermatozoa (|log2 fold change| > 1, FDR < 0.05), along with the type(s) of spermatogenic defect in which each variant was detected.
4. Discussion
In this study, we investigated the potential role of ncRNAs harboring infertility-specific variants in the context of mitochondrial dynamics and male infertility. By integrating whole-genome sequencing (WGS) with testicular RNA-seq data, we identified a subset of lncRNAs with rare variants uniquely found in infertile men, several of which also exhibited significant transcriptional deregulation in azoospermic samples. Using bioinformatics tools, we further explored their functional relevance by examining lncRNA-miRNA-mRNA interaction networks related to mitochondrial regulation. This dual-layered analysis—combining genomic variation with expression profiling—suggests that rare variants in lncRNA loci may disrupt regulatory networks affecting mitochondrial pathways essential for germ cell development. Overall, our findings provide a novel framework for understanding how non-coding genomic variation may intersect with mitochondrial biology to influence male reproductive capacity, highlighting a potential mechanistic link between lncRNA dysregulation, mitochondrial dysfunction, and impaired spermatogenesis.
More specifically, among the lncRNAs identified in this study, several have previously been associated with oxidative stress, apoptosis, or mitochondrial regulation in other biological systems. MEG3, an lncRNA significantly downregulated in our study (“High presence” vs. “No presence” and “High presence” vs. “Rare presence”) and harboring unique variants in all subcategories of infertile men (asthenozoospermia, oligozoospermia, teratozoospermia), has been shown to modulate mitochondria-dependent apoptotic pathways by interacting with p53 and regulating Bcl-2 family proteins [43]. A recent study also demonstrates that MEG3 is differentially expressed in NOA testicular tissue, where it promotes apoptosis and autophagy while inhibiting germ cell proliferation [44]. Mechanistically, MEG3 acts as a competing endogenous RNA (ceRNA) for miR-21, thereby modulating the SPRY1/ERK/mTOR signaling pathway and forming a regulatory feedback loop involving NF-κB [44]. These findings, supported by in vitro and in vivo functional assays, align with our observation that MEG3 harbors infertility-specific variants and is transcriptionally deregulated in infertile men, reinforcing its potential role in the pathogenesis of male infertility through mitochondrial and stress-response pathways.
Similarly, one of the lncRNAs identified in our study, GAS5, emerges as a multifaceted regulator. Originally characterized as a glucocorticoid receptor decoy that sensitizes cells to apoptosis and is found to accumulate in growth-arrested cells [45,46], GAS5 is now also recognized for its involvement in metabolic and oxidative stress pathways. A recent study has shown that GAS5 translocates to the mitochondria under energy stress, where it modulates the tricarboxylic acid (TCA) cycle flux by disrupting the physical association of key metabolic enzymes—fumarate hydratase (FH), malate dehydrogenase (MDH2), and citrate synthase (CS) [47]. This disruption contributes to altered mitochondrial energy output and is associated with improved survival in certain cancers, suggesting a tumor-suppressive, energy-stabilizing role. Furthermore, GAS5 has been indicated to function as a tumor-inhibitor lncRNA due to its role in stimulating growth arrest and apoptosis [48]. More specifically, GAS5 acts as a competing endogenous RNA (ceRNA), particularly by sequestering miR-21, a microRNA with broad anti-apoptotic and pro-proliferative effects, further linking GAS5 activity to mitochondrial apoptosis and indicating its association with several human diseases [49]. In reproductive biology, GAS5 has been identified in placental tissues, where its expression inversely correlates with maternal stress [50] and appears dysregulated in preeclampsia [51], suggesting broader roles in reproductive health and fetal development. Additionally, GAS5 was found to be overexpressed in patients with recurrent pregnancy loss and positively correlated with the protein levels of TNF-α in plasma and trophoblasts [52]. In our study, GAS5 was significantly downregulated (“High presence” vs. “No presence” and “High presence” vs. “Rare presence”), and unique variants in the GAS5 locus were found in asthenozoospermic men, suggesting a potential disruption in its regulatory function. Given its mitochondrial localization and its known involvement in energy metabolism and apoptosis, GAS5 deregulation may represent a mechanistic link between mitochondrial dysfunction and defective spermatogenesis, contributing to male infertility.
Another lncRNA of interest emerging from our analysis is HOTAIR (HOX transcript antisense RNA), a well-characterized epigenetic regulator with established roles in chromatin remodeling and transcriptional silencing [53]. Originally described in the context of HOX gene regulation, HOTAIR is now recognized as a significant cancer-related lncRNA, affecting a wide range of cellular functions, including apoptosis, stemness, self-renewal, proliferation, DNA repair, and epithelial–mesenchymal transition (EMT) [54,55]. Beyond its role in chromatin dynamics, HOTAIR has also been implicated in regulating mitochondrial function and oxidative stress [56]. Recent studies indicate that the deregulation of HOTAIR alters mitochondrial membrane potential and ROS production [57]. Additionally, research shows that HOTAIR enhances ROS generation by inhibiting the Nrf2 pathway, which plays a crucial role in cellular defense against oxidative stress [58]. In the context of reproduction, HOTAIR expression has been detected in both male and female gonadal tissues [59,60,61]. Specifically, it was reported that its expression was lower in asthenozoospermic and oligoasthenozoospermic men and was associated with ROS-related defects in sperm function [62]. Furthermore, another study demonstrated that low HOTAIR expression resulted in elevated malondialdehyde levels, DNA damage, and reduced sperm motility [61]. In our study, HOTAIR was found to be significantly differentially expressed in azoospermic testicular samples (upregulated in “High Presence” vs. “No Presence”, “High Presence” vs. “Rare Presence” and “Rare Presence” vs. “No Presence”), and we also identified infertility-specific genetic variants in its genomic region, pointing toward a possible regulatory disturbance. Given HOTAIR’s known involvement in epigenetic silencing, mitochondrial homeostasis, and apoptosis suppression, its deregulation may contribute to an altered testicular microenvironment, inhibiting germ cell development or promoting Sertoli cell dysfunction.
Another notable lncRNA from our analysis is PVT1 (plasmacytoma variant translocation 1). Initially characterized as an oncogene, PVT1 influences cell proliferation, autophagy, and apoptosis across multiple tissues [63]. However, several studies also demonstrate its association with mitochondrial dynamics and oxidative stress. Notably, single-cell transcriptomic studies in skeletal muscle have shown that PVT1 becomes activated during muscle atrophy, disrupting mitochondrial respiration, altering mitochondrial morphology, and activating both mitophagy and apoptotic pathways in vivo [64]. Furthermore, PVT1 has been linked to mitochondrial dynamics in cardiomyocytes, operating within a regulatory network that includes ZFP36L2, miR-21-5p, and the mitochondrial fission/fusion regulator MARCH5. Through this axis, PVT1 modulates mitochondrial morphology and function, attenuating mitochondrial fusion and fission [65]. Several other studies indicate that PVT1 is a stress-responsive lncRNA [66], but its role in reproduction remains uncharacterized. In our study, PVT1 variants were found in all subtypes of male infertility, and PVT1 was significantly dysregulated in azoospermic testicular samples (downregulated in “High Presence” vs. “No Presence” and “Rare Presence” vs. “No Presence”). Taken together, the documented roles of PVT1 in mitochondrial dynamics, oxidative stress, and autophagy support a model in which PVT1 contributes to disrupted mitochondrial homeostasis in spermatogenic failure and male infertility.
Furthermore, CDKN2B-AS1, also known as ANRIL, is a long antisense transcript that plays recognized roles in cell cycle control, apoptosis, and inflammation and is associated with cancer and several other diseases [67]. It predominantly functions through Polycomb Repressive Complexes (PRC1/2) to silence tumor suppressor genes and by sponging miRNAs, such as miR-126-5p, miR-181a-5p, and miR-15b-5p, thereby modulating apoptosis and EMT programs [68]. Although CDKN2B-AS1 has not been directly linked to mitochondrial pathways, its interactions with apoptotic regulators and inflammatory mediators suggest it may act as a potential upstream modulator of mitochondrial health. In this study, CDKN2B-AS1 variants were found in teratozoospermic men, and its expression was dysregulated in azoospermic testicular tissue (upregulated in “High Presence” vs. “No Presence” and “Rare Presence” vs. “No Presence”). These findings indicate that its disruption may influence mitochondrial-mediated germ cell death and induce inflammation in the testicular microenvironment, potentially contributing to impaired spermatogenesis.
Building on these observations, our findings align with a growing body of literature that implicates mitochondrial dysfunction as a central hallmark of male infertility. Numerous studies have reported altered expression of mitochondrial genes and compromised oxidative phosphorylation in the spermatozoa of infertile men, often associated with elevated levels of reactive oxygen species (ROS), reduced mitochondrial membrane potential, and disrupted energy homeostasis [12,13,14,15,69]. However, despite these associations, the upstream regulatory mechanisms responsible for initiating and sustaining mitochondrial dysfunction in the male reproductive system remain poorly understood. Our study adds new insight by highlighting lncRNA–mitochondrial gene interactions as a possible layer of transcriptional and post-transcriptional regulation affecting mitochondrial integrity in the testis. Specifically, we demonstrate that several lncRNAs harboring infertility-specific variants also exhibit transcriptional dysregulation in azoospermic testicular samples. These lncRNAs are predicted to interact with miRNAs and protein-coding genes involved in mitochondrial function, oxidative stress response, and apoptosis.
Therefore, based on our integrative genomic and transcriptomic analysis, we propose a hypothetical mechanism in which genetic variants in specific lncRNAs, GAS5, HOTAIR, PVT1, MEG3, and CDKN2B-AS1, lead to their transcriptional deregulation and disrupt key regulatory interactions with miRNAs and mitochondrial genes. Under physiological conditions, these lncRNAs play essential roles in maintaining mitochondrial homeostasis by fine-tuning processes such as apoptosis, oxidative stress responses, and metabolic signaling, as explained above. However, variants that affect their expression levels or their structural integrity may compromise their normal function as competing endogenous RNAs (ceRNAs) or molecular scaffolds, thereby altering downstream gene regulation. Consequently, when these lncRNAs are dysregulated, they may collectively contribute to mitochondrial membrane potential loss, increased ROS generation, oxidative damage, defective mitophagy, and metabolic insufficiency. These mitochondrial impairments can trigger apoptosis in germ cells, particularly spermatogonia, or disrupt Sertoli cell support functions, compromising the spermatogenic niche. Ultimately, these events may converge to drive the spermatogenic arrest and testicular failure observed in many forms of male infertility (Figure 4).
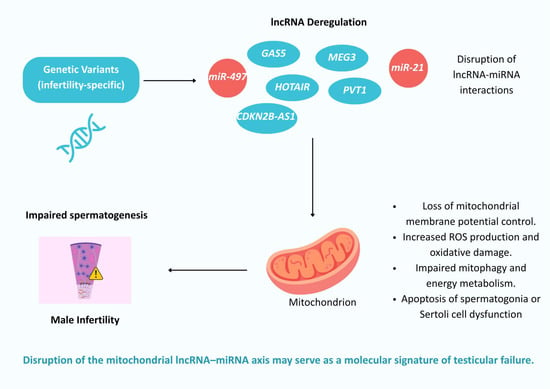
Figure 4.
Hypothetical mechanism linking infertility-specific lncRNA variants to mitochondrial dysfunction and male infertility.
Despite these promising findings, our study is not without limitations. First, functional validation experiments were not conducted, which limits the ability to directly confirm the causal role of the identified lncRNAs in mitochondrial or spermatogenic regulation. Second, the relatively small sample size in both WGS and RNA-seq datasets may limit statistical power and the generalizability of the findings. A limitation of our study is the use of different phenotypic groups in the discovery (oligozoospermia, asthenozoospermia, teratozoospermia) and validation (non-obstructive azoospermia) phases. However, this approach is widely employed in male infertility research, as various subtypes often share common underlying molecular mechanisms and disrupted biological pathways [70,71,72]. Additionally, semen parameters are known to vary significantly over time due to hormonal, environmental, and physiological factors. Men may progress from milder phenotypes, such as oligozoospermia, to more severe conditions like azoospermia [73,74]. This dynamic nature of semen quality complicates strict categorical classification and supports the concept of male infertility as a continuum. Therefore, despite differences in phenotype between the two cohorts, our findings provide valuable insight into shared pathways implicated in male reproductive dysfunction. We acknowledge this limitation and highlight the need for future studies involving matched and longitudinally followed patient cohorts to further strengthen genotype–phenotype correlations. Another limitation of this study is the use of the GRCh37 (hg19) genome assembly. Although this version was selected for compatibility with pre-existing datasets and widely used bioinformatic tools, it offers less comprehensive coverage compared to the more recent GRCh38 (hg38) assembly. Future studies utilizing updated genome assemblies may improve the detection of additional non-coding variants and enhance the overall resolution of the analyses. It should also be noted that all participants in this study were volunteers, contributing to a diverse sample representing various age groups and lifestyle habits. While this diversity enriches the dataset, it may also introduce variability that impacts the findings. Although a significant age difference was observed between the asthenozoospermic and oligozoospermic groups, no significant difference was found between the normozoospermic group and any other infertile group. Since the primary aim of this study was to compare the normozoospermic group with the various infertile groups to identify unique variants, we consider our comparisons robust. Nevertheless, we acknowledge that age remains an important biological factor in the context of male infertility and that future studies should include larger, age-matched cohorts to minimize potential confounding effects. Another important limitation of our study is the use of pooled DNA samples for whole-genome sequencing, which restricts the resolution of individual-level genetic data. Pooling was employed due to logistical and financial constraints, allowing us to include a greater number of participants and explore broader genomic differences between asthenozoospermic and normozoospermic men within the scope of an exploratory study. All samples were pooled in equimolar concentrations to minimize bias and ensure balanced representation. While this approach is widely used in hypothesis-generating studies, particularly when dealing with resource-intensive technologies like WGS, it inherently limits the ability to assess variant frequency within individuals or capture inter-individual biological variability. Larger-scale studies with individual-level sequencing and sufficient statistical power will be essential to validate and expand upon our observations. Finally, while we focused on known lncRNAs with annotated mitochondrial relevance, it is possible that additional uncharacterized or novel lncRNAs contribute to the observed phenotypes but were not captured by our selection criteria.
Despite its limitations, this study also has several strengths. It presents a novel integrative approach combining whole-genome sequencing and transcriptomic profiling of testicular tissue to uncover ncRNA-associated mechanisms contributing to male infertility. By focusing on infertility-specific rare variants located in lncRNAs and validating their transcriptional relevance in azoospermic samples, we provide dual-layered evidence supporting a potential regulatory role of these molecules in mitochondrial dysfunction. Importantly, the fact that these lncRNA variants were found exclusively in infertile men also suggests that they may hold diagnostic potential as biomarkers. Additionally, we applied bioinformatic prioritization and functional annotation strategies to link lncRNA variation with known mitochondrial pathways, offering a new perspective on the genetic and molecular complexity of male infertility. Finally, the inclusion of well-characterized clinical samples further strengthens the biological relevance of our findings.
5. Conclusions
This study provides novel insights into the molecular landscape of male infertility by identifying infertility-specific variants in ncRNAs and linking them to mitochondrial dysfunction and impaired spermatogenesis. Through an integrative analysis of whole-genome sequencing and testicular RNA-seq data, we highlight a subset of lncRNAs—GAS5, HOTAIR, PVT1, MEG3, and CDKN2B-AS1—that are not only genetically altered in infertile men but also transcriptionally deregulated in the testicular tissue of infertile men. Bioinformatic predictions and known interactions further support their roles in maintaining mitochondrial membrane stability, regulating oxidative stress, mitophagy, and apoptosis. These findings propose a hypothetical yet biologically plausible mechanism in which disruption of the lncRNA–miRNA–mitochondria axis contributes to the pathophysiology of male infertility. However, as a preliminary study, it is limited by the absence of functional validation. The findings presented here should be considered hypothesis-generating and serve as a framework for future research. Follow-up experimental studies are necessary to validate the proposed pathways and interactions and to elucidate their biological significance. Such efforts will be critical for translating these preliminary observations into mechanistic understanding and, potentially, clinical applications. Overall, our study underscores the significance of non-coding genomic elements in reproductive biology and provides a new framework for understanding the molecular underpinnings of male infertility through the lens of lncRNA-driven mitochondrial regulation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/dna5030038/s1, Table S1a: For each lncRNA included in the publications by Gusic & Prokisch (2020) [34] and Sun et al. (2022) [35], the gene name provided by Ensembl was recorded, along with the Ensembl ID, which was used to identify variants mapped in these lncRNA genes in the annotation file derived from comparisons between normozoospermic and non-normozoospermic individuals.; Table S1b: For each miRNA included in the publications by Gusic & Prokisch (2020) [34] and Sun et al. (2022) [35], the gene name provided by Ensembl was recorded, along with the Ensembl ID, which was used to identify variants mapped in these miRNA genes in the annotation file derived from comparisons between normozoospermic and non-normozoospermic individuals.; Table S2: Unique variants in lncRNAs associated with mitochondrial dynamics identified in asthenozoospermic men after filtering. Variants shared between two subcategories of male infertility are marked with double asterisks (**), while those common to all three subcategories are marked with triple asterisks (***); Table S3: Unique variants in lncRNAs associated with mitochondrial dynamics identified in oligozoospermic men after filtering. Variants shared between two subcategories of male infertility are marked with double asterisks (**), while those common to all three subcategories are marked with triple asterisks (***); Table S4: Unique variants in lncRNAs associated with mitochondrial dynamics identified in teratozoospermic men after filtering. Variants shared between two subcategories of male infertility are marked with double asterisks (**), while those common to all three subcategories are marked with triple asterisks (***).
Author Contributions
Conceptualization, M.-A.K. and Z.M.; methodology, G.S. and M.-A.K.; validation, G.S. and M.-A.K.; formal analysis, G.S., M.-A.K. and A.K.; investigation, G.S., M.-A.K. and A.K.; data curation, G.S. and M.-A.K.; writing—original draft preparation, G.S. and M.-A.K.; writing—review and editing, A.K. and Z.M.; visualization, G.S. and M.-A.K.; supervision, Z.M.; project administration, Z.M.; funding acquisition, Z.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Spermogene project, which is co-financed by the European Regional Development Fund of the European Union and Greek national funds, through the Operational Program Competitiveness, Entrepreneurship, and Innovation, under the call RESEARCH–CREATE–INNOVATE (grant number Τ1ΕΔK-02787).
Institutional Review Board Statement
This study was conducted in accordance with the Declaration of Helsinki and approved by the ethics committee of the Medical Faculty of the University of Thessaly on 20 April 2016, with approval code 20.04/2016, in response to request number 1, 15 April 2016.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Whole-genome sequencing data of teratozoospermic and normozoospermic men presented in this study are available through SRA (BioProject ID PRJNA875412, http://www.ncbi.nlm.nih.gov/bioproject/875412, accessed on 9 June 2025).
Acknowledgments
The authors wish to thank all the men for their participation in this study.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| lncRNA | Long noncoding RNA |
| ncRNA | Noncoding RNA |
| NOA | Nonobstructive Azoospermia |
| ROS | Reactive Oxygen Species |
| WGS | Whole Genome Sequencing |
| WHO | World Health Organization |
References
- Zegers-Hochschild, F.; Adamson, G.D.; Dyer, S.; Racowsky, C.; de Mouzon, J.; Sokol, R.; Rienzi, L.; Sunde, A.; Schmidt, L.; Cooke, I.D.; et al. The International Glossary on Infertility and Fertility Care, 2017. Fertil. Steril. 2017, 108, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Schlegel, P.N.; Sigman, M.; Collura, B.; De Jonge, C.J.; Eisenberg, M.L.; Lamb, D.J.; Mulhall, J.P.; Niederberger, C.; Sandlow, J.I.; Sokol, R.Z.; et al. Diagnosis and Treatment of Infertility in Men: AUA/ASRM Guideline Part I. Fertil. Steril. 2021, 115, 54–61. [Google Scholar] [CrossRef]
- Agarwal, A.; Baskaran, S.; Parekh, N.; Cho, C.L.; Henkel, R.; Vij, S.; Arafa, M.; Panner Selvam, M.K.; Shah, R. Male Infertility. Lancet 2021, 397, 319–333. [Google Scholar] [CrossRef]
- Kothandaraman, N.; Agarwal, A.; Abu-Elmagd, M.; Al-Qahtani, M.H. Pathogenic Landscape of Idiopathic Male Infertility: New Insight towards Its Regulatory Networks. npj Genom. Med. 2016, 1, 16023. [Google Scholar] [CrossRef]
- Boeri, L.; Kandil, H.; Ramsay, J. Idiopathic Male Infertility—What Are We Missing? Arab J. Urol. 2024, 23, 215–229. [Google Scholar] [CrossRef]
- Lv, M.Q.; Ge, P.; Zhang, J.; Yang, Y.Q.; Zhou, L.; Zhou, D.X. Temporal Trends in Semen Concentration and Count among 327 373 Chinese Healthy Men from 1981 to 2019: A Systematic Review. Hum. Reprod. 2021, 36, 1751–1775. [Google Scholar] [CrossRef]
- Levine, H.; Jørgensen, N.; Martino-Andrade, A.; Mendiola, J.; Weksler-Derri, D.; Jolles, M.; Pinotti, R.; Swan, S.H. Temporal Trends in Sperm Count: A Systematic Review and Meta-Regression Analysis of Samples Collected Globally in the 20th and 21st Centuries. Hum. Reprod. Update 2023, 29, 157–176. [Google Scholar] [CrossRef]
- Slade, P.; O’Neill, C.; Simpson, A.J.; Lashen, H. The Relationship between Perceived Stigma, Disclosure Patterns, Support and Distress in New Attendees at an Infertility Clinic. Hum. Reprod. 2007, 22, 2309–2317. [Google Scholar] [CrossRef]
- Wu, A.K.; Odisho, A.Y.; Washington, S.L.; Katz, P.P.; Smith, J.F. Out-of-Pocket Fertility Patient Expense: Data from a Multicenter Prospective Infertility Cohort. J. Urol. 2014, 191, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Moraes, C.R.; Meyers, S. The Sperm Mitochondrion: Organelle of Many Functions. Anim. Reprod. Sci. 2018, 194, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Vertika, S.; Singh, K.K.; Rajender, S. Mitochondria, Spermatogenesis, and Male Infertility—An Update. Mitochondrion 2020, 54, 26–40. [Google Scholar] [CrossRef] [PubMed]
- Shamsi, M.B.; Kumar, R.; Bhatt, A.; Bamezai, R.N.K.; Kumar, R.; Gupta, N.P.; Das, T.K.; Dada, R. Mitochondrial DNA Mutations in Etiopathogenesis of Male Infertility. Indian J. Urol. 2008, 24, 150–154. [Google Scholar] [CrossRef]
- Amor, H.; Hammadeh, M.E. A Systematic Review of the Impact of Mitochondrial Variations on Male Infertility. Genes 2022, 13, 1182. [Google Scholar] [CrossRef]
- Hermann Ayekoue, J.E.; Sylvère N’Zi, K.G.; Berenger Ako, A.A.; N’Guessan, M.F.; Guillaume Yayé, Y.; Amadou Coulibaly, F.; Joseph Djaman, A. Polymorphism of Mitochondrial DNA Genes Involved in Asthenozoospermia in Infertile Patients of Côte d’Ivoire. Reprod. Dev. Med. 2023, 7, 38–43. [Google Scholar] [CrossRef]
- Kyrgiafini, M.A.; Giannoulis, T.; Chatziparasidou, A.; Mamuris, Z. Elucidating the Role of OXPHOS Variants in Asthenozoospermia: Insights from Whole Genome Sequencing and an In Silico Analysis. Int. J. Mol. Sci. 2024, 25, 4121. [Google Scholar] [CrossRef]
- Boguenet, M.; Bouet, P.E.; Spiers, A.; Reynier, P.; May-Panloup, P. Mitochondria: Their Role in Spermatozoa and in Male Infertility. Hum. Reprod. Update 2021, 27, 697–719. [Google Scholar] [CrossRef]
- Takeshima, T.; Usui, K.; Mori, K.; Asai, T.; Yasuda, K.; Kuroda, S.; Yumura, Y. Oxidative Stress and Male Infertility. Reprod. Med. Biol. 2020, 20, 41. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Koli, S.; Reddy, K.V.R. Regulatory Non-Coding Transcripts in Spermatogenesis: Shedding Light on “Dark Matter”. Andrology 2014, 2, 360–369. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Wang, K.; Gao, Y.; Wang, C.; Li, L.; Liao, Y.; Hu, K.; Liang, M. Roles of Noncoding RNA in Reproduction. Front. Genet. 2021, 12, 2513. [Google Scholar] [CrossRef]
- Aliakbari, F.; Eshghifar, N.; Mirfakhraie, R.; Pourghorban, P.; Azizi, F. Coding and Non-Coding RNAs, as Male Fertility and Infertility Biomarkers. Int. J. Fertil. Steril. 2021, 15, 158. [Google Scholar] [CrossRef]
- Kyrgiafini, M.A.; Sarafidou, T.; Mamuris, Z. The Role of Long Noncoding RNAs on Male Infertility: A Systematic Review and In Silico Analysis. Biology 2022, 11, 1510. [Google Scholar] [CrossRef] [PubMed]
- Kyrgiafini, M.A.; Mamuris, Z. Circular RNAs and Their Role in Male Infertility: A Systematic Review. Biomolecules 2023, 13, 1046. [Google Scholar] [CrossRef]
- Shi, Z.; Yu, M.; Guo, T.; Sui, Y.; Tian, Z.; Ni, X.; Chen, X.; Jiang, M.; Jiang, J.; Lu, Y.; et al. MicroRNAs in Spermatogenesis Dysfunction and Male Infertility: Clinical Phenotypes, Mechanisms and Potential Diagnostic Biomarkers. Front. Endocrinol. 2024, 15, 1293368. [Google Scholar] [CrossRef]
- Salas-Huetos, A.; James, E.R.; Aston, K.I.; Carrell, D.T.; Jenkins, T.G.; Yeste, M. The Role of MiRNAs in Male Human Reproduction: A Systematic Review. Andrology 2020, 8, 7–26. [Google Scholar] [CrossRef]
- Yu, P.; Zhao, X.; Zhou, D.; Wang, S.; Hu, Z.; Lian, K.; Zhang, N.; Duan, P.; Yu, P.; Zhao, X.; et al. The MicroRNA-Mediated Apoptotic Signaling Axis in Male Reproduction: A Possible and Targetable Culprit in Male Infertility. Cell Biol. Toxicol. 2025, 41, 54. [Google Scholar] [CrossRef]
- Du, L.; Chen, W.; Zhang, D.; Cui, Y.; He, Z. The Functions and Mechanisms of PiRNAs in Mediating Mammalian Spermatogenesis and Their Applications in Reproductive Medicine. Cell Mol. Life Sci. 2024, 81, 379. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 9 July 2025).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Dyer, S.C.; Austine-Orimoloye, O.; Azov, A.G.; Barba, M.; Barnes, I.; Barrera-Enriquez, V.P.; Becker, A.; Bennett, R.; Beracochea, M.; Berry, A.; et al. Ensembl 2025. Nucleic Acids Res. 2025, 53, D948–D957. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Garrison, E.; Marth, G. Haplotype-Based Variant Detection from Short-Read Sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- Gusic, M.; Prokisch, H. NcRNAs: New Players in Mitochondrial Health and Disease? Front. Genet. 2020, 11, 95. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Lu, Y.; Zhang, H.; Zhang, J.; Fang, X.; Wang, J.; Li, M. Mitochondrial Non-Coding RNAs Are Potential Mediators of Mitochondrial Homeostasis. Biomolecules 2022, 12, 1863. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinforma. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef]
- Auton, A.; Abecasis, G.R.; Altshuler, D.M.; Durbin, R.M.; Bentley, D.R.; Chakravarti, A.; Clark, A.G.; Donnelly, P.; Eichler, E.E.; Flicek, P.; et al. A Global Reference for Human Genetic Variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]
- Pierre, A.S.; Génin, E. How Important Are Rare Variants in Common Disease? Brief. Funct. Genom. 2014, 13, 353–361. [Google Scholar] [CrossRef]
- Rentzsch, P.; Witten, D.; Cooper, G.M.; Shendure, J.; Kircher, M. CADD: Predicting the Deleteriousness of Variants throughout the Human Genome. Nucleic Acids Res. 2019, 47, D886–D894. [Google Scholar] [CrossRef] [PubMed]
- Oscanoa, J.; Sivapalan, L.; Gadaleta, E.; Dayem Ullah, A.Z.; Lemoine, N.R.; Chelala, C. SNPnexus: A Web Server for Functional Annotation of Human Genome Sequence Variation (2020 Update). Nucleic Acids Res. 2020, 48, W185–W192. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.R.; Liu, W.; Zhang, Q.; Guo, A.Y. LncRNASNP2: An Updated Database of Functional SNPs and Mutations in Human and Mouse LncRNAs. Nucleic Acids Res. 2018, 46, D276–D280. [Google Scholar] [CrossRef]
- Chatziparasidou, A.; Sarafidou, T.; Kyrgiafini, M.-A.; Moutou, K.; Markantoni, M.; Giannoulis, T.; Papatheodorou, A.; Oraiopoulou, C.; Samolada, G.; Christoforidis, N.; et al. Unraveling the Genetic Basis of Azoospermia: Transcriptome Profiling Analyses in a Greek Population. FS Sci. 2024, 6, 16–29. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhong, Y.; Wang, Y.; Zhang, X.; Batista, D.L.; Gejman, R.; Ansell, P.J.; Zhao, J.; Weng, C.; Klibanski, A. Activation of P53 by MEG3 Non-Coding RNA. J. Biol. Chem. 2007, 282, 24731–24742. [Google Scholar] [CrossRef]
- Fang, X.; Lu, X.; Ma, Y.; Sun, N.; Jiao, Y.; Meng, H.; Song, M.; Jin, H.; Yao, G.; Song, N.; et al. Possible Involvement of a MEG3-MiR-21-SPRY1-NF-ΚB Feedback Loop in Spermatogenic Cells Proliferation, Autophagy, and Apoptosis. iScience 2024, 27, 110904. [Google Scholar] [CrossRef]
- Kino, T.; Hurt, D.E.; Ichijo, T.; Nader, N.; Chrousos, G.P. Noncoding RNA Gas5 Is a Growth Arrest- and Starvation-Associated Repressor of the Glucocorticoid Receptor. Sci. Signal. 2010, 3, ra8. [Google Scholar] [CrossRef]
- Garabedian, M.J.; Logan, S.K. Glucocorticoid Receptor DNA Binding Decoy Is a Gas. Sci. Signal. 2010, 3, pe5. [Google Scholar] [CrossRef] [PubMed]
- Sang, L.; Ju, H.-Q.; Yang, Z.; Ge, Q.; Zhang, Z.; Liu, F.; Yang, L.; Gong, H.; Shi, C.; Qu, L.; et al. Mitochondrial Long Non-Coding RNA GAS5 Tunes TCA Metabolism in Response to Nutrient Stress. Nat. Metab. 2021, 3, 90–106. [Google Scholar] [CrossRef] [PubMed]
- Mourtada-Maarabouni, M.; Pickard, M.R.; Hedge, V.L.; Farzaneh, F.; Williams, G.T. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 2009, 28, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.N.T.; Pyburn, J.S.; Nguyen, N.L.; Schank, M.B.; Zhao, J.; Wang, L.; Leshaodo, T.O.; El Gazzar, M.; Moorman, J.P.; Yao, Z.Q. Epigenetic Regulation by LncRNA GAS5/MiRNA/MRNA Network in Human Diseases. Int. J. Mol. Sci. 2025, 26, 1377. [Google Scholar] [CrossRef]
- Mparmpakas, D.; Zachariades, E.; Sotiriadis, G.; Goumenou, A.; Harvey, A.J.; Gidron, Y.; Karteris, E. Differential Expression of Placental Glucocorticoid Receptors and Growth Arrest-Specific Transcript 5 in Term and Preterm Pregnancies: Evidence for Involvement of Maternal Stress. Obstet. Gynecol. Int. 2014, 2014, 239278. [Google Scholar] [CrossRef]
- Zheng, D.; Hou, Y.; Li, Y.; Bian, Y.; Khan, M.; Li, F.; Huang, L.; Qiao, C. Long Non-Coding RNA Gas5 Is Associated With Preeclampsia and Regulates Biological Behaviors of Trophoblast via MicroRNA-21. Front. Genet. 2020, 11, 188. [Google Scholar] [CrossRef]
- Wang, M.M.; Zhong, J.X.; Xiang, Y.Y. LncRNA-GAS5 Related to the Processes of Recurrent Pregnancy Loss by Regulating Th1/Th2 Balance. Kaohsiung J. Med. Sci. 2021, 37, 479. [Google Scholar] [CrossRef] [PubMed]
- Bhan, A.; Mandal, S.S. LncRNA HOTAIR: A Master Regulator of Chromatin Dynamics and Cancer. Biochim. Biophys. Acta 2015, 1856, 151. [Google Scholar] [CrossRef]
- Zhu, C.; Wang, X.; Wang, Y.; Wang, K. Functions and Underlying Mechanisms of LncRNA HOTAIR in Cancer Chemotherapy Resistance. Cell Death Discov. 2022, 8, 383. [Google Scholar] [CrossRef]
- Hajjari, M.; Salavaty, A. HOTAIR: An Oncogenic Long Non-Coding RNA in Different Cancers. Cancer Biol. Med. 2015, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, M.; Chen, W.; Wang, R.; Ye, Z.; Wang, Y.; Li, X.; Cai, C. LncRNA-HOTAIR Inhibition Aggravates Oxidative Stress-Induced H9c2 Cells Injury through Suppression of MMP2 by MiR-125. Acta Biochim. Biophys. Sin. 2018, 50, 996–1006. [Google Scholar] [CrossRef]
- Kong, L.; Zhou, X.; Wu, Y.; Wang, Y.; Chen, L.; Li, P.; Liu, S.; Sun, S.; Ren, Y.; Mei, M.; et al. Targeting HOTAIR Induces Mitochondria Related Apoptosis and Inhibits Tumor Growth in Head and Neck Squamous Cell Carcinoma in Vitro and in Vivo. Curr. Mol. Med. 2015, 15, 952–960. [Google Scholar] [CrossRef]
- You, H.; Li, H.; Gou, W. LncRNA HOTAIR Promotes ROS Generation and NLRP3 Inflammasome Activation by Inhibiting Nrf2 in Diabetic Retinopathy. Medicine 2023, 102, E35155. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Wei, L.; Qian, F.; Bo, L.; Gao, S.; Yang, G.; Mao, C. LncRNA HOTAIR Regulates Autophagy and Proliferation Mechanisms in Premature Ovarian Insufficiency through the MiR-148b-3p/ATG14 Axis. Cell Death Discov. 2024, 10, 44. [Google Scholar] [CrossRef]
- Banikazemi, Z.; Heidar, Z.; Rezaee, A.; Taghavi, S.P.; Zadeh Modarres, S.; Asemi, Z.; Goleij, P.; Jahed, F.; Mazaheri, E.; Taghizadeh, M. Long Non-Coding RNAs and Female Infertility: What Do We Know? Pathol. Res. Pract. 2023, 250, 154814. [Google Scholar] [CrossRef]
- Asl, A.J.; Sharifi, M.; Dashti, A.; Reza Dashti, G. Relationship between Long Non-Coding RNA MALAT1 and HOTAIR Expression with Sperm Parameters, DNA and Malondialdehyde Levels in Male Infertility. Tissue Cell 2023, 85, 102248. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, Z.; Li, X.; Zhang, P.; Wang, J.; Zhu, D.; Chen, X.; Ye, L. Low Long Non-Coding RNA HOTAIR Expression Is Associated with down-Regulation of Nrf2 in the Spermatozoa of Patients with Asthenozoospermia or Oligoasthenozoospermia. Int. J. Clin. Exp. Pathol. 2015, 8, 14198–14205. [Google Scholar]
- Li, R.; Wang, X.; Zhu, C.; Wang, K. LncRNA PVT1: A Novel Oncogene in Multiple Cancers. Cell Mol. Biol. Lett. 2022, 27, 84. [Google Scholar] [CrossRef]
- Alessio, E.; Buson, L.; Chemello, F.; Peggion, C.; Grespi, F.; Martini, P.; Massimino, M.L.; Pacchioni, B.; Millino, C.; Romualdi, C.; et al. Single Cell Analysis Reveals the Involvement of the Long Non-Coding RNA Pvt1 in the Modulation of Muscle Atrophy and Mitochondrial Network. Nucleic Acids Res. 2019, 47, 1653–1670. [Google Scholar] [CrossRef]
- Wu, F.; Huang, W.; Tan, Q.; Guo, Y.; Cao, Y.; Shang, J.; Ping, F.; Wang, W.; Li, Y. ZFP36L2 Regulates Myocardial Ischemia/Reperfusion Injury and Attenuates Mitochondrial Fusion and Fission by LncRNA PVT1. Cell Death Dis. 2021, 12, 614. [Google Scholar] [CrossRef]
- Tabury, K.; Monavarian, M.; Listik, E.; Shelton, A.K.; Choi, A.S.; Quintens, R.; Arend, R.C.; Hempel, N.; Ryan Miller, C.; Gyorrfy, B.; et al. PVT1 Is a Stress-Responsive LncRNA That Drives Ovarian Cancer Metastasis and Chemoresistance. Life Sci. Alliance 2022, 5, e202201370. [Google Scholar] [CrossRef]
- Song, C.; Qi, Y.; Zhang, J.; Guo, C.; Yuan, C. CDKN2B-AS1: An Indispensable Long Non-Coding RNA in Multiple Diseases. Curr. Pharm. Des. 2020, 26, 5335–5346. [Google Scholar] [CrossRef] [PubMed]
- Hjazi, A.; Ghaffar, E.; Asghar, W.; Khalaf, H.A.; Ullah, M.I.; Romero-Parra, R.M.; Hussien, B.M.; Alazbjee, A.A.A.; Bisht, Y.S.; Mustafa, Y.F.; et al. CDKN2B-AS1 as a Novel Therapeutic Target in Cancer: Mechanism and Clinical Perspective. Biochem. Pharmacol. 2023, 213, 115627. [Google Scholar] [CrossRef] [PubMed]
- Vahedi Raad, M.; Firouzabadi, A.M.; Tofighi Niaki, M.; Henkel, R.; Fesahat, F. The Impact of Mitochondrial Impairments on Sperm Function and Male Fertility: A Systematic Review. Reprod. Biol. Endocrinol. 2024, 22, 83. [Google Scholar] [CrossRef] [PubMed]
- Razavi, S.M.; Sabbaghian, M.; Jalili, M.; Divsalar, A.; Wolkenhauer, O.; Salehzadeh-Yazdi, A. Comprehensive Functional Enrichment Analysis of Male Infertility. Sci. Rep. 2017, 7, 15778. [Google Scholar] [CrossRef]
- Chatziparasidou, A.; Kyrgiafini, M.A.; Sarafidou, T.; Moutou, K.A.; Mamuris, Z. Genetic Insights into Azoospermia and Severe Oligozoospermia: Discovering Seven SNPs through GWAS and In Silico Analysis. Curr. Issues Mol. Biol. 2024, 46, 6522–6532. [Google Scholar] [CrossRef] [PubMed]
- Salman, A.; Radwan, A.F.; Shaker, O.G.; Adel, A.; Sayed, G.A. A Comparison of the Expression Patterns and Diagnostic Capability of the NcRNAs NEAT1 and MiR-34a in Non-Obstructive Azoospermia and Severe Oligospermia. Hum. Genom. 2025, 19, 35. [Google Scholar] [CrossRef] [PubMed]
- Bak, C.W.; Song, S.H.; Yoon, T.K.; Lim, J.J.; Shin, T.E.; Sung, S. Natural Course of Idiopathic Oligozoospermia: Comparison of Mild, Moderate and Severe Forms. Int. J. Urol. 2010, 17, 937–943. [Google Scholar] [CrossRef] [PubMed]
- Scott, M.; Rodriguez, A.; Marciano, O.; Nordgren, R.; Lundy, S.D.; Raheem, O.A. A 10-Year Longitudinal Analysis of the Impact of Demographic, Lifestyle, and Medical Factors on Semen Qualities in Men in a City in the Midwestern Region of the United States of America. Asian J. Androl. 2025, 27, 464–469. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).