DNA Replication in Time and Space: The Archaeal Dimension
Abstract
1. Introduction: The Where, When, and How of DNA Replication
1.1. The DNA Replication Problem
1.2. The Polymerase Puzzle
“exact copies can be made from it so that this information can be used again and elsewhere in time and space.”
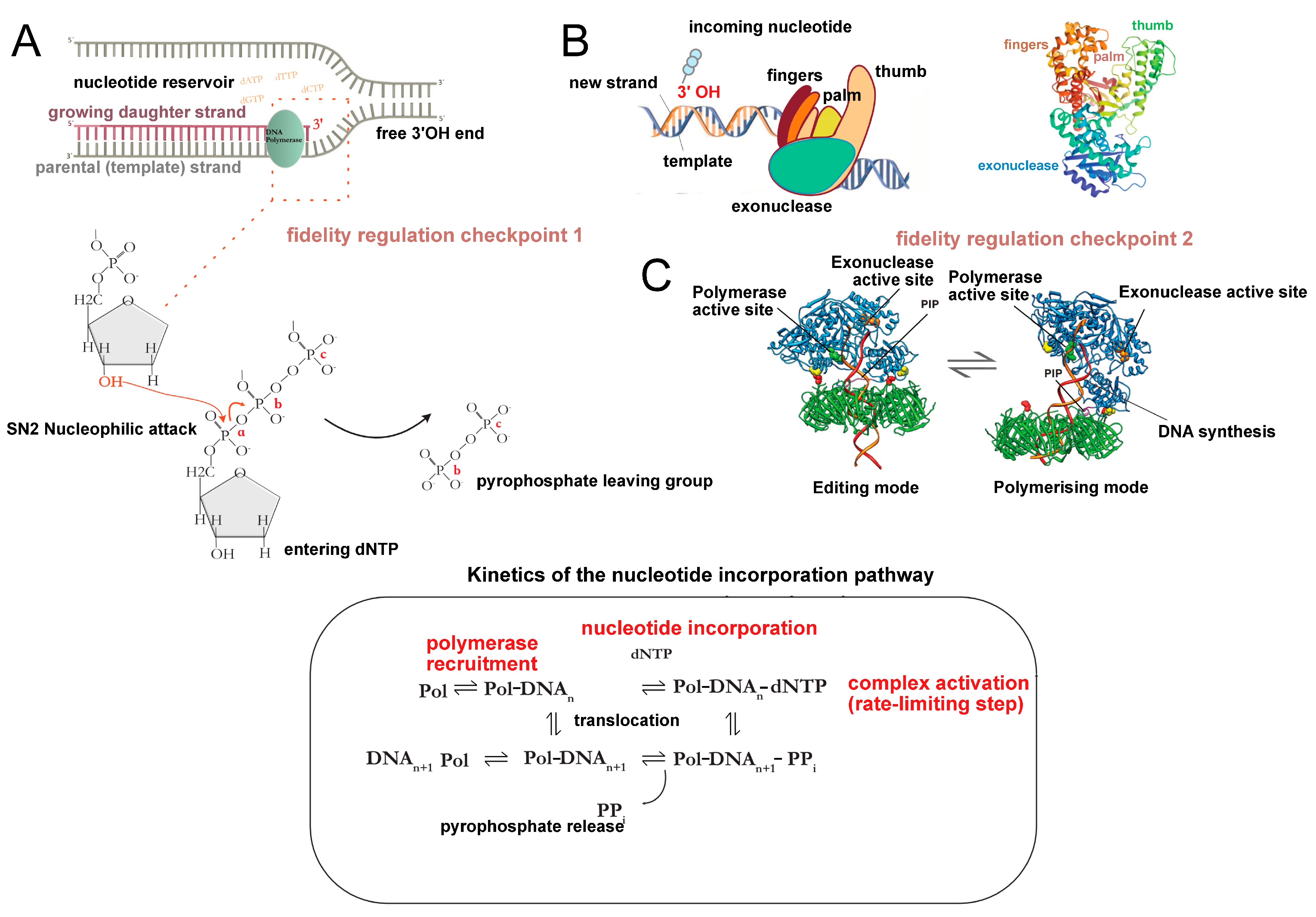
2. The Replicon Model: Leading Paradigm for the Study of DNA Replication
“we still know very little about the general system which integrates cellular controls, the regulation of DNA replication, the formation of bacterial membrane, and the process of cellular division with its equipartition of the DNA copies”
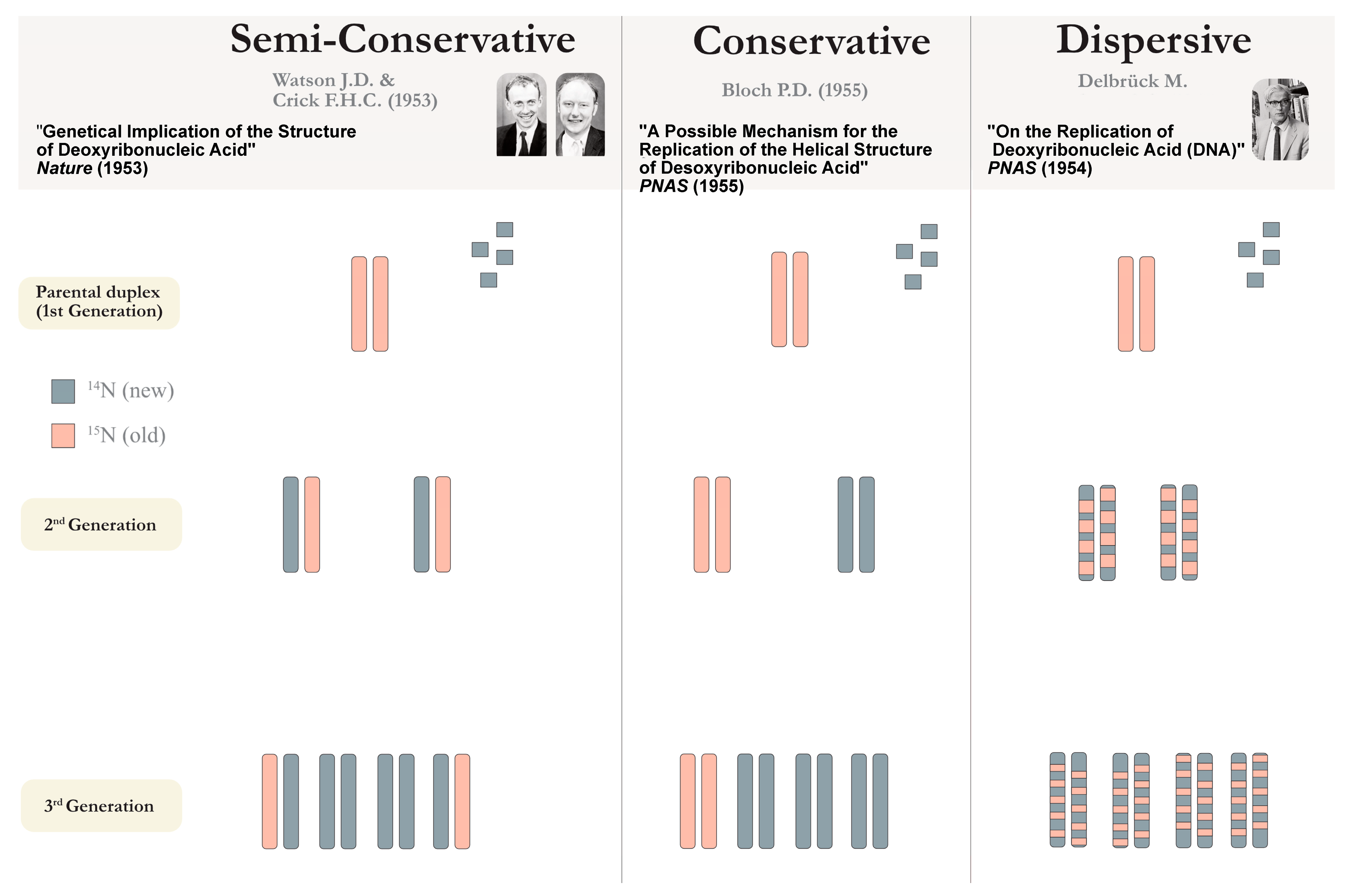
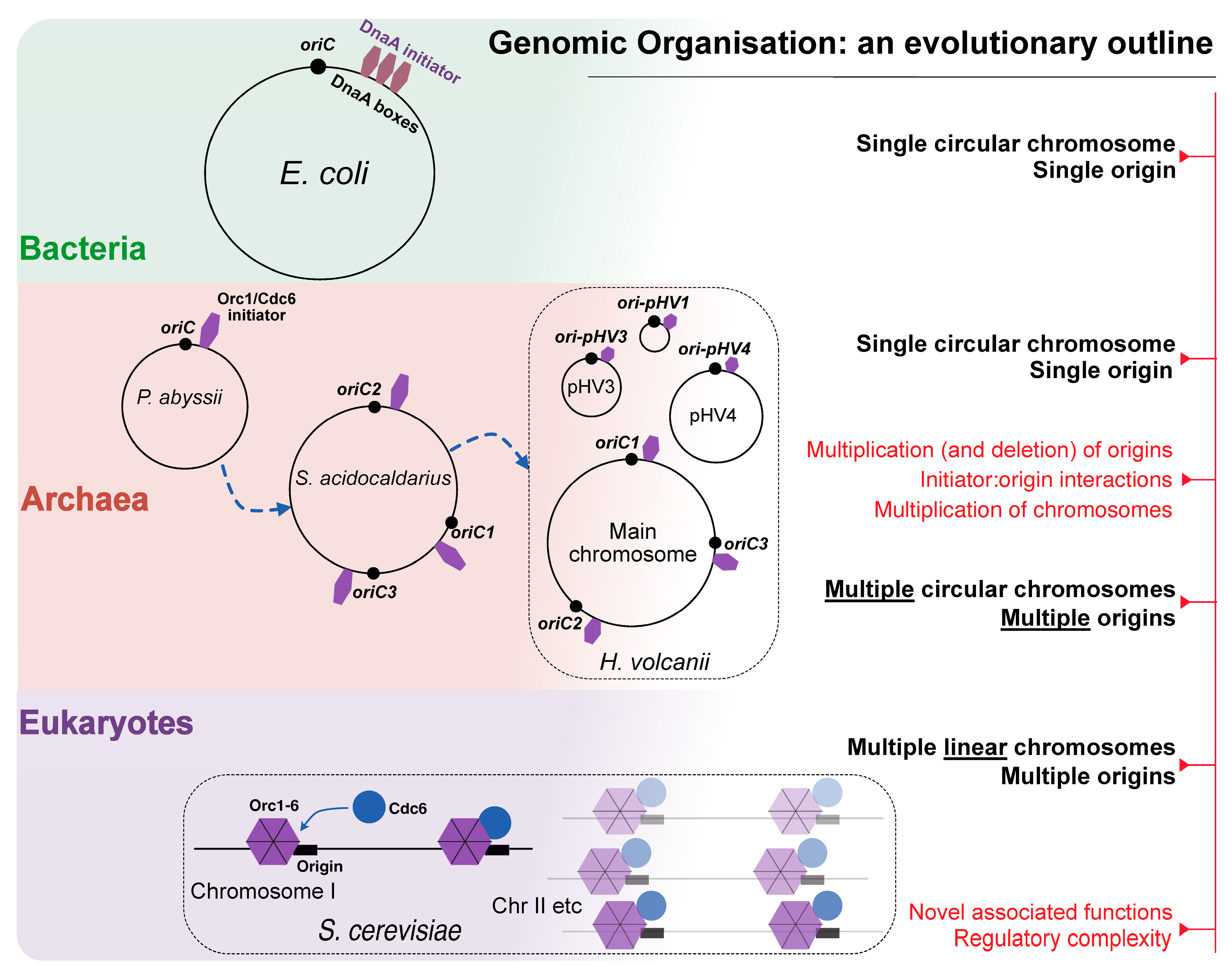
3. The Divided Genome: Nature’s Riddle
3.1. The Diversity of Replication Factors
3.2. Many Origins, One Chromosome: Time to Revisit the Single Replicon Model?
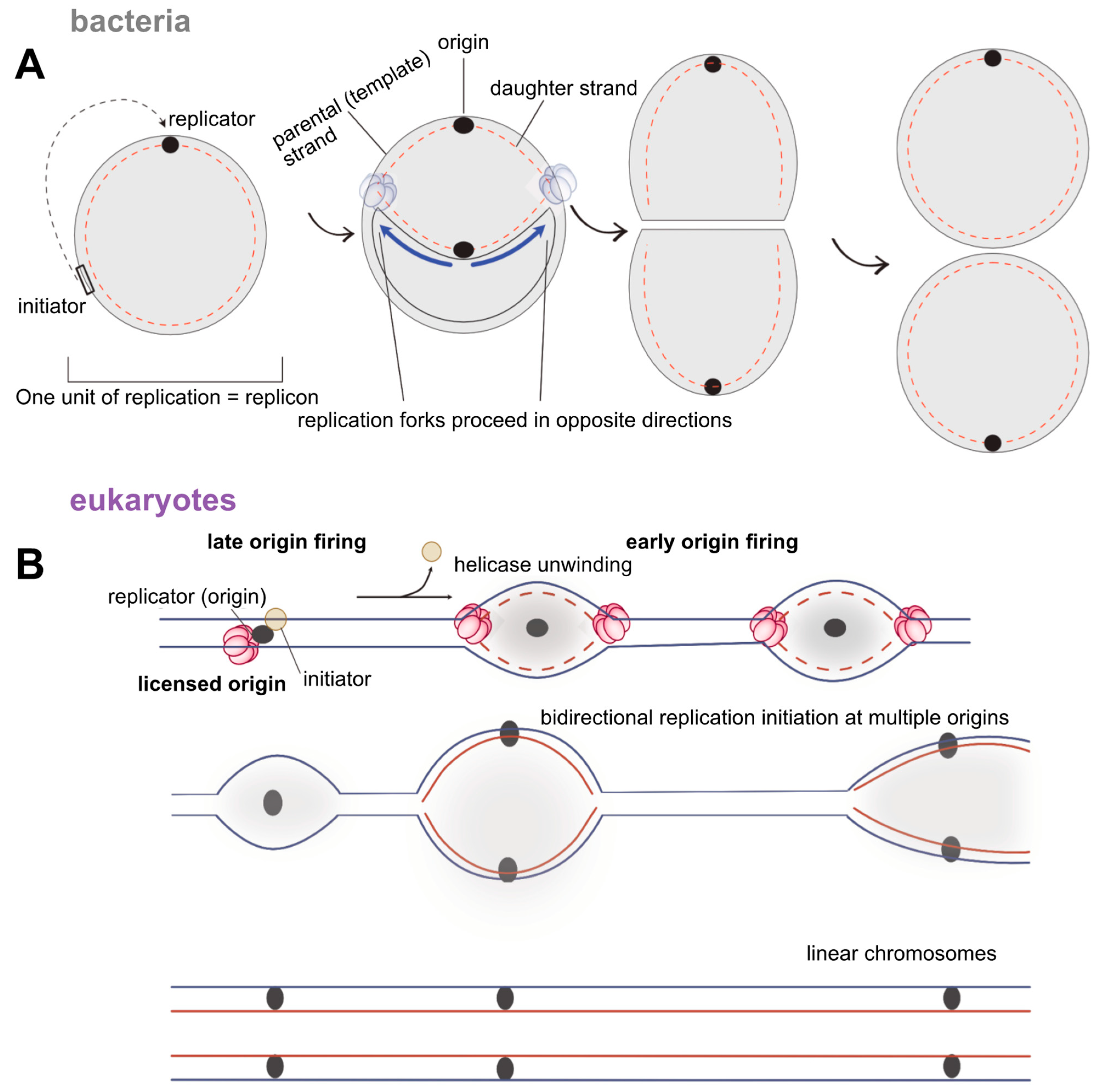
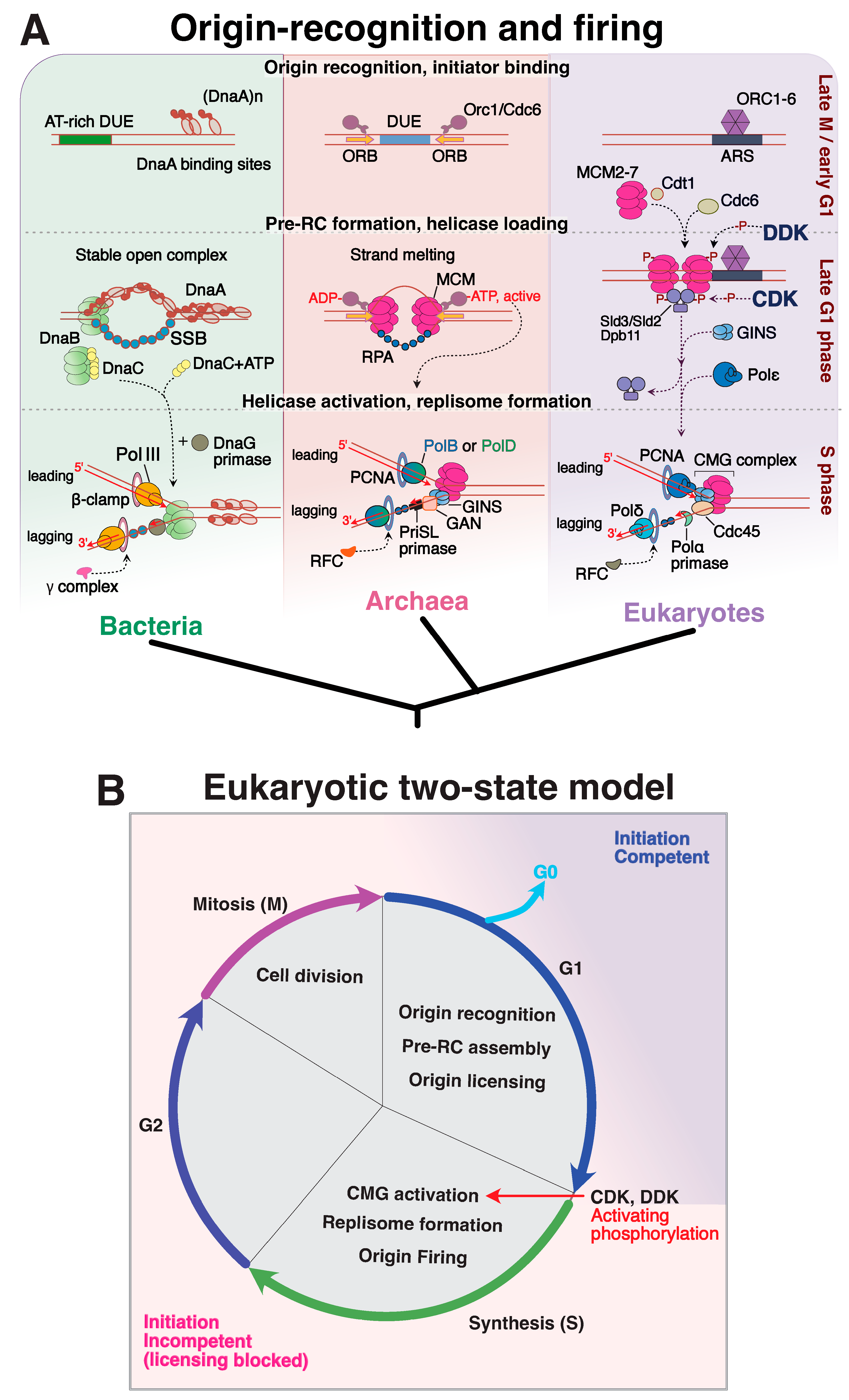
4. Where Do We Start? DNA Replication Initiation Across the Three Domains of Life
4.1. Bacteria
4.2. Eukaryotes
4.3. Archaea
| Replisome Assembly Step | Eukaryotes | Archaea | Bacteria |
|---|---|---|---|
| STAGE I Origin Recognition | ORC (Orc 1, 2, 3, 4, 5, 6) | Orc/Cdc6 a,b | DnaA |
| STAGE II Pre-RC formation | Cdc6/Cdt1 | Orc/Cdc6 a,b WhiP b | DnaA |
| STAGE III DNA duplex melting | Orc/Cdc6 MCM helicase (Mcm 2, 3, 4, 5, 6, 7 heterohexamer) | Orc/Cdc6 a,b MCM helicase a,b (homohexamer) | (DnaA)n |
| STAGE IV Helicase Loading | Cdc6 Cdt1 helicase loader | Orc/Cdc6 a,b helicase loader | DnaC helicase loader (DnaI in gram positive bacteria) DnaB helicase (DnaC in gram negative bacteria) |
| STAGE V Polymerase— Replisome assembly | CMG complex PCNA clamp RFC clamp loader PriSL primase B-family polymerases (Pol ε, Pol δ) | GINS a,b PCNA clamp RFC clamp loader PriSL a,b/PriX b primase B-family polymerase a,b, D-family polymerase a | β clamp DnaG primase C-Family polymerase (Pol III) |
| Recommended Literature | [106] | [111] [129] (focus on H. volcanii) | [93] |
5. DNA Replication and Recombination: A Dynamic Interplay
Adding a Level of Complexity: The Asymmetry of DNA Replication
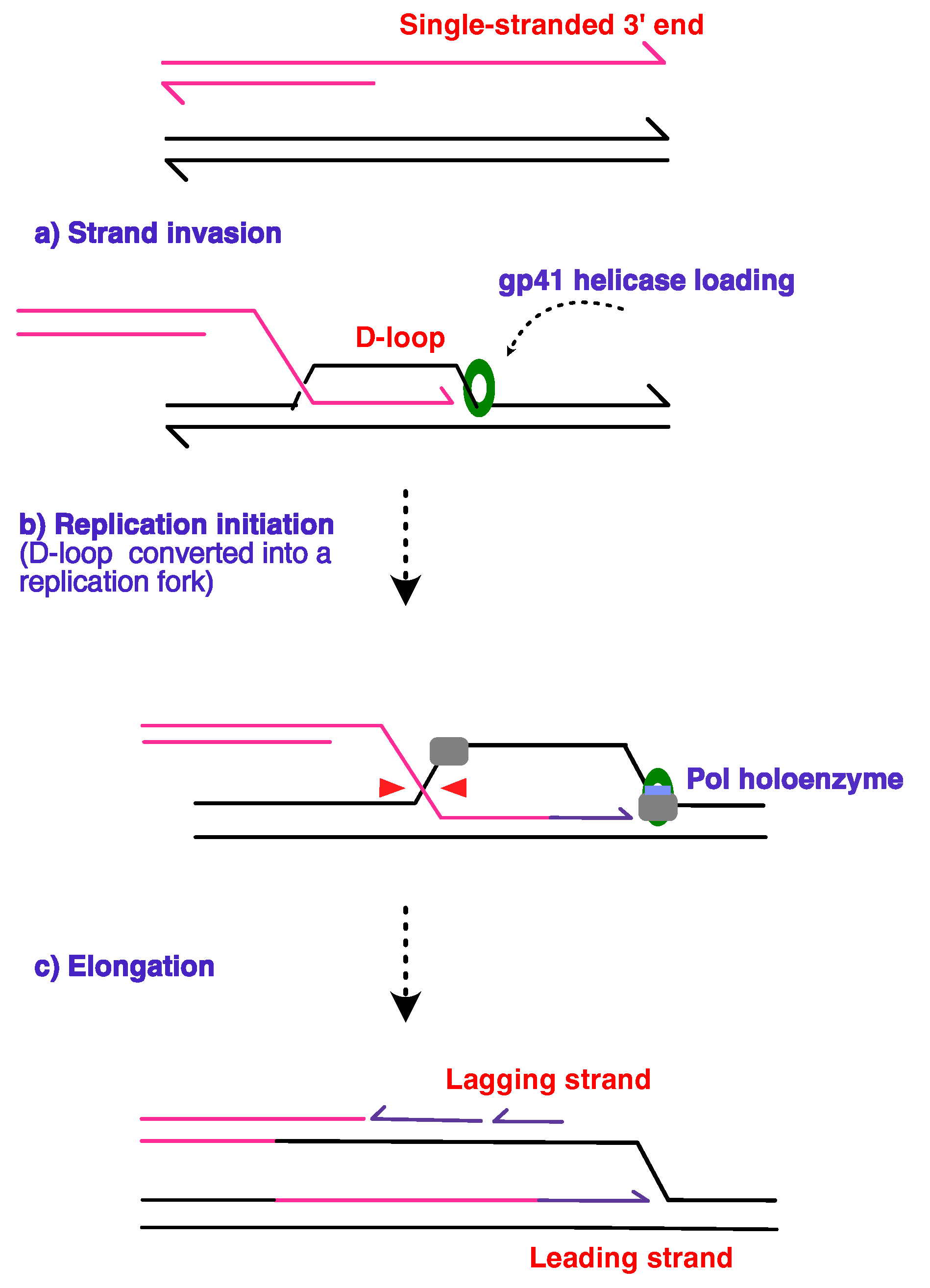
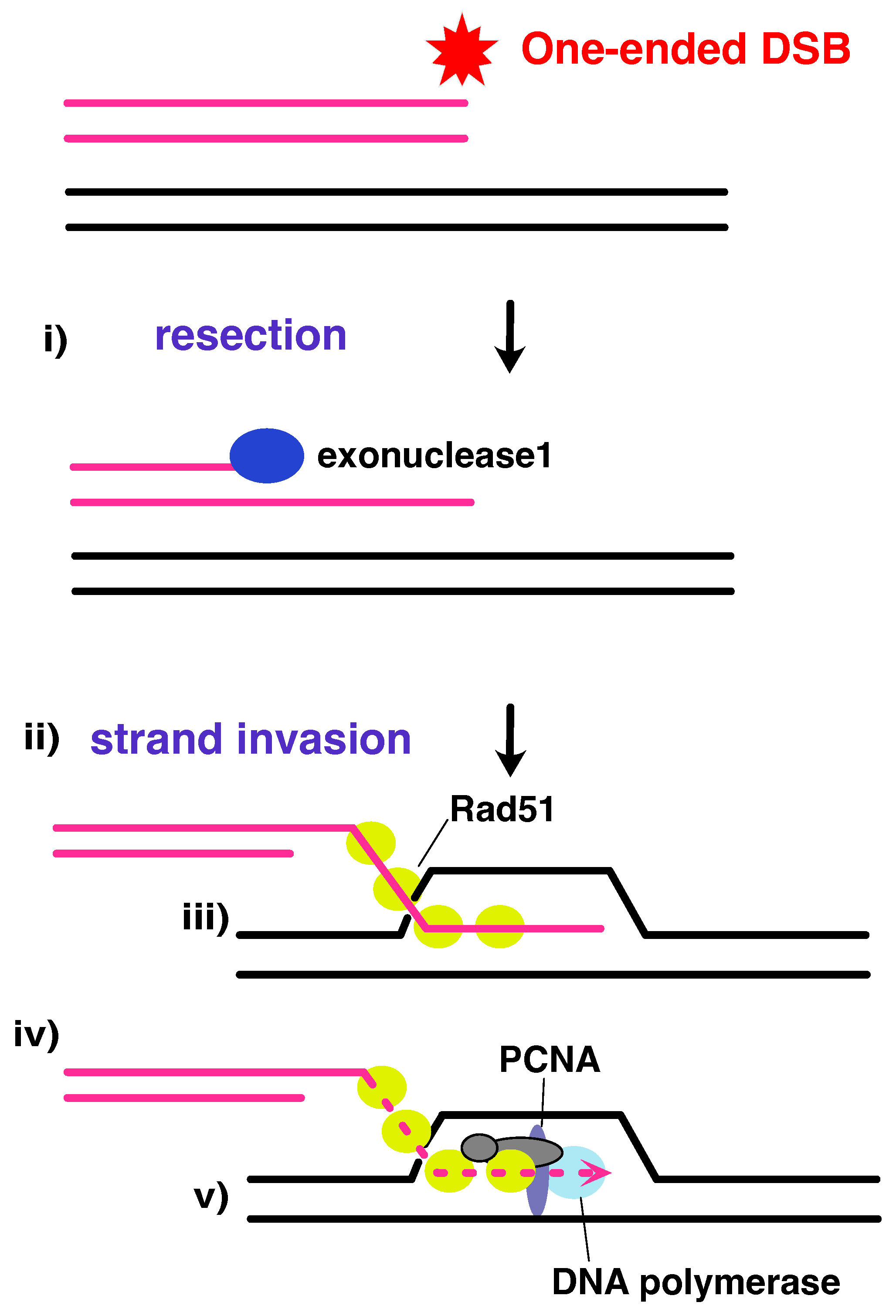
6. Recombination Dependent Replication
6.1. Clue No.1: Lessons from Viral Models
6.2. Clue No.2: Break-Induced DNA Replication in Eukaryotes
6.3. Clue No.3: Origin-Independent Replication Initiation in Bacteria and Archaea
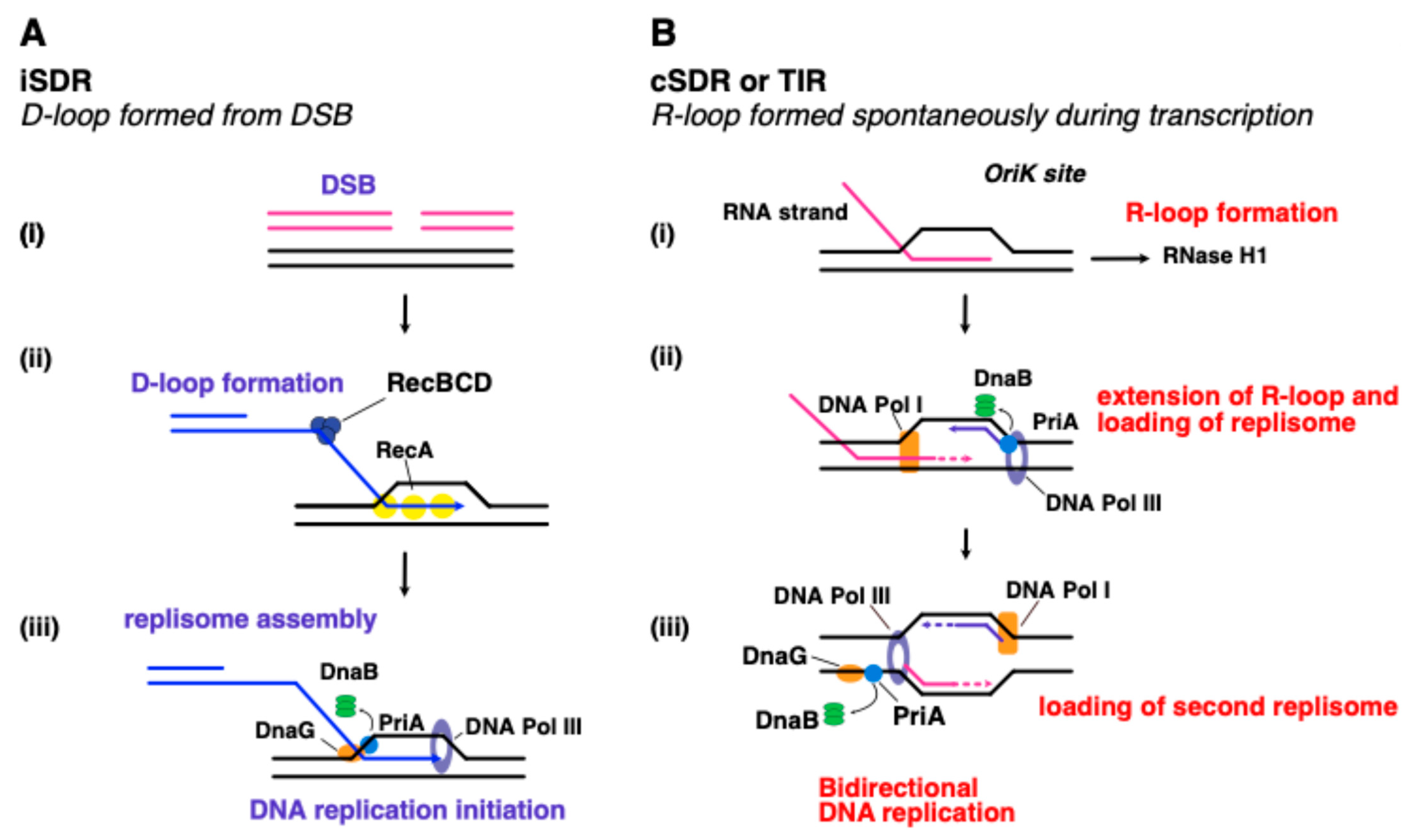
7. Conclusions: The Archaeal Domain as a Window into Our Evolutionary Past
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gaillard, H.; García-Muse, T.; Aguilera, A. Replication Stress and Cancer. Nat. Rev. Cancer 2015, 15, 276–289. [Google Scholar] [CrossRef] [PubMed]
- Avery, O.T.; Macleod, C.M.; McCarthy, M. Studies on the Chemical Nature of the Substance Inducing Transformation of Pneumococcal Types. J. Exp. Med. 1944, 79, 137–158. [Google Scholar] [CrossRef]
- Cobb, M. Oswald Avery, DNA, and the Transformation of Biology. Curr. Biol. 2014, 24, R55–R60. [Google Scholar] [CrossRef] [PubMed]
- Stent, G.S. Prematurity and Uniqueness in Scientific Discovery. Sci. Am. 1972, 227, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Mirsky, A.E.; Pollister, A.W.; Chromosin, A. Desoxyribose Nucleoprotein Complex of the Cell Nucleus. J. Gen. Physiol. 1946, 30, 117–148. [Google Scholar] [CrossRef] [PubMed]
- Levene, P.A.; La Forge, F.B. On Chondrosamine. Proc. Natl. Acad. Sci. USA 1915, 1, 190–191. [Google Scholar] [CrossRef] [PubMed]
- Chargaff, E. Chemical Specificity of Nucleic Acids and Mechanism of Their Enzymatic Degradation. Experientia 1950, 6, 201–209. [Google Scholar] [CrossRef]
- Hershey, A.D.; Chase, M. Independent Functions of Viral Protein and Nucleic Acid in Growth of Bacteriophage. J. Gen. Physiol. 1952, 36, 39–56. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, H.V. How History Has Blended. Nature 1974, 249, 803–805. [Google Scholar] [CrossRef]
- Northrop, J. Growth and Phage Production of Lysogenic B. Megatherium. J. Gen. Physiol. 1951, 34, 715–735. [Google Scholar] [CrossRef]
- Swift, H. The Constancy of Desoxyribose Nucleic Acid in Plant Nuclei. Proc. Natl. Acad. Sci. USA 1950, 36, 643–654. [Google Scholar] [CrossRef]
- Watson, J.D.; Crick, F.H.C. Genetical Implications of the Structure of Deoxyribonucleic Acid. Nature 1953, 171, 964–967. [Google Scholar] [CrossRef]
- Franklin, R.; Gosling, R. Molecular Configuration in Sodium Thymonucleate. Nature 1953, 171, 740–741. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.; Stokes, A.; Wilson, H. Molecular Structure of Nucleic Acids: Molecular Structure of Deoxypentose Nucleic Acids. Nature 1953, 171, 738–740. [Google Scholar] [CrossRef] [PubMed]
- Gulland, J.M.; Jordan, D.O.; Taylor, H.F.W. Deoxypentose Nucleic Acids. Part II. Electrometric Titration of the Acidic and the Basic Groups of the Deoxypentose Nucleic Acid of Calf Thymus. J. Chem. Soc. Resumed 1947, 25, 1131–1141. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.D.; Crick, F.H.C. Molecular Structure of Nucleic Acids; a Structure for Deoxyribose Nucleic Acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- Bloch, D.P. A Possible Mechanism for the Replication of the Helical Structure of Desoxyribonucleic Acid. Proc. Natl. Acad. Sci. USA 1955, 41, 1058–1064. [Google Scholar] [CrossRef]
- Taylor, J.H.; Woods, P.S.; Hughes, W.L. The Organization and Duplication Of Chromosomes as Revealed by Autoradiographic Studies Using Tritium-Labeled Thymidine. Proc. Natl. Acad. Sci. USA 1957, 43, 122–128. [Google Scholar] [CrossRef]
- Meselson, M.; Stahl, F.W. The Replication of DNA in Escherichia coli. Proc. Natl. Acad. Sci. USA 1958, 44, 671–682. [Google Scholar] [CrossRef]
- Holmes, F.L. The DNA Replication Problem, 1953–1958. Cell 1998, 23, 117–120. [Google Scholar] [CrossRef]
- Levinthal, C. The Mechanism of DNA Replication and Genetic Recombination in Phage. Proc. Natl. Acad. Sci. USA 1956, 42, 394–404. [Google Scholar] [CrossRef] [PubMed]
- Stent, G.S.; Jerne, N.K. The Distribution of Parental Phosphorus Atoms Among Bacteriophage Progeny. Proc. Natl. Acad. Sci. USA 1955, 41, 704–709. [Google Scholar] [CrossRef]
- Lehman, I.R.; Bessman, M.J.; Simms, E.S.; Kornberg, A. Enzymatic Synthesis of Deoxyribonucleic Acid. I. Preparation of Substrates and Partial Purification of an Enzyme from Escherichia coli. J. Biol. Chem. 1958, 233, 163–170. [Google Scholar] [CrossRef]
- Bessman, M.J.; Lehman, I.R.; Simms, E.S.; Kornberg, A. Enzymatic Synthesis of Deoxyribonucleic Acid. II. General Properties of the Reaction. J. Biol. Chem. 1958, 233, 171–177. [Google Scholar] [CrossRef]
- Lieberman, I.; Kornberg, A.; Simms, E.S. Enzymatic Syntheses of Pyrimidine and Purine Nucleotides. II.1 Orotidine-5′-phosphate Pyrophosphorylase and Decarboxylase. J. Am. Chem. Soc. 1954, 76, 2844–2845. [Google Scholar] [CrossRef]
- Cori, G.T.; Cori, C.F. Crystalline Muscle Phosphorylase. J. Biol. Chem. 1943, 151, 57–63. [Google Scholar] [CrossRef]
- Brutlag, D.; Kornberg, A. Enzymatic Synthesis of Deoxyribonucleic Acid. 36. A Proofreading Funtion for the 3′ Leads to 5′ Exonuclease Activity in the Deoxyribonucleic Acid Polymerases. J. Biol. Chem. 1972, 247, 241–248. [Google Scholar] [CrossRef]
- Delarue, M.; Poch, O.; Tordo, N.; Moras, D.; Argos, P. An Attempt to Unify the Structure of Polymerases. Protein Eng. Des. Sel. 1990, 3, 461–467. [Google Scholar] [CrossRef]
- Joyce, C.M.; Steitz, T.A. Function and Structure Relationships in DNA Polymerases. Annu. Rev. Biochem. 1994, 63, 777–822. [Google Scholar] [CrossRef]
- Bebenek, K.; Kunkel, T.A. Functions of DNA Polymerases. In Advances in Protein Chemistry; Elsevier: Amsterdam, The Netherlands, 2004; Volume 69, pp. 137–165. ISBN 978-0-12-034269-3. [Google Scholar]
- Braithwaite, D.K.; Ito, J. Compilation, Alignment, and Phylogenetic Relationships of DNA Polymerases. Nucleic Acids Res. 1993, 21, 787–802. [Google Scholar] [CrossRef]
- Beese, L.S.; Derbyshire, V.; Steitz, T.A. Structure of DNA Polymerase I Klenow Fragment Bound to Duplex DNA. Science 1993, 260, 352–355. [Google Scholar] [CrossRef] [PubMed]
- Morin, J.A.; Cao, F.J.; Lázaro, J.M.; Arias-Gonzalez, J.R.; Valpuesta, J.M.; Carrascosa, J.L.; Salas, M.; Ibarra, B. Active DNA Unwinding Dynamics during Processive DNA Replication. Proc. Natl. Acad. Sci. USA 2012, 109, 8115–8120. [Google Scholar] [CrossRef]
- Bérut, A.; Arakelyan, A.; Petrosyan, A.; Ciliberto, S.; Dillenschneider, R.; Lutz, E. Experimental Verification of Landauer’s Principle Linking Information and Thermodynamics. Nature 2012, 483, 187–189. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, J.; Nozawa, Y. Self-Reproducing System Can Behave as Maxwell’s Demon: Theoretical Illustration under Prebiotic Conditions. J. Theor. Biol. 1998, 194, 205–221. [Google Scholar] [CrossRef]
- Forterre, P.; Filée, J.; Myllykallio, H. Origin and Evolution of DNA and DNA Replication Machineries. In The Genetic Code and the Origin of Life; Springer: Boston, MA, USA, 2004; pp. 145–168. ISBN 978-0-306-47843-7. [Google Scholar]
- Rothwell, P.J.; Waksman, G. Structure and Mechanism of DNA Polymerases. In Advances in Protein Chemistry; Elsevier: Amsterdam, The Netherlands, 2005; Volume 71, pp. 401–440. ISBN 978-0-12-034271-6. [Google Scholar]
- Steitz, T.A. DNA- and RNA-Dependent DNA Polymerases. Curr. Opin. Struct. Biol. 1993, 3, 31–38. [Google Scholar] [CrossRef]
- Bębenek, A.; Ziuzia-Graczyk, I. Fidelity of DNA Replication—A Matter of Proofreading. Curr. Genet. 2018, 64, 985–996. [Google Scholar] [CrossRef]
- Ekundayo, B.; Bleichert, F. Origins of DNA Replication. PLoS Genet. 2019, 15, e1008320. [Google Scholar] [CrossRef]
- Jacob, F.; Monod, J. Genetic Regulatory Mechanisms in the Synthesis of Proteins. J. Mol. Biol. 1961, 3, 318–356. [Google Scholar] [CrossRef]
- Ryter, A.; Hirota, Y.; Jacob, F. DNA-Membrane Complex and Nuclear Segregation in Bacteria. Cold Spring Harb. Symp. Quant. Biol. 1968, 33, 669–676. [Google Scholar] [CrossRef]
- Morange, M. What History Tells Us XXXI. The Replicon Model: Between Molecular Biology and Molecular Cell Biology. J. Biosci. 2013, 38, 225–227. [Google Scholar] [CrossRef]
- Jacob, F.; Brenner, S.; Cuzin, F. On the Regulation of DNA Replication in Bacteria. Cold Spring Harb. Symp. Quant. Biol. 1963, 28, 329–348. [Google Scholar] [CrossRef]
- Huberman, J.A.; Riggs, A.D. On the Mechanism of DNA Replication in Mammalian Chromosomes. J. Mol. Biol. 1968, 32, 327–341. [Google Scholar] [CrossRef] [PubMed]
- Novick, R.P. Plasmid Incompatibility. Microbiol. Rev. 1987, 51, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Kohiyama, M.; Hiraga, S.; Matic, I.; Radman, M. Bacterial Sex: Playing Voyeurs 50 Years Later. Science 2003, 301, 802–803. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, S.; Hirota, Y. Cloning and Mapping of the Replication Origin of Escherichia Coli. Proc. Natl. Acad. Sci. USA 1977, 74, 5458–5462. [Google Scholar] [CrossRef]
- Mackiewicz, P.; Zakrzewska-Czerwińska, J.; Zawilak, A.; Dudek, M.; Cebrat, M. Where Does Bacterial Replication Start? Rules for Predicting the oriC Region. Nucleic Acids Res. 2004, 32, 3781–3791. [Google Scholar] [CrossRef]
- Chakraborty, T.; Yoshinaga, K.; Lother, H.; Messer, W. Purification of the E. Coli dnaA Gene Product. EMBO J. 1982, 1, 1545–1549. [Google Scholar] [CrossRef]
- Fujita, M.Q.; Yoshikawa, H. Structure of the dnaA Region of Pseudomonas putida: Conservation among Three Bacteria, Bacillus subtilis, Escherichia coli and P. putida. Mol. Gen. Genet. MGG 1989, 215, 381–387. [Google Scholar]
- Chan, C.S.; Tye, B.K. Autonomously Replicating Sequences in Saccharomyces Cerevisiae. Proc. Natl. Acad. Sci. USA 1980, 77, 6329–6333. [Google Scholar] [CrossRef]
- Broach, J.R.; Li, Y.-Y.; Feldman, J.; Jayaram, M.; Abraham, J.; Nasmyth, K.A.; Hicks, J.B. Localization and Sequence Analysis of Yeast Origins of DNA Replication. Cold Spring Harb. Symp. Quant. Biol. 1983, 47, 1165–1173. [Google Scholar] [CrossRef]
- Bell, S.P.; Stillman, B. ATP-Dependent Recognition of Eukaryotic Origins of DNA Replication by a Multiprotein Complex. Nature 1992, 357, 128–134. [Google Scholar] [CrossRef]
- Kogoma, T. Stable DNA Replication: Interplay between DNA Replication, Homologous Recombination, and Transcription. Microbiol. Mol. Biol. Rev. 1997, 61, 212–238. [Google Scholar] [PubMed]
- Gavin, K.A.; Hidaka, M.; Stillman, B. Conserved Initiator Proteins in Eukaryotes. Science 1995, 270, 1667–1671. [Google Scholar] [CrossRef]
- Taylor, J.H. Increase in DNA Replication Sites in Cells Held at the Beginning of S Phase. Chromosoma 1977, 62, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Hamlin, J.L.; Mesner, L.D.; Lar, O.; Torres, R.; Chodaparambil, S.V.; Wang, L. A Revisionist Replicon Model for Higher Eukaryotic Genomes. J. Cell. Biochem. 2008, 105, 321–329. [Google Scholar] [CrossRef]
- diCenzo, G.C.; Finan, T.M. The Divided Bacterial Genome: Structure, Function, and Evolution. Microbiol. Mol. Biol. Rev. 2017, 81, e00019-17. [Google Scholar] [CrossRef] [PubMed]
- Harrison, P.W.; Lower, R.P.J.; Kim, N.K.D.; Young, J.P.W. Introducing the Bacterial ‘Chromid’: Not a Chromosome, Not a Plasmid. Trends Microbiol. 2010, 18, 141–148. [Google Scholar] [CrossRef]
- Krawiec, S.; Riley, M. Organization of the Bacterial Chromosome. Microbiol. Rev. 1990, 54, 502–539. [Google Scholar] [CrossRef]
- Suwanto, A.; Kaplan, S. Physical and Genetic Mapping of the Rhodobacter sphaeroides 2.4.1 Genome: Presence of Two Unique Circular Chromosomes. J. Bacteriol. 1989, 171, 5850–5859. [Google Scholar] [CrossRef]
- Woese, C.R.; Fox, G.E. Phylogenetic Structure of the Prokaryotic Domain: The Primary Kingdoms. Proc. Natl. Acad. Sci. USA 1977, 74, 5088–5090. [Google Scholar] [CrossRef]
- Bult, C.J.; White, O.; Olsen, G.J.; Zhou, L.; Fleischmann, R.D.; Sutton, G.G.; Blake, J.A.; FitzGerald, L.M.; Clayton, R.A.; Gocayne, J.D.; et al. Complete Genome Sequence of the Methanogenic Archaeon, Methanococcus jannaschii. Science 1996, 273, 1058–1073. [Google Scholar] [CrossRef]
- Myllykallio, H.; Lopez, P.; López-García, P.; Heilig, R.; Saurin, W.; Zivanovic, Y.; Philippe, H.; Forterre, P. Bacterial Mode of Replication with Eukaryotic-Like Machinery in a Hyperthermophilic Archaeon. Science 2000, 288, 2212–2215. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, F.; Forterre, P.; Ishino, Y.; Myllykallio, H. In Vivo Interactions of Archaeal Cdc6/Orc1 and Minichromosome Maintenance Proteins with the Replication Origin. Proc. Natl. Acad. Sci. USA 2001, 98, 11152–11157. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, F.; Norais, C.; Forterre, P.; Myllykallio, H. Identification of Short ‘Eukaryotic’ Okazaki Fragments Synthesized from a Prokaryotic Replication Origin. EMBO Rep. 2003, 4, 154–158. [Google Scholar] [CrossRef]
- Robinson, N.P.; Dionne, I.; Lundgren, M.; Marsh, V.L.; Bernander, R.; Bell, S.D. Identification of Two Origins of Replication in the Single Chromosome of the Archaeon Sulfolobus solfataricus. Cell 2004, 116, 25–38. [Google Scholar] [CrossRef]
- Lundgren, M.; Andersson, A.; Chen, L.; Nilsson, P.; Bernander, R. Three Replication Origins in Sulfolobus Species: Synchronous Initiation of Chromosome Replication and Asynchronous Termination. Proc. Natl. Acad. Sci. USA 2004, 101, 7046–7051. [Google Scholar] [CrossRef] [PubMed]
- She, Q.; Peng, X.; Zillig, W.; Garrett, R.A. Gene Capture in Archaeal Chromosomes. Nature 2001, 409, 478. [Google Scholar] [CrossRef]
- Robinson, N.P.; Bell, S.D. Extrachromosomal Element Capture and the Evolution of Multiple Replication Origins in Archaeal Chromosomes. Proc. Natl. Acad. Sci. USA 2007, 104, 5806–5811. [Google Scholar] [CrossRef]
- Zhang, R.; Zhang, C.-T. Multiple Replication Origins of the Archaeon Halobacterium Species NRC-1. Biochem. Biophys. Res. Commun. 2003, 302, 728–734. [Google Scholar] [CrossRef]
- Wu, Z.; Liu, H.; Liu, J.; Liu, X.; Xiang, H. Diversity and Evolution of Multiple Orc/Cdc6-Adjacent Replication Origins in Haloarchaea. BMC Genomics 2012, 13, 478. [Google Scholar] [CrossRef]
- Hawkins, M.S. DNA Replication Origins in Haloferax volcanii. Ph.D. Thesis, University of Nottingham, Nottingham, UK, 2009. [Google Scholar]
- Gehring, A.M.; Astling, D.P.; Matsumi, R.; Burkhart, B.W.; Kelman, Z.; Reeve, J.N.; Jones, K.L.; Santangelo, T.J. Genome Replication in Thermococcus kodakarensis Independent of Cdc6 and an Origin of Replication. Front. Microbiol. 2017, 8, 2084. [Google Scholar] [CrossRef]
- Hawkins, M.; Malla, S.; Blythe, M.J.; Nieduszynski, C.A.; Allers, T. Accelerated Growth in the Absence of DNA Replication Origins. Nature 2013, 503, 544–547. [Google Scholar] [CrossRef] [PubMed]
- Liman, G.L.S.; Lennon, C.W.; Mandley, J.L.; Galyon, A.M.; Zatopek, K.M.; Gardner, A.F.; Santangelo, T.J. Intein Splicing Efficiency and RadA Levels Can Control the Mode of Archaeal DNA Replication. Sci. Adv. 2024, 10, eadp4995. [Google Scholar] [CrossRef]
- Mc Teer, L.; Moalic, Y.; Cueff-Gauchard, V.; Catchpole, R.; Hogrel, G.; Lu, Y.; Laurent, S.; Hemon, M.; Aubé, J.; Leroy, E.; et al. Cooperation between Two Modes for DNA Replication Initiation in the Archaeon Thermococcus barophilus. mBio 2024, 15, e03200-23. [Google Scholar] [CrossRef]
- Dulermo, R. Archaeal DNA Replication Initiation: Bridging LUCA’s Legacy and Modern Mechanisms. Front. Microbiol. 2025, 16, 1561973. [Google Scholar] [CrossRef] [PubMed]
- Kreuzer, K.N.; Brister, J.R. Initiation of Bacteriophage T4 DNA Replication and Replication Fork Dynamics: A Review in the Virology Journal Series on Bacteriophage T4 and Its Relatives. Virol. J. 2010, 7, 358. [Google Scholar] [CrossRef] [PubMed]
- Richter, S.; Hagemann, M.; Messer, W. Transcriptional Analysis and Mutation of a dnaA-Like Gene in Synechocystis sp. Strain PCC 6803. J. Bacteriol. 1998, 180, 4946–4949. [Google Scholar] [CrossRef]
- Ohbayashi, R.; Hirooka, S.; Onuma, R.; Kanesaki, Y.; Hirose, Y.; Kobayashi, Y.; Fujiwara, T.; Furusawa, C.; Miyagishima, S. Evolutionary Changes in DnaA-Dependent Chromosomal Replication in Cyanobacteria. Front. Microbiol. 2020, 11, 786. [Google Scholar] [CrossRef]
- Masai, H. Replicon Hypothesis Revisited. Biochem. Biophys. Res. Commun. 2022, 633, 77–80. [Google Scholar] [CrossRef]
- Dyall-Smith, M.L.; Pfeiffer, F.; Klee, K.; Palm, P.; Gross, K.; Schuster, S.C.; Rampp, M.; Oesterhelt, D. Haloquadratum walsbyi: Limited Diversity in a Global Pond. PLoS ONE 2011, 6, e20968. [Google Scholar] [CrossRef]
- Xu, Y.; Bremer, H. Chromosome Replication in Escherichia coli Induced by Oversupply of DnaA. Mol Gen Genet 1988, 211, 138–142. [Google Scholar] [CrossRef]
- Grimwade, J.E.; Ryan, V.T.; Leonard, A.C. IHF Redistributes Bound Initiator Protein, DnaA, on Supercoiled oriC of Escherichia coli. Mol. Microbiol. 2000, 35, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Rozgaja, T.A.; Grimwade, J.E.; Iqbal, M.; Czerwonka, C.; Vora, M.; Leonard, A.C. Two Oppositely Oriented Arrays of Low-Affinity Recognition Sites in oriC Guide Progressive Binding of DnaA during Escherichia coli Pre-RC Assembly: DnaA-oriC Interaction at Arrayed Sites. Mol. Microbiol. 2011, 82, 475–488. [Google Scholar] [CrossRef] [PubMed]
- Matsui, M.; Oka, A.; Takanami, M.; Yasuda, S.; Hirota, Y. Sites of dnaA Protein-Binding in the Replication Origin of the Escherichia coli K-12 Chromosome. J. Mol. Biol. 1985, 184, 529–533. [Google Scholar] [CrossRef]
- Erzberger, J.P. The Structure of Bacterial DnaA: Implications for General Mechanisms Underlying DNA Replication Initiation. EMBO J. 2002, 21, 4763–4773. [Google Scholar] [CrossRef]
- Erzberger, J.P.; Berger, J.M. Evolutionary Relationships and Structural Mechanisms of AAA+ Proteins. Annu. Rev. Biophys. Biomol. Struct. 2006, 35, 93–114. [Google Scholar] [CrossRef] [PubMed]
- Katayama, T.; Kasho, K.; Kawakami, H. The DnaA Cycle in Escherichia Coli: Activation, Function and Inactivation of the Initiator Protein. Front. Microbiol. 2017, 8, 2496. [Google Scholar] [CrossRef]
- Kowalski, D.; Eddy, M.J. The DNA Unwinding Element: A Novel, Cis-Acting Component That Facilitates Opening of the Escherichia coli Replication Origin. EMBO J. 1989, 8, 4335–4344. [Google Scholar] [CrossRef]
- Wegrzyn, K.; Konieczny, I. Toward an Understanding of the DNA Replication Initiation in Bacteria. Front. Microbiol. 2024, 14, 1328842. [Google Scholar] [CrossRef]
- Robinson, N.P.; Bell, S.D. Origins of DNA Replication in the Three Domains of Life. FEBS J. 2005, 272, 3757–3766. [Google Scholar] [CrossRef]
- Schwob, E. Flexibility and Governance in Eukaryotic DNA Replication. Curr. Opin. Microbiol. 2004, 7, 680–690. [Google Scholar] [CrossRef]
- Segurado, M.; Gómez, M.; Antequera, F. Increased Recombination Intermediates and Homologous Integration Hot Spots at DNA Replication Origins. Mol. Cell 2002, 10, 907–916. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, H.; Chetlangia, N.; Prasanth, S.G. Chromatin’s Influence on Pre-Replication Complex Assembly and Function. Biology 2024, 13, 152. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, D.M.; Takebayashi, S.-I.; Ryba, T.; Lu, J.; Pope, B.D.; Wilson, K.A.; Hiratani, I. Space and Time in the Nucleus: Developmental Control of Replication Timing and Chromosome Architecture. Cold Spring Harb. Symp. Quant. Biol. 2010, 75, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Labib, K. How Do Cdc7 and Cyclin-Dependent Kinases Trigger the Initiation of Chromosome Replication in Eukaryotic Cells? Genes Dev. 2010, 24, 1208–1219. [Google Scholar] [CrossRef]
- O’Donnell, M.; Langston, L.; Stillman, B. Principles and Concepts of DNA Replication in Bacteria, Archaea, and Eukarya. Cold Spring Harb. Perspect. Biol. 2013, 5, a010108. [Google Scholar] [CrossRef]
- Hartwell, L.H. Sequential Function of Gene Products Relative to DNA Synthesis in the Yeast Cell Cycle. J. Mol. Biol. 1976, 104, 803–817. [Google Scholar] [CrossRef]
- Hofmann, J.F.X.; Beach, D. Cdt 1 Is an Essential Target of the Cdc 1 O/Sct 1 Transcription Factor: Requirement for DNA Replication and Inhibition of Mitosis. EMBO J. 1994, 13, 425–434. [Google Scholar] [CrossRef]
- Nishitani, H.; Lygerou, Z.; Nishimoto, T.; Nurse, P. The Cdt1 Protein Is Required to License DNA for Replication in fission Yeast. Nature 2000, 404, 625–628. [Google Scholar] [CrossRef]
- Schmidt, J.M.; Yang, R.; Kumar, A.; Hunker, O.; Seebacher, J.; Bleichert, F. A Mechanism of Origin Licensing Control through Autoinhibition of S. cerevisiae ORC·DNA·Cdc6. Nat. Commun. 2022, 13, 1059. [Google Scholar] [CrossRef]
- Randell, J.C.W.; Bowers, J.L.; Rodríguez, H.K.; Bell, S.P. Sequential ATP Hydrolysis by Cdc6 and ORC Directs Loading of the Mcm2-7 Helicase. Mol. Cell 2006, 21, 29–39. [Google Scholar] [CrossRef]
- Costa, A.; Diffley, J.F.X. The Initiation of Eukaryotic DNA Replication. Annu. Rev. Biochem. 2022, 91, 107–131. [Google Scholar] [CrossRef] [PubMed]
- Ilves, I.; Petojevic, T.; Pesavento, J.J.; Botchan, M.R. Activation of the MCM2-7 Helicase by Association with Cdc45 and GINS Proteins. Mol. Cell 2010, 37, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Zegerman, P.; Diffley, J.F.X. Phosphorylation of Sld2 and Sld3 by Cyclin-Dependent Kinases Promotes DNA Replication in Budding Yeast. Nature 2007, 445, 281–285. [Google Scholar] [CrossRef]
- Ausiannikava, D.; Allers, T. Diversity of DNA Replication in the Archaea. Genes 2017, 8, 56. [Google Scholar] [CrossRef]
- Wu, Z.; Liu, J.; Yang, H.; Xiang, H. DNA Replication Origins in Archaea. Front. Microbiol. 2014, 5, 179. [Google Scholar] [CrossRef]
- Greci, M.D.; Bell, S.D. Archaeal DNA Replication. Annu. Rev. Microbiol. 2020, 74, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Lopez, P.; Philippe, H.; Myllykallio, H.; Forterre, P. Identification of Putative Chromosomal Origins of Replication in Archaea. Mol. Microbiol. 1999, 32, 883–886. [Google Scholar] [CrossRef]
- Liu, J.; Smith, C.L.; DeRyckere, D.; DeAngelis, K.; Martin, G.S.; Berger, J.M. Structure and Function of Cdc6/Cdc18: Implications for Origin Recognition and Checkpoint Control. Mol. Cell 2000, 6, 637–648. [Google Scholar] [CrossRef]
- Matsunaga, F.; Glatigny, A.; Mucchielli-Giorgi, M.-H.; Agier, N.; Delacroix, H.; Marisa, L.; Durosay, P.; Ishino, Y.; Aggerbeck, L.; Forterre, P. Genomewide and Biochemical Analyses of DNA-Binding Activity of Cdc6/Orc1 and Mcm Proteins in Pyrococcus sp. Nucleic Acids Res. 2007, 35, 3214–3222. [Google Scholar] [CrossRef]
- Matsunaga, F.; Takemura, K.; Akita, M.; Adachi, A.; Yamagami, T.; Ishino, Y. Localized Melting of Duplex DNA by Cdc6/Orc1 at the DNA Replication Origin in the Hyperthermophilic Archaeon Pyrococcus furiosus. Extremophiles 2010, 14, 21–31. [Google Scholar] [CrossRef]
- Gaudier, M.; Schuwirth, B.S.; Westcott, S.L.; Wigley, D.B. Structural Basis of DNA Replication Origin Recognition by an ORC Protein. Science 2007, 317, 1213–1216. [Google Scholar] [CrossRef] [PubMed]
- Grainge, I. Biochemical Analysis of Components of the Pre-Replication Complex of Archaeoglobus fulgidus. Nucleic Acids Res. 2003, 31, 4888–4898. [Google Scholar] [CrossRef] [PubMed]
- Samson, R.Y.; Xu, Y.; Gadelha, C.; Stone, T.A.; Faqiri, J.N.; Li, D.; Qin, N.; Pu, F.; Liang, Y.X.; She, Q.; et al. Specificity and Function of Archaeal DNA Replication Initiator Proteins. Cell Rep. 2013, 3, 485–496. [Google Scholar] [CrossRef]
- Dueber, E.L.C.; Corn, J.E.; Bell, S.D.; Berger, J.M. Replication Origin Recognition and Deformation by a Heterodimeric Archaeal Orc1 Complex. Science 2007, 317, 1210–1213. [Google Scholar] [CrossRef] [PubMed]
- Grainge, I.; Gaudier, M.; Schuwirth, B.S.; Westcott, S.L.; Sandall, J.; Atanassova, N.; Wigley, D.B. Biochemical Analysis of a DNA Replication Origin in the Archaeon Aeropyrum pernix. J. Mol. Biol. 2006, 363, 355–369. [Google Scholar] [CrossRef]
- Akita, M.; Adachi, A.; Takemura, K.; Yamagami, T.; Matsunaga, F.; Ishino, Y. Cdc6/Orc1 from Pyrococcus furiosus May Act as the Origin Recognition Protein and Mcm Helicase Recruiter. Genes Cells 2010, 15, 537–552. [Google Scholar] [CrossRef]
- Kelman, L.M.; O’Dell, W.B.; Kelman, Z. Unwinding 20 Years of the Archaeal Minichromosome Maintenance Helicase. J. Bacteriol. 2020, 202, e00729-19. [Google Scholar] [CrossRef]
- Kasiviswanathan, R.; Shin, J.-H.; Kelman, Z. Interactions between the Archaeal Cdc6 and MCM Proteins Modulate Their Biochemical Properties. Nucleic Acids Res. 2005, 33, 4940–4950. [Google Scholar] [CrossRef]
- Samson, R.Y.; Abeyrathne, P.D.; Bell, S.D. Mechanism of Archaeal MCM Helicase Recruitment to DNA Replication Origins. Mol. Cell 2016, 61, 287–296. [Google Scholar] [CrossRef]
- Samson, R.Y.; Bell, S.D. MCM Loading—An Open-and-Shut Case? Mol. Cell 2013, 50, 457–458. [Google Scholar] [CrossRef]
- Mohammed Khalid, A.A.; Parisse, P.; Medagli, B.; Onesti, S.; Casalis, L. Atomic Force Microscopy Investigation of the Interactions between the MCM Helicase and DNA. Materials 2021, 14, 687. [Google Scholar] [CrossRef] [PubMed]
- Kelman, Z.; Lee, J.-K.; Hurwitz, J. The Single Minichromosome Maintenance Protein of Methanobacterium Thermoautotrophicum DeltaH Contains DNA Helicase Activity. Proc. Natl. Acad. Sci. USA 1999, 96, 14783–14788. [Google Scholar] [CrossRef]
- Haugland, G.T.; Shin, J.-H.; Birkeland, N.-K.; Kelman, Z. Stimulation of MCM Helicase Activity by a Cdc6 Protein in the Archaeon Thermoplasma acidophilum. Nucleic Acids Res. 2006, 34, 6337–6344. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Arnaiz, P.; Dattani, A.; Smith, V.; Allers, T. Haloferax volcanii—A Model Archaeon for Studying DNA Replication and Repair. Open Biol. 2020, 10, 200293. [Google Scholar] [CrossRef]
- Ishino, S.; Kelman, L.M.; Kelman, Z.; Ishino, Y. The Archaeal DNA Replication Machinery: Past, Present and Future. Genes Genet. Syst. 2013, 88, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Sauguet, L.; Raia, P.; Henneke, G.; Delarue, M. Shared Active Site Architecture between Archaeal PolD and Multi-Subunit RNA Polymerases Revealed by X-Ray Crystallography. Nat. Commun. 2016, 7, 12227. [Google Scholar] [CrossRef] [PubMed]
- Čuboňová, L.; Richardson, T.; Burkhart, B.W.; Kelman, Z.; Connolly, B.A.; Reeve, J.N.; Santangelo, T.J. Archaeal DNA Polymerase D but Not DNA Polymerase B Is Required for Genome Replication in Thermococcus kodakarensis. J. Bacteriol. 2013, 195, 2322–2328. [Google Scholar] [CrossRef]
- Oki, K.; Yamagami, T.; Nagata, M.; Mayanagi, K.; Shirai, T.; Adachi, N.; Numata, T.; Ishino, S.; Ishino, Y. DNA Polymerase D Temporarily Connects Primase to the CMG-like Helicase before Interacting with Proliferating Cell Nuclear Antigen. Nucleic Acids Res. 2021, 49, 4599–4612. [Google Scholar] [CrossRef]
- Oki, K.; Nagata, M.; Yamagami, T.; Numata, T.; Ishino, S.; Oyama, T.; Ishino, Y. Family D DNA Polymerase Interacts with GINS to Promote CMG-Helicase in the Archaeal Replisome. Nucleic Acids Res. 2022, 50, 3601–3615. [Google Scholar] [CrossRef]
- Martínez-Carranza, M.; Vialle, L.; Madru, C.; Cordier, F.; Tekpinar, A.D.; Haouz, A.; Legrand, P.; Le Meur, R.A.; England, P.; Dulermo, R.; et al. Communication between DNA Polymerases and Replication Protein A within the Archaeal Replisome. Nat. Commun. 2024, 15, 10926. [Google Scholar] [CrossRef]
- Cairns, J. The Bacterial Chromosome and Its Manner of Replication as Seen by Autoradiography. J. Mol. Biol. 1963, 6, 208–213. [Google Scholar] [CrossRef]
- Wu, C.A.; Zechner, E.L.; Marians, K.J. Coordinated Leading- and Lagging-Strand Synthesis at the Escherichia coli DNA Replication Fork. I. Multiple Effectors Act to Modulate Okazaki Fragment Size. J. Biol. Chem. 1992, 267, 4030–4044. [Google Scholar] [CrossRef] [PubMed]
- Cha, T.A.; Alberts, B.M. The Bacteriophage T4 DNA Replication Fork. J. Biol. Chem. 1989, 264, 12220–12225. [Google Scholar] [CrossRef]
- Nakai, H.; Richardson, C.C. Leading and Lagging Strand Synthesis at the Replication Fork of Bacteriophage T7. Distinct Properties of T7 Gene 4 Protein as a Helicase and Primase. J. Biol. Chem. 1988, 263, 9818–9830. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, K.; Okazaki, T.; Okazaki, R. Mechanism of DNA Chain Growth, II. Accumulation of Newly Synthesized Short Chains in E. coli Infected with Ligase-Defective T4 Phages. Proc. Natl. Acad. Sci. USA 1968, 60, 1356–1362. [Google Scholar] [CrossRef] [PubMed]
- Cronan, G.E.; Kouzminova, E.A.; Kuzminov, A. Near-Continuously Synthesized Leading Strands in Escherichia coli Are Broken by Ribonucleotide Excision. Proc. Natl. Acad. Sci. USA 2019, 116, 1251–1260. [Google Scholar] [CrossRef]
- Kelman, L.M.; Kelman, Z. Archaea: An Archetype for Replication Initiation Studies? Mol. Microbiol. 2003, 48, 605–615. [Google Scholar] [CrossRef]
- Shen, W.; Wang, Z.; Ning, K.; Cheng, F.; Engelhardt, J.F.; Yan, Z.; Qiu, J. Hairpin Transfer-Independent Parvovirus DNA Replication Produces Infectious Virus. J. Virol. 2021, 95, e01108-21. [Google Scholar] [CrossRef]
- Lujan, S.A.; Williams, J.S.; Kunkel, T.A. DNA Polymerases Divide the Labor of Genome Replication. Trends Cell Biol. 2016, 26, 640–654. [Google Scholar] [CrossRef]
- Iyer, L.M. Origin and Evolution of the Archaeo-Eukaryotic Primase Superfamily and Related Palm-Domain Proteins: Structural Insights and New Members. Nucleic Acids Res. 2005, 33, 3875–3896. [Google Scholar] [CrossRef]
- Forterre, P. Why Are There So Many Diverse Replication Machineries? J. Mol. Biol. 2013, 425, 4714–4726. [Google Scholar] [CrossRef]
- Georgescu, R.; Langston, L.; O’Donnell, M. A Proposal: Evolution of PCNA’s Role as a Marker of Newly Replicated DNA. DNA Repair 2015, 29, 4–15. [Google Scholar] [CrossRef]
- Leipe, D.D.; Aravind, L.; Koonin, E.V. Did DNA Replication Evolve Twice Independently? Nucleic Acids Res. 1999, 27, 3389–3401. [Google Scholar] [CrossRef]
- Sinha, N.K.; Morris, C.F.; Alberts, B.M. Efficient in Vitro Replication of Double-Stranded DNA Templates by a Purified T4 Bacteriophage Replication System. J. Biol. Chem. 1980, 255, 4290–4303. [Google Scholar] [CrossRef]
- Alberts, B.M.; Barry, J.; Bedinger, P.; Formosa, T.; Jongeneel, C.V.; Kreuzer, K.N. Studies on DNA Replication in the Bacteriophage T4 In Vitro System. Cold Spring Harb Symp Quant Biol 1983, 47, 655–668. [Google Scholar] [CrossRef]
- Kreuzer, K.N. Recombination-Dependent DNA Replication in Phage T4. Trends Biochem. Sci. 2000, 25, 165–173. [Google Scholar] [CrossRef]
- Mosig, G.; Gewin, J.; Luder, A.; Colowick, N.; Vo, D. Two Recombination-Dependent DNA Replication Pathways of Bacteriophage T4, and Their Roles in Mutagenesis and Horizontal Gene Transfer. Proc. Natl. Acad. Sci. USA 2001, 98, 8306–8311. [Google Scholar] [CrossRef]
- Belanger, K.G.; Kreuzer, K.N. Bacteriophage T4 Initiates Bidirectional DNA Replication through a Two-Step Process. Mol. Cell 1998, 2, 693–701. [Google Scholar] [CrossRef]
- Syeda, A.H.; Hawkins, M.; McGlynn, P. Recombination and Replication. Cold Spring Harb. Perspect. Biol. 2014, 6, a016550. [Google Scholar] [CrossRef]
- Yang, S.; VanLoock, M.S.; Yu, X.; Egelman, E.H. Comparison of Bacteriophage T4 UvsX and Human Rad51 Filaments Suggests That RecA-like Polymers May Have Evolved independently. J. Mol. Biol. 2001, 312, 999–1009. [Google Scholar] [CrossRef]
- Liu, J.; Marians, K.J. PriA-Directed Assembly of a Primosome on D Loop DNA. J. Biol. Chem. 1999, 274, 25033–25041. [Google Scholar] [CrossRef]
- Jones, J.M. The Phi X174-Type Primosome Promotes Replisome Assembly at the Site of Recombination in Bacteriophage Mu Transposition. EMBO J. 1997, 16, 6886–6895. [Google Scholar] [CrossRef]
- Ferreira-Cerca, S. (Ed.) Archaea: Methods and Protocols; Methods in Molecular Biology; Springer: New York, NY, USA, 2022; Volume 2522, ISBN 978-1-07-162444-9. [Google Scholar]
- Epum, E.A.; Haber, J.E. DNA Replication: The Recombination Connection. Trends Cell Biol. 2022, 32, 45–57. [Google Scholar] [CrossRef]
- Anand, R.P.; Lovett, S.T.; Haber, J.E. Break-Induced DNA Replication. Cold Spring Harb. Perspect. Biol. 2013, 5, a010397. [Google Scholar] [CrossRef]
- He, L.; Lever, R.; Cubbon, A.; Tehseen, M.; Jenkins, T.; Nottingham, A.O.; Horton, A.; Betts, H.; Fisher, M.; Hamdan, S.M.; et al. Interaction of Human HelQ with DNA Polymerase Delta Halts DNA Synthesis and Stimulates DNA Single-Strand Annealing. Nucleic Acids Res. 2023, 51, 1740–1749. [Google Scholar] [CrossRef]
- Malkova, A.; Ira, G. Break-Induced Replication: Functions and Molecular Mechanism. Curr. Opin. Genet. Dev. 2013, 23, 271–279. [Google Scholar] [CrossRef]
- Ravoitytė, B.; Wellinger, R. Non-Canonical Replication Initiation: You’re Fired! Genes 2017, 8, 54. [Google Scholar] [CrossRef]
- Rossi, M.J.; DiDomenico, S.F.; Patel, M.; Mazin, A.V. RAD52: Paradigm of Synthetic Lethality and New Developments. Front. Genet. 2021, 12, 780293. [Google Scholar] [CrossRef]
- Kogoma, T.; Lark, K.G. Characterization of the Replication of Escherichia Coli DNA in the Absence of Protein Synthesis: Stable DNA Replication. J. Mol. Biol. 1975, 94, 243–256. [Google Scholar] [CrossRef]
- Masai, H.; Asai, T.; Kubota, Y.; Arai, K.; Kogoma, T. Escherichia Coil PriA Protein Is Essential for Inducible and Constitutive Stable DNA Replication. EMBO J. 1994, 13, 5338–5345. [Google Scholar] [CrossRef]
- Crossley, M.P.; Bocek, M.; Cimprich, K.A. R-Loops as Cellular Regulators and Genomic Threats. Mol. Cell 2019, 73, 398–411. [Google Scholar] [CrossRef] [PubMed]
- Zaitsev, E.N.; Kowalczykowski, S.C. A Novel Pairing Process Promoted by Escherichia Coli RecA Protein: Inverse DNA and RNA Strand Exchange. Genes Dev. 2000, 14, 740–749. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, M.; Clikeman, J.A.; Bates, D.B.; Kogoma, T. RecA Protein-Dependent R-Loop Formation in Vitro. Genes Dev. 2000, 14, 360–365. [Google Scholar] [CrossRef]
- Kogoma, T. Recombination by Replication. Cell 1996, 85, 625–627. [Google Scholar] [CrossRef] [PubMed]
- Rosenshine, I.; Tchelet, R.; Mevarech, M. The Mechanism of DNA Transfer in the Mating System of an Archaebacterium. Science 1989, 245, 1387–1389. [Google Scholar] [CrossRef]
- Yang, H.; Wu, Z.; Liu, J.; Liu, X.; Wang, L.; Cai, S.; Xiang, H. Activation of a Dormant Replication Origin Is Essential for Haloferax Mediterranei Lacking the Primary Origins. Nat. Commun. 2015, 6, 8321. [Google Scholar] [CrossRef]
- Pace, N.R.; Sapp, J.; Goldenfeld, N. Phylogeny and beyond: Scientific, Historical, and Conceptual Significance of the First Tree of Life. Proc. Natl. Acad. Sci. USA 2012, 109, 1011–1018. [Google Scholar] [CrossRef]
- Sanger, F.; Brownlee, G.G.; Barrell, B.G. A Two-Dimensional Fractionation Procedure for Radioactive Nucleotides. J. Mol. Biol. 1965, 13, 373–398. [Google Scholar] [CrossRef]
- Albers, S.-V.; Forterre, P.; Prangishvili, D.; Schleper, C. The Legacy of Carl Woese and Wolfram Zillig: From Phylogeny to Landmark Discoveries. Nat. Rev. Microbiol. 2013, 11, 713–719. [Google Scholar] [CrossRef]
- Zillig, W.; Klenk, H.-P.; Palm, P.; Pühler, G.; Gropp, F.; Garrett, R.A.; Leffers, H. The Phylogenetic Relations of DNA-Dependent RNA Polymerases of Archaebacteria, Eukaryotes, and Eubacteria. Can. J. Microbiol. 1989, 35, 73–80. [Google Scholar] [CrossRef]
- Zillig, W. Comparative Biochemistry of Archaea and Bacteria. Curr. Opin. Genet. Dev. 1991, 1, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Woese, C.R. Towards a Natural System of Organisms: Proposal for the Domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA 1990, 87, 4576–4579. [Google Scholar] [CrossRef]
- Forterre, P.; Elie, C.; Kohiyama, M. Aphidicolin Inhibits Growth and DNA Synthesis in Halophilic Arachaebacteria. J. Bacteriol. 1984, 159, 800–802. [Google Scholar] [CrossRef] [PubMed]
- Ishino, Y.; Komori, K.; Cann, I.K.O.; Koga, Y. A Novel DNA Polymerase Family Found in Archaea. J. Bacteriol. 1998, 180, 2232–2236. [Google Scholar] [CrossRef] [PubMed]
- Baum, B.; Spang, A. On the Origin of the Nucleus: A Hypothesis. Microbiol. Mol. Biol. Rev. 2023, 87, e00186-21. [Google Scholar] [CrossRef]
- Rinke, C.; Chuvochina, M.; Mussig, A.J.; Chaumeil, P.-A.; Davín, A.A.; Waite, D.W.; Whitman, W.B.; Parks, D.H.; Hugenholtz, P. A Standardized Archaeal Taxonomy for the Genome Taxonomy Database. Nat. Microbiol. 2021, 6, 946–959. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Serdyuk, A.; Allers, T. DNA Replication in Time and Space: The Archaeal Dimension. DNA 2025, 5, 24. https://doi.org/10.3390/dna5020024
Serdyuk A, Allers T. DNA Replication in Time and Space: The Archaeal Dimension. DNA. 2025; 5(2):24. https://doi.org/10.3390/dna5020024
Chicago/Turabian StyleSerdyuk, Anastasia, and Thorsten Allers. 2025. "DNA Replication in Time and Space: The Archaeal Dimension" DNA 5, no. 2: 24. https://doi.org/10.3390/dna5020024
APA StyleSerdyuk, A., & Allers, T. (2025). DNA Replication in Time and Space: The Archaeal Dimension. DNA, 5(2), 24. https://doi.org/10.3390/dna5020024









