Abstract
The ability of a nucleic acid molecule to self-replicate is the driving force behind the evolution of cellular life and the transition from RNA to DNA as the genetic material. Thus, the physicochemical properties of genome replication, such as the requirement for a terminal hydroxyl group for de novo DNA synthesis, are conserved in all three domains of life: eukaryotes, bacteria, and archaea. Canonical DNA replication is initiated from specific chromosomal sequences termed origins. Early bacterial models of DNA replication proposed origins as regulatory points for spatiotemporal control, with replication factors acting on a single origin on the chromosome. In eukaryotes and archaea, however, replication initiation usually involves multiple origins, with complex spatiotemporal regulation in the former. An alternative replication initiation mechanism, recombination-dependent replication, is observed in every cellular domain (and viruses); DNA synthesis is initiated instead from the 3′ end of a recombination intermediate. In the domain archaea, species including Haloferax volcanii are not only capable of initiating DNA replication without origins but grow faster without them. This raises questions about the necessity and nature of origins. Why have archaea retained such an alternative DNA replication initiation mechanism? Might recombination-dependent replication be the ancestral mode of DNA synthesis that was used during evolution from the primordial RNA world? This review provides a historical overview of major advancements in the study of DNA replication, followed by a comparative analysis of replication initiation systems in the three domains of life. Our current knowledge of origin-dependent and recombination-dependent DNA replication in archaea is summarised.
1. Introduction: The Where, When, and How of DNA Replication
The core genetic information processing pathways and associated machinery—which promote cellular life and its propagation—are conserved across the three domains of life: eukarya, bacteria, and archaea. It is now widely accepted that the genomic content of every organism is contained as DNA within the chromosomes of its cells. Before these cells can divide, the entire genome must undergo accurate and timely duplication for its two identical copies to be segregated into the daughter cells. Faithful duplication requires that the genome is replicated only once per cell division cycle. Errors in DNA replication present a threat to not only the viability of an individual cell (i.e., chromosome rearrangements and breakage leading to cell apoptosis) but also to the entire organism—where DNA lesions impede replication progression, resulting in stalled or blocked replication forks. Eventually, the accumulation of sustained DNA damage leads to genomic instability (i.e., global replication stress)—a hallmark of cancer aetiology [1]. Thus, proper genome maintenance is dependent on the cooperation of several tightly linked processes termed the ‘Three Rs’: Replication, Recombination, and Repair.
Much scientific effort has been directed at understanding how the assembly of replication machinery is coordinated in space and time, and what safety mechanisms are activated should any errors arise. Given the prevalence of genomic instability in human disease, it is unsurprising that DNA replication constitutes one of the most active research areas in today’s field of molecular biology. Therefore, this section aims to provide a historical perspective on the period of uncertainty (i.e., the ‘Replication Problem’) and the subsequent burst of research (i.e., the ‘Molecular Biology Revolution’) that has led us to the latest guiding paradigm—a theoretical framework—for DNA replication mechanisms.
1.1. The DNA Replication Problem
The breakthrough marking the beginning of molecular biology was when the ‘transforming principle’ from pneumococcal bacteria was discovered to be made of DNA, rather than proteins [2,3]. Avery and colleagues provided the initial evidence of the genetic material’s chemical composition. The implication of DNA playing a role in the transmission of genetic information—regarded as an axiom today—presented a theoretical challenge against the existing protein-centred hypothesis [4,5]. Despite the isolated substance being resistant to trypsin, chymotrypsin, and ribonuclease agents, Mirsky argued that there was a possibility of protein impurities in Avery’s samples, thus igniting a long debate on the true nature of the transforming principle. Because there was no counter-evidence, the ability of DNA to transform all organisms was regarded as a working hypothesis for almost a decade.
The main obstacle to the acceptance of Avery’s work was that the DNA polymer was thought of as ‘too simple’: how can a single molecular entity consisting of a four-base nucleotide sequence permit such diversity in genes across all kingdoms of life? The prevailing idea at that time—promulgated by Levene in the early 1920s as the ‘tetranucleotide hypothesis’—was that DNA was composed of a linear sequence of four repeating nucleotides found in equal amounts: adenosine, thymine, guanine, and cytosine [6]. Chargaff experimentally established that the amount of adenine was equal to thymine, and cytosine to guanine [7]—with the respective ratios differing across species—thereby building upon Levene’s tetranucleotide hypothesis. Hershey-Chase’s experiments supporting DNA’s genetic role [8] were readily accepted despite the 25% of protein contamination [9], and provided an essential clue. Prior to the defining series of experiments, the role of nucleic acid in phages as the “essential, autocatalytic part”, with the protein being only necessary for cell entry, had already been suspected [10]. This assertion was supported by previous studies of quantification of DNA in plant nuclei, which varied in specified amounts across different strains, as well as during the cell division cycle. The authors therein have prematurely described DNA as the “component of a gene” [11]. All what was needed, at that time, was an experiment that would show just that. It was in fact Roger Herriott, who had written a letter to Hershey containing a lucid prediction of the nucleic acid being injected by the virus as the transforming principle, who influenced Hershey to devise the ‘blender ‘experiment [3]. The following year, genes—previously seen as a hypothetical abstraction—were ‘rediscovered’ as concrete, structural entities. It was common knowledge that the key to understanding the biological processes of heredity was contained in the structure of DNA. Watson and Crick [12] were the first in the race to apply the available crystallographic [13,14] and biochemical data [15], and assign a right-handed double helix model [16] to the enigmatic molecule. Assuming that ‘form follows function’, they proposed that the complementary nucleotide bases between coiled helices were held by hydrogen bonds (eventually termed Watson–Crick interactions).
Another important consequence of the model was that it implied a self-duplicating mechanism, whereby one strand of the helix acts as a template to direct the synthesis of the new strand through phosphodiester bond formation between the sugars and the nitrogenous bases, arranged in an antiparallel orientation. Their suggestion raised contention among other leaders in the field who noticed a problem with their model; namely, the mechanism of helical unwinding through hydrogen bond breaking before synthesis [17,18]. Results from autoradiographic studies—consistent with Levinthal’s labelled phage replication models—have further shown the same semi-conservative mode of DNA replication to be present in higher eukaryotes [19].
Delbrück, however, drew attention to the plectonemic coiling of the helix and argued against the semi-conservative replication by suggesting an alternative dispersive mode (see Figure 1). The first hint at the replication debate was settled when Meselson and Stahl provided evidence of semi-conservative replication [20]. Watson concluded: “Nor does the need to untwist the DNA molecule to separate the two intertwined strands represent a real problem”(for a review see [21]).
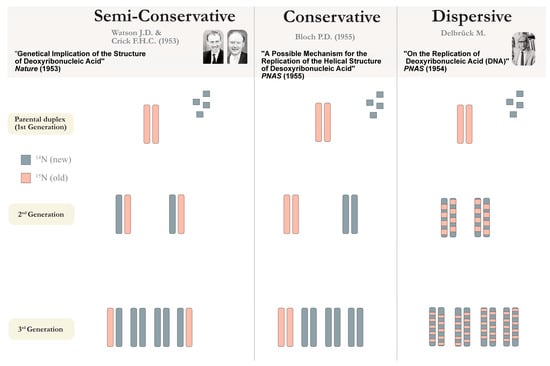
Figure 1.
Proposed mechanisms of DNA replication: Semi-conservative, conservative, and dispersive. The schematic represents the expected outcomes according to each mode of replication, represented by the pioneering groups during the molecular biology revolution, as proposed by Levinthal in 1956 [17,21]. In semi-conservative replication, the two parental strands separate, with each strand acting as a template to direct the synthesis through complementary base pairing, with the resulting daughter duplex consisting of the newly synthesised, and parental strands. In conservative replication, the original duplex is conserved, and in dispersive mode, the double helix remains unwound, while segments break and re-join through crossing over, thus the newly synthesised DNA appears ‘dispersed’ in the daughter strands. Meselson and Stahl demonstrated semi-conservative replication by taking an alternative approach to radioactive labelling (in contrast to the Phage group’s use of bacteriophage, rendering inconclusive results [22])—and growing E. coli cells in 14NH4Cl/15NH4Cl media containing 15N (‘heavy’) and 14N (‘light’) nitrogen isotopes to measure the gradient densities every generation. This elegant experiment was conducted using a combination of Avery’s DNA isolation, density labelling, and density-gradient centrifugation techniques.
1.2. The Polymerase Puzzle
The key evidence for DNA’s genetic role, and its semi-conservative mode of replication, came from Kornberg’s lab, where the chemical process of DNA synthesis was reconstituted in vitro, followed by the purification of the “catalytic extracts”, which contained the enzyme required for phosphodiester bond formation and chain elongation—discovered as DNA polymerase—in Escherichia coli [23,24]. Previous analyses have shown that the precursor to strand formation must be an activated nucleoside 5′-phosphate [25]. Analogous to glucose-1-phosphate being activated to uridine-diphosphate glucose in glycogen synthesis [26], Kornberg’s group generated four 32P labelled nucleotide bases—dATP, dCTP, dTTP, and dGTP—to serve as starting units for the synthetic DNA strand extension reaction. From that, they formed their initial hypotheses regarding the enzymatic mechanisms and the chemical composition of the replication products.
(1) Is the synthesised DNA strand identical to its template?
Does DNA synthesis proceed in a template-directed manner like Watson–Crick’s model would suggest, and is the newly synthesised DNA therefore a complementary copy of its template? The ‘nearest neighbour’ technique of 32P labelled nucleotides revealed that the frequency of nucleotide pairs, and the complementary base ratios between the ‘starting’, and the synthesised strand remained identical, serving as corroboratory evidence for the antiparallel orientation outlined in the double helix model. During enzymatic DNA synthesis, a new strand complementary to the existing template strand is synthesised from the 5′ terminus of the existing RNA primer at the beginning of the nascent strand. The authors were surprised to find that all four nucleotide bases, as well as DNA polymerase and Mg2+, were required; if the template substrate served as a simple primer, why were all four nucleotides a necessity? This has prompted further questioning:
(2) Does replication proceed in a template-directed manner as predicted by Watson and Crick, catalysed by DNA polymerase?
When “DNA primers” containing differing ratios (i.e., 0.5 to 1.9) of nucleotide base pairs were used—the synthesised product maintained the initial nucleotide pair ratios and was independent of the concentrations of the individual bases, thus indicating template-directed replication.
These conclusions have laid the foundation for DNA replication research using bacterial models that occupied scientists for the next 70 years and counting. During the Nobel Prize acceptance lecture, Kornberg compared DNA synthesis to a “tape recording” in that:
“exact copies can be made from it so that this information can be used again and elsewhere in time and space.”
But how, during DNA polymerase-directed synthesis, are these newly synthesised duplex copies made faithful to the original duplex? And how is primer strand synthesis made complementary to the template strand?
Considering that the nucleotide pool contains an unequal proportion of the four bases, what are the regulatory mechanisms that ensure accurate nucleotide selectivity? At the base-pairing selection step: (1) the correct nucleotide must be selected for the polymerisation reaction through correct geometric pairing with the polymerase, and (2) the preceding nucleotide in the primer terminus is “proofread” for accurate base pairing before the addition of the second nucleotide (see Figure 2).
While preparing their manuscript, Kornberg’s group faced a problem: they were unable to remove deoxyribonuclease activity from the polymerase. It was later found that the culprit was the activity of the forward exonuclease (5′ to 3′), which is active throughout DNA synthesis. After removing the enzyme with proteolysis [27], this revealed a second 3′-5′ exonuclease, which carried out proofreading, as well as nucleotide excision (i.e., editing) mechanisms, by excising the mispaired nucleotide at the 3′- end of the primer. When the true replicative enzyme—called DNA polymerase III (Pol III)—was isolated from E. coli, it took on the role as the primary enzyme responsible for the elongation of the majority of the bacterial chromosome. This leads to another universal hallmark of replicative (i.e., proofreading-capable) DNA polymerases—that is, the inability of de novo synthesis. Replicative polymerases are incapable of performing the initial phosphodiester bond formation between two dNTPs—in contrast to RNA polymerases—thus they must add nucleotides to a pre-existing RNA primer site at the template, synthesised by a specialised RNA polymerase called primase, and extend it from 3′-OH end of the single-stranded DNA template strand (n.b., implications of the 3′ prime end requirement form a recurrent theme throughout this review, and is made relevant in various systems). RNA polymerases contain a single-nucleotide long primer within an internal site, which eradicates the need for a primer, at the expense of their proofreading abilities.
At that time, whether DNA synthesis was template-dependent was a contested topic. Kornberg’s mentor, Severo Ochoa, had reported that polynucleotide phosphorylase (i.e., a type of RNase) polymerises NDPs into random polymers under non-physiological conditions; the result of which had halted Kornberg’s initial progress in his in vitro DNA synthesis experiments. Nonetheless, the joint recipients of the 1959 Nobel Prize award for “their discovery of the mechanisms in the biological synthesis of ribonucleic acid and deoxyribonucleic acid” were Arthur Kornberg and Severo Ochoa. Contrary to Kornberg, Ochoa demonstrated in vitro RNA synthesis by RNA polymerase to be template-independent.
Since then, sequence conservation [28] and biochemical [29] studies have led to the classification of DNA polymerases from all three domains of life into six families: A, B, C, D, X and Y—with the first four polymerases responsible for high fidelity DNA replication, while X and Y are more specialised forms of lesion bypass, and translesion synthesis polymerases involved in DNA Repair [30]. PolA, PolB, and PolC are homologous to Pol I, Pol II, and Pol III families in E. coli [31], respectively, with family B most commonly found in eukaryotes, families A and C in bacteria, and families B and D in archaea.
Despite differences in function between polymerases, the archetypal DNA-dependent polymerase (Figure 2B) is composed of a core polymerisation catalytic site, which itself is composed of fingers, palm, and thumb subdomains, as well as a separate 3′–5′ exonuclease domain that proceeds in the opposing direction to DNA synthesis. In case of a mismatched base pair, the catalytic step is slowed down, and the nascent strand terminus is ‘shuttled’ from the polymerisation to the exonuclease active site of the DNA polymerase (Figure 2C) for the excision of the incorrect nucleotide through bond hydrolysis. Such structural distribution of enzymatic function is exemplified by the crystal structure of the multidomain E. coli Pol I Klenow fragment [32], which retains 3′–5′ exonuclease (proofreading) and 5′–3′ polymerisation activities, thereby contributing to DNA synthesis fidelity through intrinsic proofreading and strand displacement synthesis abilities [33].
It is worthwhile to note that the polymerase is a molecular motor capable of translocation along the template strand, which proceeds chiefly in terms of chemical thermodynamics. In other words, DNA polymerase acts as a “channel” for the copying of genetic information, by the “reading” of each nucleotide on the template strand, and “writing in” of the complementary nucleotide through a nucleotidyl transfer reaction, where the paired nucleotides are stabilised by hydrogen bonds and base stacking interactions. This ability to convert “information” through a physical reaction or “work” has led some authors to propose that the polymerase functions analogously to Maxwell’s demon [34,35]. The “memory” of an organism’s genetic information is embedded within the DNA polymer’s structure, where DNA replication is the reversible process of “retrieving” and “storing” of this information—with information processing and assimilation being the defining features of a complex system or a living organism. The RNA-first scenario proposes that while the modern genetic apparatus is encoded by proteins, in the early pre-DNA environments (i.e., the RNA world), the ancient ribozyme harboured the ability to self-replicate; the DNA molecule—due to its inherent stability—has replaced RNA as the main genetic material. The structural similarities of polymerase families A and B, as well as viral RNA polymerases all suggest a common origin [36].
Following this reasoning, the polymerase is the earliest form of a self-reproducing system that has evolved from prebiotic conditions; whose ability to harbour both “information” and “function” has been the driving factor of evolution itself. Thus, it can be assumed that the basic physicochemical forces underpinning DNA replication are both conserved and fundamental in all living systems. Various kinetic studies (for a review see [37], and citations therein) using DNA polymerases have therefore led to a minimal model [38] of the polymerisation process; its mechanics are outlined in Figure 2A.
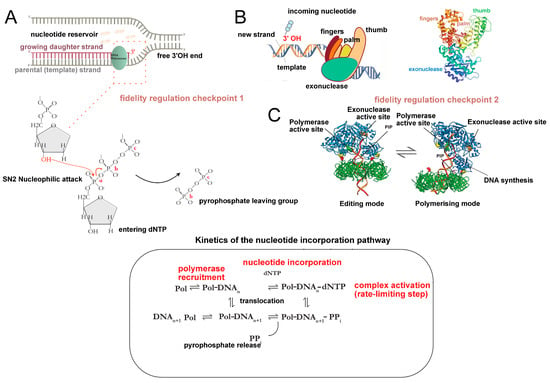
Figure 2.
(A) Universal mechanism of nucleotide incorporation during the polymerisation step in DNA replication. Among all studied replicative polymerases, phosphodiester bond formation occurs via a conserved stepwise mechanism. (B) The side chains of the ‘fingers’ domain (refer to diagram (B); schematic of the polymerase multidomain organisation (left); crystal structure of E. coli Pol I Klenow fragment (right), adapted from [28] (PBD ID:1KFD)) bind the incoming dNTP, and position it in the conserved palm domain (i.e., the catalytic unit). The active site of the palm contains two essential aspartic acid residues that coordinate the two divalent ions necessary for the nucleotidyl transfer reaction: the activated 3′OH on the nascent strand terminus performs a nucleophilic attack on the α-phosphate of the dNTP, thus resulting in phosphodiester bond formation through a condensation reaction. The inorganic pyrophosphate group (PPi) bond is hydrolysed, and the free energy change ensures forward translocation of the polymerase along the template. (C) The dNTP substrate can only undergo activation in its 5′ position, which is what imposes the strict unidirectionality of DNA replication. What is the reason behind this universal requirement if the 5′-OH is just as capable of a nucleophilic attack? The answer lies in the proofreading function of the polymerase; the addition of one nucleotide per synthesis step ensures fidelity, and polymerase repurposing for multiple enzymatic reactions without dissociating from the DNA is bioenergetically convenient. If the energy-carrying 5′triphosphate had been on the nascent strand, rather than the incoming nucleotide, then an additional pyrophosphate-recharging step would be required to activate the 5′ terminus before the next synthesis step. The thumb domain assists in the switching of the polymerase between polymerisation to editing modes ((C); tertiary structure of the Pyrococcus furiosus PCNA-PolB-DNA (PCNA = Proliferating Cell Nuclear Antigen) complex switching from pol and exo states, taken from [34]).
Polymerase selectivity is one of the major contributors to the overall fidelity of replication, with the proofreading function increasing the accuracy of copying by 102–103 fold at the nucleotide level [39]. On a more global scale, however, the order of replication events must be regulated both temporally and spatially. The genome must be replicated during the synthesis (S-phase) stage before cell division, and at the same time, replication must occur only once per cell division cycle to avoid over-replication. Aberrant replication initiation events can lead to chromosome copy number alterations (i.e., aneuploidy or polyploidy) and promote genomic instability through the accumulation of mutations. Thus, the formation of the replication bubble must occur at a specific locus of the chromosome—dictated by the location of the replication origin—and proceed in a timely manner in accordance with the cell division cycle, as well as transcription and DNA repair events [40]. It is therefore unsurprising that the main regulatory step through which this is imposed is replication initiation.
2. The Replicon Model: Leading Paradigm for the Study of DNA Replication
It is helpful to think of the initiation of any biological event as a result of the direct or combined action of regulatory elements; on specific substrates, as well as the negative or positive effects these elements elicit upon binding. Early models of gene expression control were centred around its repression—for example, the (lac) operon model of bacterial gene regulation, as proposed by Jacob and Monod [41], states that gene expression is controlled by a regulatory circuit formed through specific interaction between a trans-acting repressor factor and a cis-acting operator. The authors reached a—what may currently seem rather short-sighted—conclusion that these genetic control mechanisms operate solely through inhibition and that the removal of these repressive effects is the main event that activates protein synthesis. With the lack of integrative approaches, progress in bacterial cell biology research had come to an impasse; there was a fundamental gap in knowledge on the integrative action of molecular mechanisms within the cell. Jacob and colleagues had expressed this growing sentiment at the 1969 Cold Spring Harbor symposium [42]:
“we still know very little about the general system which integrates cellular controls, the regulation of DNA replication, the formation of bacterial membrane, and the process of cellular division with its equipartition of the DNA copies”
Following the discovery of extra-chromosomal, self-replicating genetic elements—called episomes, a term now used interchangeably with plasmids [43]—Jacob et al. [44] proposed a simple replicon model for replication initiation in E. coli circular chromosome. In their model, an individual unit of replication—the replicon—is defined by the specific chromosomal sequence called a replicator (i.e., replication origin or ori; ‘operator of replication’) [45], from which replication is initiated upon the interaction with the trans-acting, diffusible initiator protein (whose own structural gene is found frequently in proximity to the native origin) in a sequence-specific manner. This, in turn, triggers the recruitment of a helix-unwinding enzyme called helicase that acts as a stable platform for the assembly of the replication machinery—collectively referred to as the replisome—in a concerted manner forming a replication bubble from the single strands for elongation to occur (see Figure 2A,B). A defining feature of the replicon unit is that it encodes specific determinants (that is, the replicator and the initiator), which allow it to process control signals allowing it to autonomously replicate as one whole. In fact, a phenomenon called plasmid incompatibility arises when both plasmids cannot coexist in one cell, as they possess replicons with specificity for the same initiation factors, thus leading to unstable inheritance, with one or both of those plasmids eventually eliminated from the cell line [46].
An observant reader may notice that the replicon is a reworking of the earlier lac operon model, combined with the idea of a diffusible factor interacting with the membrane during bacterial conjugation [47]—the operon repressor is analogous to the initiator, and the operator to the replicator, with one critical distinction being that the initiator acts as an activator in a positive interaction with the origin. However, due to the nature of replication being inherently autocatalytic, regulation cannot be complete without the reciprocal actions of both activation and repression mechanisms that occur during distinct stages of the cell cycle. If the rate of replication is determined by the frequency of initiation events, what are the distinct factors that regulate origin firing in space and time?
3. The Divided Genome: Nature’s Riddle
The following section discusses the limitations of the single replicon model—the findings that stimulated its subsequent reworkings, and a revision of the commonly accepted terms, such as replicon unit, origin of replication, etc.
3.1. The Diversity of Replication Factors
The replicon model (see Figure 3) was shown to be highly adaptable to most bacterial systems, with limitations arising when extended to more complex genomes, such as the ones of higher eukaryotes. Due to genetic simplicity (i.e., a circular chromosome with a single bidirectional origin) and ease of culture, E. coli served as the leading model for the identification of ARS (Autonomously Replicating Sequence) elements through the cloning of candidate replicator fragments into a marked plasmid vector, selected for their ability to self-replicate and remain as a separate unit within the host cell. Using this simple ARS assay, the E. coli replicator—Origin of Chromosomal Replication (oriC)—was identified [48], thus making the replicon model a guiding paradigm for replication regulation and origin prediction [49] in bacterial systems. Supporting evidence for the replicon model came from the isolation of the E. coli initiator—a 473 amino acid protein called DnaA [46,47]—which was shown to bind with high affinity to DNA containing the sequence for oriC in an ATP-dependent manner [50]. The DnaA initiator protein, which binds to the specific 9 bp consensus sequence called DnaA box (clustered within the 250 bp oriC region; the DnaA gene itself is usually found adjacent to the origin), and which controls the replication of the entire chromosome, was found to be highly conserved among bacterial species [51]. In eukaryotes, such as budding (Saccharomyces cerevisiae) and fission (Schizosaccharomyces pombe) yeast, the ARS technique developed through bacterial genetics led to the isolation [52] and sequence analysis [53] of yeast ARS elements—100bp long, with a characteristic AT-rich consensus sequence (5′-[A/T]TTTAT[A/G]TTT[A/T]-3′)—that serve as putative replicators. From that, the eukaryotic initiator multiprotein complex ORC (Origin Recognition Complex) was purified from budding yeast in 1992 [54].
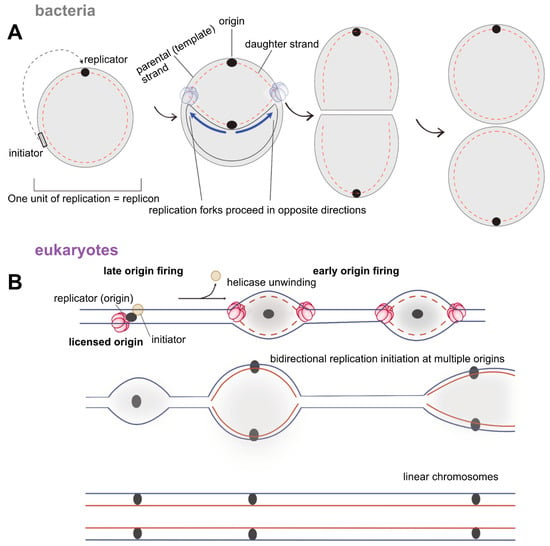
Figure 3.
(A) Early model of the replicon hypothesis in bacterial systems. (B) Adaptation of the replicon model to eukaryotic genomes. The earlier model was reworked to accommodate the multiple origin organisation in eukaryotes, from studies in ARS elements in budding yeast. In eukaryotes, origins are fired asynchronously during S-phase. For an origin to be ‘activated’, it must first be licensed through the recruitment of various replication factors. The licensing of origins during G1 is what prevents over-replication or aberrant re-replication events. Thus, a single set of initiation factors activates hundreds to thousands of replication origins on a single eukaryotic linear chromosome. The initiation signal itself is generated by the cell cycle machinery; namely with the increase of the cyclin dependent kinase or CDK levels.
One may think of a replicator as a specific initiation site or control point for an individual event of bidirectional replication; the single origin model in bacteria served as a useful starting point for the identification of several initiator proteins under set physiological growth conditions. However, E. coli cells were shown to undergo sustained DNA replication, despite the arrested protein synthesis during the DNA damage response (e.g., thymine starvation). This occurs in an origin-independent manner—both oriC and DnaA are shown to be dispensable—and is termed stable DNA Replication or SDR [55].
While the hetero-hexameric ORC initiator is conserved in eukaryotes, with orthologues found from yeast to humans [56], the cis-acting replicators or multiple origins are highly diverse among different species. In the majority of bacteria, the dual DnaA-oriC interaction occurs in a sequence-specific manner to replicate the single circular chromosome. This is in contrast to eukaryotic replication systems, which typically possess many linear chromosomes that are larger in size, and on which there are multiple origins—where one round of replication may initiate from hundreds to thousands of origins, as depicted in early autoradiography studies [57]. The way the replicon model falls short is that it fails to address the spatial and temporal regulation of initiation, which occurs in eukaryotes such as fission yeast, where there is an excess of activation-capable origins and more fluid control mechanisms [58].
3.2. Many Origins, One Chromosome: Time to Revisit the Single Replicon Model?
The replicon model was constructed on the dogma that bacterial domain members can be defined by the possession of a single, circular chromosome that encodes a conserved set of essential genes [59,60,61] (see glossary). However, this paradigm was overturned when alphaproteobacteria containing a secondary replicon carrying essential genes were discovered [62], and the expansion to other members of the bacterial domain stimulated a revision of these historically used terms. Moreover, 10% of bacteria contain more than one chromosome, in addition to the primary chromosome carrying the essential genes—termed a ‘chromid’ (see Box 1 for definition) [60].
Box 1. Glossary of revised terms used in this review. For more detailed descriptions see [59,60].

Carl Woese utilised Sanger’s rRNA fingerprinting technique to compare the sequences of 16S rRNA of different organisms in the 1970s. His tremendous efforts led to the identification of the third domain of life—the archaea [63]—which provided a novel platform for comparative molecular biology. Search similarity techniques to previously known origins in other domains for bona fide origins in archaea have not given results; the nature of the archaeal origin, or if replication was initiated through origins at all, remained unknown long after the archaeal genomes were first sequenced [64]. Since archaea bear a morphological resemblance to bacteria in terms of their chromosomal structure, it was initially proposed that they contain a single replication origin. Indeed, using cumulative oligomer skew analysis, Myllykalio and coworkers [65] have identified the first replication origin (i.e, oriC) in the hyperthermophile Pyrococcus abyssi, corroborated with experimental evidence from two-dimensional gel [66] and RIP (Replication Initiation Point) mapping [67]. The first archaea with multiple origins to be mapped using gel analysis came from the Sulfolobus genus [68], stimulating a major shift in thinking at the time. Through the use of MFA (Marker Frequency Analysis) techniques [69], it was also shown that bidirectional replication occurs from each of the three origins. These three origins were also found to be involved in complex cross-interaction with the adjacently encoded initiator proteins, Orc 1-1 and Orc1-3 [70], as well as a WhiP (winged-helix initiation protein) [71]. However, what makes the Sulfolobus genome especially intriguing is that the genomic region adjacent to oriC3 appears to be ‘captured’ from a virus or an extra-chromosomal element of viral origin (see glossary for extra-chromosomal element). The staggering sequence diversity of the Sulfolobus origins (oriC3) also hints at independent derivation through horizontal gene transfer (HGT) [68].
Similarly to Sulfolobus, other archaeal genomes [71,72,73] were also found to be composed of multiple replicons, with each replicon containing more than one replication origin. What is the exact definition of a replicon or a single replication control point, given the divided genomic architecture and the cross-interaction between multiple replicator–initiator systems? DiCenzo and Finan [59] suggest that classical terms such as ‘replicon’ should be used with caution—if not at all discarded—when describing genomes that fall outside the canonical E. coli model. It was also assumed a priori that genome replication cannot be initiated without replication origins. However some archaeal species, such as Haloferax volcanii, demonstrate that replication without origins occurs faster, and without any phenotypic deficits [74]. In this case, replication proceeds in a stochastic manner; without fixed initiation points on the genome, which appear to be randomly dispersed.
In other archaeal species, experimental deletion or inactivation of origins or initiator proteins results in the activation of secondary replication pathways, or the activation of dormant origins. For example, in the thermophile Thermococcus kodakarensis, deletion of the origin (as well as the Cdc6 initiator protein) resulted in strains that are still capable of DNA replication [75], enabled through an origin-independent mechanism. Moreover, RadA and RadB were shown to be essential in origin-deleted cells, consistent with the Haloferax model, where Recombination-Dependent Replication (RDR) based mechanisms of initiation are employed as well. But what was particularly surprising in the T. kodakarensis study [75] was the apparent failure to detect a defined origin of replication in both wildtype and Δcdc6 cells, evidenced by the flattened MFA peaks. In contrast, the same MFA technique to map origins in Haloferax shows three distinct peaks, denoting origins oriC1, oriC2, and oriC3 [76]. This indicates that (under laboratory conditions) the origin of T. kodakarensis is not used, even when present. However, when levels of the recombinase RadA are reduced, thereby impairing the efficiency of homologous recombination, the origin is then used to initiate DNA replication [77]; similar findings have been made with the related archaeal species Thermococcus barophilus [78], reviewed in [79]. Origin-independent replication is not a newfound phenomenon and has been periodically observed in all three domains of life. In fact, the first genome in which RDR was demonstrated was T4 bacteriophage, which initiates replication from RNA-DNA intermediate structures (i.e., R-loops) upon infection, to then progress to initiate replication through RDR in the later stages of the life cycle [80].
While canonical DnaA-oriC-dependent initiation is a highly conserved mechanism in bacteria, as indicated by the proximity of DnaA box clusters to the oriC region [49], in cyanobacteria, DnaA dependency varies between species. Synechococcus elongatus, for instance, replicates its genome through a DnaA-dependent mechanism, displaying a regular GC skew profile. Conversely, Synechocystis sp. PCC 6803, displays irregular GC skew profiles, thus, suggesting asynchronous replication initiation from multiple chromosomal sites. Moreover, deletion of DnaA in the latter species did not result in any growth defects, nor halt DNA replication [81,82]. Building on the fact that most species of cyanobacteria possess multiple copies of the genome, the authors proposed that DnaA-oriC-independent replication has evolved independently in free-living bacteria, with DnaA being lost from symbionts. It is notable that H. volcanii, T. kodakarensis, and Synechococcus sp. PCC 6803, which can all replicate their genomes in an origin-independent way while suffering no growth defects, belong to different domains—and yet, they exhibit one interesting similarity: all three species have polyploid genomes.
The above cases challenge conventional ideas behind replication initiation, as well as the use of origins themselves, raising the possibility that initiation mechanisms are more flexible than purported in textbooks. Masai, in his review [83], has asked us to reevaluate the orthodox model of replication initiation; he speculates that the ancestral form of DNA replication may have initiated directly from an R-loop, bypassing the need for any initiator–replicator control mechanism.
Thus, we arrive at another critical juncture; what is then the initial evolutionary purpose for replication origins? Are origins of replication ancestral genetic elements or were they recently obtained through HGT? If so, at what point in evolutionary history have origins been captured? And perhaps more importantly, could the extra-chromosomal elements capture postulate be extended to explain the evolution of multiple initiation sites and the linearisation of the chromosome in eukaryotes? The diversity of replication factors (Orc/Cdc6 proteins) and ORB sequences in archaeal species (notably, differences between S. solfataricus, H. volcanii, and Halobacterium sp. NRC-1) suggests independent evolutionary diversification via extrachromosomal element capture [73]. Namely, some origins were recently acquired and had already been present on extrachromosomal elements before they were inserted into the main chromosome. This raises the possibility that the ancient archaeal chromosome did not replicate from fixed sequences, but in an origin-independent manner.
Given the above lines of evidence, the reader might then arrive at the conclusion that the organisation of multireplicon genomes in prokaryotes is far from stochastic—that their maintenance must hold some functional or evolutionary purpose [59]. In fact, genome rearrangements such as insertion–deletion events from mobile genetic elements [84], and origin transfer [73] between species were the driving force that shaped genomic organisation in the Haloarchaea class of archaea. What existing studies have failed to resolve is the reasoning behind the ‘hidden cost’ of the multipartite genome—that is, increased complexity. What are the genetic events that led to the expansion into multiple replicons, and do they confer any advantage to the cell?
Taking the conjecture that the modern eukaryotic cell evolved from a lineage of archaea containing multiple origins, the study of archaeal replication origins can therefore provide an understanding of the complex mechanisms in eukaryotes and potentially give insight into some of the selection pressures present at the primordial times of the Last Universal Common Ancestor(s) or LUCA. For this task, the ideal model would be an archaeon that is easy to culture within laboratory conditions and one which would be amenable to genetic manipulation.
Therefore, the next sections aim to familiarise the reader with the events starting from origin–recognition, leading up in stages to full replisome assembly—with a special focus on the archaeal domain—before continuing into some exceptional cases of replication (e.g., RDR) and their implications.
4. Where Do We Start? DNA Replication Initiation Across the Three Domains of Life
The initiatory steps leading up to replisome formation can be broadly classified into five distinct stages: (I) origin recognition, (II) pre-RC (pre-Replicative Complex) assembly, (III) replicative helicase activation and DNA unwinding, and (IV) loading of replicative DNA polymerases along with other enzymes that support the replisome (V) to ensure high processivity (see Table 1). Stages of replication have been separated for comparative analysis between the three domains of life. Each model organism therein was purposefully chosen to demonstrate the evolutionary transitions in genomic organisation.
4.1. Bacteria
Before the DNA polymerase can associate and extend the DNA strand, the double helix must first be unwound. This requires the assembly of a higher-order nucleoprotein complex (i.e., pre-RC), which will then recruit the helicase. As previously discussed, the typical bacterial origin, oriC, appears once per chromosome for most bacteria, and its sequence is typically encoded adjacent to the initiator protein DnaA. The clusters of high- and low-affinity DnaA
Boxes are contained within the oriC region, which, together with the concentration of the initiator DnaA, are involved in regulating the frequency of initiation (i.e., origin firing) and initiation synchrony [85,86]. The classic mechanism describes a single monomer of DnaA binding to the consensus sequence—consequently named the DnaA box—to induce a ‘bend’ in the interaction site, thereby facilitating DNA melting. However, as with most biological systems, the molecular reality is much more complex. In E. coli oriC, there have been a total of 12 characterised DnaA boxes to this date [87]—all with varying degrees of conservation to the original consensus. Each DnaA protein monomer binds to the respective DNA box: R1, R2, and R4 (high-affinity sites), or I, τ, and C (low-affinity sites, which lie between the R-sites) [40,88].
The DnaA protein is composed of four structural domains: domain I (N-terminal module, which facilitates the recruitment of DnaB), domain II (linker segment), domain III (largest domain, containing the AAA+ ATPase fold), and domain IV (C-terminal DnaA box-binding domain). This brings us to the hallmark feature of the protein: its multimodular structure is what confers it with multifunctionality and the ability to coordinate entire replisome assembly. DnaA belongs to the AAA+ superfamily of ATPases (that is, ATPases associated with various cellular activities), and thus shares an evolutionary relationship with the eukaryotic (Orc1) and archaeal (Orc1/Cdc6; Cdc6 = Cell Division Cycle 6) initiator proteins [89], which bear structural similarities [90]. The levels of DnaA-ATP are regulated in accordance with the cell cycle; during initiation, the active form of DnaA-ATP can bind to low-affinity 9-mer DnaA boxes and oligomerize. After initiation, DnaA-ATP is subsequently hydrolysed into its inactive form, DnaA-ADP—an autoregulatory mechanism that prevents over-initiation of bacterial replication [80,81].
The binding of the integration host factor to its binding site causes a sharp bend on the dsDNA, thereby facilitating DnaA binding with the DnaA-initiator-associating protein at the DnaA oligomerization region, leading to the unwinding of the adjacent AT-rich region—termed the DUE (DNA unwinding element) [91,92]. This creates a stable open complex structure or ‘bubble’ (i.e., the pre-RC) to which the helicase loading protein, DnaC, through its interaction with domain I of DnaA, binds two hexamers of DnaB helicase and loads them onto the ssDNA (single-stranded DNA) region, at opposite orientations. The helicase then recruits DnaG primase, which itself binds to the DnaB-DnaC complex, thus leading to ATP-ADP hydrolysis, stimulating helicase activation. The DnaB pair of helicases unwind with the directionality of 5′ to 3′—hence the helicases work in opposing directions, and establish a bidirectional replication fork to which replication machinery can be loaded (for the latest overview of bacterial initiation, see [93]).
4.2. Eukaryotes
Contrary to their bacterial counterpart, progress to fully characterise the eukaryotic origins and initiation process was lagging. This points to an obvious difference: the size of the genome. Take a simple model eukaryote—S. cerevisiae or budding yeast—and compare it to the bacterial model of E. coli: the genomes are 12.2 Mb v. 4.6 Mb, respectively. As the DNA strands are unwound and chromatin is disassembled with every replication cycle, genotoxins have greater access during these phases of the S-stage of the cycle. The speed of accurate DNA replication thus becomes an important point. While E. coli, with its small genome, single point of origin, as well as fast-moving replication forks of 30 kb per min, shortens the duration of this stage, eukaryotes have evolved to have a chromatinised genome composed of multiple replicons.
Hence, the first point of contrast in eukaryotic initiation is increased spatiotemporal control to ensure accurate replication of a larger genome. There are multiple origins on a single chromosome—with increased flexibility of initiator interaction as origins become less defined and have less sequence conservation (with the notable exception being S. pombe, which compared to S. cerevisiae, lacks distinct sequences, apart from rich AT regions [94]). Thus, we witness another emerging trend: with the increasing number of origins, there is an overall increase in their flexibility, when it comes to origin selection for activation (see Figure 4). With each origin activation cycle, there is a ‘pool’ of dormant origins, which are reserved for cases when the ‘primary’ origins become inactivated, or during specific growth conditions (i.e., as a DNA damage response). Thus, only a subset of origins becomes activated in a stochastic manner, with origin selection largely governed by changes in the chromatin structure. Schwob [95] posits an intriguing explanation: accumulation of recombination intermediates at replication origins in fission yeast drives genomic instability, which in turn may have promoted replicator diversification and redundancy as a counteractive mechanism [96]. Although pre-RC origin selection displays flexibility, the tight regulation of origin firing, corresponding to the stages of the cell cycle and influenced by additional epigenetic factors, is an ongoing question of current investigations (for a recent review, see [97]). Once per mitotic cell cycle, the genome must be replicated with utmost precision due to the selective pressure of genomic instability and cell death as a result of over- or under-replication. This is reflected in the tight control mechanisms that couple the process of initiation to the stages of the cell cycle, which are centred around preventing re-replication.
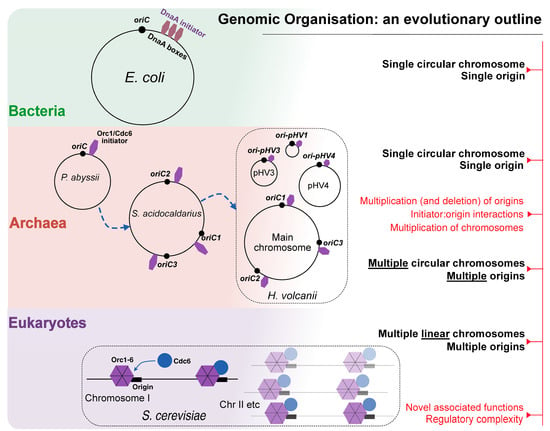
Figure 4.
Evolutionary timeline of replicator diversification across the three domains of life. Figure adapted from [95].
The major difficulty that came in characterising eukaryotic origins is that there are multiple origins on a single chromosome that lack discernible sequence motifs and that the origins in higher eukaryotes are largely defined through complex chromatin interactions (i.e., a subset of origins, termed a ‘cluster’, can be activated according to the developmental phase [98]). This led to the development of the two-state model of initiation (refer to Figure 5), which corresponds to the levels of CDK (cyclin dependent kinase) activity: the origins are ‘licensed’ and the pre-RC established during the G1 phase of low CDK and increased DDK (DBF4-dependent kinase or Cdc7) levels, and then subsequently activated during S-phase [99]. Analogously to previously defined bacterial systems, the ORC—a six-subunit AAA+ ATPase—binds to the ARS sequence in an ATP-dependent manner. However, unlike DnaA, ORC-ATP binding cannot directly unwind the DNA region [100]. Upon ORC binding, Cdc6 [101]—a factor displaying sequence homology to the ORC subunit Orc1, suggesting common ancestry, is recruited to form a ring-shaped structure. Concomitantly, the Cdt1 (chromatin licensing and DNA replication factor 1) initiator protein [102,103] acts as a chaperone to recruit the MCM2-7 helicase; together, this forms the intermediate ORC-Cdc6-Cdt1-MCM2-7 of the pre-RC, where the dsDNA can feed into the pore of the resulting MCM double hexamer (MCM = Minichromosome Maintenance Complex) [104]. Like DnaB, the MCM molecule must also be activated through ATP hydrolysis reaction. Many MCM hexamers are loaded following ATP hydrolysis by Cdc6 and ORC, with Cdt1 release in an iterative fashion [105] (for an excellent review on MCM loading, see [106]). We hence arrive at another control point: the activation of the MCM2-7 helicase (the ‘core’) depends on the additional proteins Cdc45 and GINS, and together, they form the CMG (Cdc45-MCM-GINS) complex, which acts as a replicative helicase [107]. The transition from the G to S-phase of the cell cycle is guarded by the increased activity of Cdc7 and CDKs. Cdc7 directly phosphorylates the N-terminus of the MCM2-7, alongside a tripartite complex consisting of Sld2-Sid3-Dbp11 factors (SDS complex) [108], which mediates CMG formation and activates the helicase. The above stepwise model is what constitutes the ‘origin firing’ step; the duplex is unwound, and replicative polymerase ε (pol-ε), alongside other replisome components, is loaded.
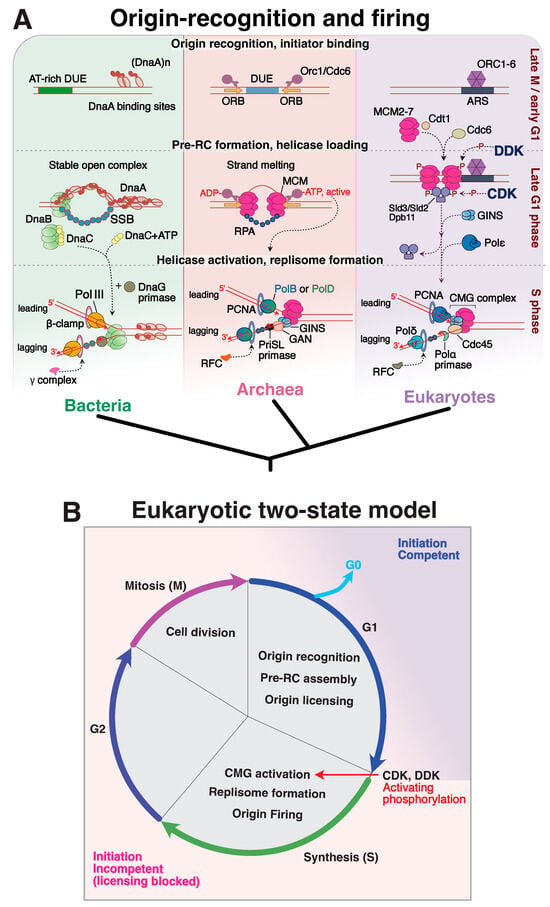
Figure 5.
(A) Replication initiation mechanism and associated replication factors—from origin recognition to full replisome assembly—across the three domains of life. (B) A schematic diagram representing the temporal control of DNA replication stages in eukaryotes. Origin licensing through phosphorylation by various CDKs serves as a major control point for the transition between the G1 to S stage of the cell cycle; hence the two-state model provides a temporal window in which origins are ‘initiation competent’ in blue, and initiation incompetent in pink.
4.3. Archaea
Archaeal chromosomes are circular and small, akin to bacteria, yet share homology with eukaryotic replication factors. The archaeal domain hence represents a unique fusion of bacterial and eukaryotic features. Identification of replication origins revealed a diverse picture of archaeal genomic architecture, ranging from one (e.g., Pyrococcus abyssi) to as many as four origins on a single chromosome (e.g., Pyrobaculum calidifontis), alongside extrachromosomal elements such as megaplasmids and multiples homologues for Cdc6 (for a review, see [109,110]. In 1997, archaeal genes homologous to the eukaryotic cdc18+/CDC6 gene family were first discovered to be transcribed along with pol genes encoding a novel polymerase (i.e., DNA Polymerase II or Pol II) in the hyperthermophile Pyrococcus furiosus. An intriguing suggestion was made by the authors: the genes encoding the polymerase were found to be arranged in tandem with the eukaryotic initiator homologues, as well as the Dmc1/Rad51A gene family, which play a role in genetic recombination, thus suggesting a potential role of Pol II as the mediator between the linked processes of replication and recombination. Subsequent studies of the Pyrococcus genome have confirmed the region of the archaeal origin to contain archaeal homologues of Orc1/Cdc6 initiator genes, as well as confirmation of the linkage between oriC and cdc6 genes [65,66].
This led some authors to speculate that the eukaryotic Cdc6 and archaeal Orc1 have diversified from a gene duplication leading back to a common ancestor [111]. Interestingly, the same study found that the promoter region for the DNA polymerase subunit genes (i.e., DP1 and DP2) overlapped with the Pyrococcus oriC sequence, providing a first hint at the replication initiation control through transcription [112]. The same year, a mutational analysis and sequence alignment study proposed a structure of archaeal Cdc6 ortholog, and its functional implications in pre-RC assembly [113]. The crystal structure of Pyrococcus cdc6 protein reveals its multidomain organisation; with domains I and II having an AAA+ ATPase module and domain III being composed of a winged-helix (WH) fold. Soon after the initial discovery, the postulated mechanism of origin-binding was confirmed in vitro [114]. The purified Orc protein was shown to bind to the origin-recognition sequences termed Origin Recognition Boxes or ORBs (a conserved 13-base repeat), as well as mini-ORB elements in Sulfolobus [68] flanking the AT-rich DUE (DNA Unwinding Element) within the origin region [67].
The inverted position of the ORBs on either side of the DUE is what precisely determines the polarity of Orc binding. The Orc initiators bend the DNA through their N-terminal AAA+ domain; an extra layer of complexity is added through varying binding affinities between Orc proteins determined by its WH domain. Initially, in vitro studies [115] in P. furiosus led authors to prematurely conclude that Orc binds in an ATP-independent manner, with the resulting structural distortion [116] of the binding site leading to the unwinding of duplex. Intuitively, one would presume that the binding mechanism is analogous to that of DnaA within the bacterial domain. While the initiator–origin recognition motif interaction is conserved, duplex unwinding upon Orc binding, helicase recruitment mechanisms, and higher-order complex assembly remain a contested topic. This is partly due to the differing methods used to study initiator–origin binding mechanisms. Biochemical studies [40,115,117,118] support strand unwinding upon Orc binding, leading to higher-order assembly, while early structural analyses present an obvious conflict. Some authors support DNA unwinding following strand distortion due to the topological stress induced by AAA+ domain binding [116], while others assert that the base pairing is maintained even after strand distortion [116,119]. This discrepancy persists within other species of archaea: biochemical analysis in Methanothermobacter thermautotrophicus [120] and Aeropyrum pernix [120] support higher order complex assembly, while Sulfolobus appear to be in contradiction (reviewed in [109], p. 60). Nevertheless, it became apparent that Orc binding and the subsequent topological changes serve as an important step in initiation; yet again, we see that the archaeal initiator mirrors the eukaryotic ORC in its main role of helicase recruitment rather than the direct origin melting of DnaA.
Contrary to early Pyrococcus studies [115], Orc needs to be ATP-bound for its activation; however, in vitro studies in the same species have shown that the loading of the helicase itself occurs via an ATP-independent mechanism [121]. Soon after MCM2-7 emerged as a candidate for the eukaryotic helicase, a number of MCM homologues were identified in archaea, with each species containing at least one homologue (for a review, see [122]). Although the biochemical properties of the archaeal MCM were known—that is, 3′ to 5′ DNA translocation capabilities, ssDNA and dsDNA binding, and ATPase activities [123]—the mechanism of MCM loading by Orc remained to be elucidated. Work from Bell lab—consistent with earlier chromatin immunoprecipitation studies [66,114]—has shown that the homohexameric open-ring MCM directly binds to the ATP-bound Orc protein in vitro [124,125]. Here, ATP binding and MCM release following ATP hydrolysis serve as a regulatory switch to confer MCM loading to a particular temporal window: a primitive version of spatiotemporal control observed in eukaryotes. Recent atomic force microscopy techniques provided further experimental verification that MCM from Methanothermobacter can interact with DNA in a variety of conformations under physiological conditions [126]. An important distinction from the eukaryotic MCM2-7, which is only active as part of the CMG (Cdc45-MCM-GINS) complex, is that the archaeal MCM displays intrinsic helicase activity in some species [127]. In others, paradoxically, MCM requires the binding of cdc6 homologues to be activated [128].

Table 1.
Overview of replication machinery found across the three domains of life. The assembly of the replisome is separated into 5 stages (stages I–V), and the relevant replisome components active in each stage are listed according to the domain of life it is found in; that is, in Archaea, Bacteria, or Eukaryota.
Table 1.
Overview of replication machinery found across the three domains of life. The assembly of the replisome is separated into 5 stages (stages I–V), and the relevant replisome components active in each stage are listed according to the domain of life it is found in; that is, in Archaea, Bacteria, or Eukaryota.
| Replisome Assembly Step | Eukaryotes | Archaea | Bacteria |
|---|---|---|---|
| STAGE I Origin Recognition | ORC (Orc 1, 2, 3, 4, 5, 6) | Orc/Cdc6 a,b | DnaA |
| STAGE II Pre-RC formation | Cdc6/Cdt1 | Orc/Cdc6 a,b WhiP b | DnaA |
| STAGE III DNA duplex melting | Orc/Cdc6 MCM helicase (Mcm 2, 3, 4, 5, 6, 7 heterohexamer) | Orc/Cdc6 a,b MCM helicase a,b (homohexamer) | (DnaA)n |
| STAGE IV Helicase Loading | Cdc6 Cdt1 helicase loader | Orc/Cdc6 a,b helicase loader | DnaC helicase loader (DnaI in gram positive bacteria) DnaB helicase (DnaC in gram negative bacteria) |
| STAGE V Polymerase— Replisome assembly | CMG complex PCNA clamp RFC clamp loader PriSL primase B-family polymerases (Pol ε, Pol δ) | GINS a,b PCNA clamp RFC clamp loader PriSL a,b/PriX b primase B-family polymerase a,b, D-family polymerase a | β clamp DnaG primase C-Family polymerase (Pol III) |
| Recommended Literature | [106] | [111] [129] (focus on H. volcanii) | [93] |
a euryarchaeaota. b crenarchaeaota (n.b., old nomenclature, now referred to as thermoproteota).
The rest of the replisome is then loaded: GAN or GINS-associated nuclease (i.e., Cdc45 or RecJ), and GINS factors, which modulate the helicase activity, as well as the PCNA, RFC, primase (PriSL), Replication Protein A, and the polymerases (B/D) [129]. To this date, there has been no successful reconstruction of the archaeal replication machinery in vitro, and archaeal initiation, particularly regulation mechanisms between multiple origins, continues to be an underexplored topic in the DNA replication field (for an excellent review of the history of the archaeal replisome, see [130]. Nonetheless, there has been some interesting progress made in elucidating the full interactome during initiation. The first study to experimentally confirm a functional connection between the archaeal DNA Polymerase D (Pol D) and CMG helicase was from 2022. The subject of the study was T. kodakarensis, which has both Pol B (Family B DNA Polymerase) and Pol D (Family D DNA Polymerase). Pol D is composed of two catalytic subunits—that is, DP1 and DP2. However, what is interesting about DP2 in particular, is that it has been found to share a homologous ‘double-psi β-barrel’ catalytic core with RNA polymerase [131]. Moreover, as Pol B in T. kodakarensis has been shown to be nonessential, it has been proposed that Pol D is the main replicative polymerase that initiates replication on both leading and lagging strands [132]. After their earlier confirmation of Pol D interacting with primase through its DP2 subunit, and switching from de novo synthesis to elongation state [133], Oki et al. (2022) then reconstructed the functional replisome assembly. Two Pol D molecules interact with GINS, through two Gins2 subunits. The authors also speculate that two Pol D molecules could form a complex with GINS, and therefore, can be coordinated with CMG helicase to synthesise the leading and lagging strands simultaneously. Inactivated MCM is first loaded onto the replisome, with the PolD2–GINS1–GAN2 being subsequently recruited—the activated helicase then translocates in the 3′ to 5′ direction along the leading strand template through its ATPase activity [134]. Another study has used several structural imaging techniques to demonstrate the interactions of RPA with PriSL and Pol D, revealing RPA to be one of the central players in replisome assembly [135].
Taking the above evidence together, it becomes apparent that the distribution of functions of replication proteins is highly diverse among archaeal species, as studies reveal a complex interactome leading up to full replisome assembly—an understanding of which still remains fragmentary.
5. DNA Replication and Recombination: A Dynamic Interplay
Adding a Level of Complexity: The Asymmetry of DNA Replication
When the first autoradiograph of the E. coli chromosome in the act of replication [136] was presented—just before the 1963 Cold Spring Harbor Symposium—Monod raised a critical question: How is simultaneous bidirectional replication achieved, given that DNA Polymerase I can only add nucleotides to the hydroxyl end of the strand (i.e., from 5′ to 3′)? The answer arrived decades later, confirming the asymmetric nature of DNA replication in vitro: one strand is replicated continuously (i.e., the leading stand—from 5′ to 3′), while the lagging strand is replicated in the opposite direction, and in segments called Okazaki fragments; that is, semi-discontinuously [137,138,139]. In vivo studies, however, have strikingly shown that both strands were synthesised as pieces when ligase was deactivated. It was only recently resolved that the seemingly discontinuous leading strand synthesis observed in vivo was only due to the ribonucleotide excision repair reactions, which fragmented the nascent DNA into Okazaki-like pieces [140,141]. In bacteria, it was observed that there are more guanine nucleotides compared to cytosines within the leading strand; these strand-specific biases (a technique termed GC skew analysis) can thus be exploited to not only distinguish the leading from lagging strands but also locate putative origins of replication and termination sites in archaea [142]. The lagging strand differs vastly by its enzymology: primase (DnaG in bacteria, PriSL in eukaryotes, and its homologue PriSL in archaea) is required to synthesise the RNA primers (providing a 3′ end for new DNA synthesis), SSB protein to protect the exposed ssDNA, with DNA repair polymerase/flap endonuclease to remove the RNA primers from the 5′-ends of Okazaki fragments, and finally DNA ligase to seal the synthesised fragments together. It becomes apparent that this universal requirement for a terminal 3′-OH group for DNA polymerase-mediated extension is observed in all living forms across all three domains (as well as viruses). Some special cases of replication involve mechanisms that bypass the need for an additional protein primer to initiate synthesis; instead, a continuous mode of replication is adapted, with the 3′OH strand, usually generated through a nick, being used as a direct primer. In fact, some viruses with linear genomes—such as vaccinia and parvoviruses—are able to utilize the 3′OH terminal hairpin sequence as a direct primer for replication through a unidirectional, strand-displacement mechanism [143]. Similarly, simple replicators like plasmids employ rolling circle replication; whereby the nick generated by rolling circle replication endonuclease replaces the need for a primase, thus representing the simplest strategies of replication initiation [144,145].
Taken together, one could simplify strand extension to three fundamental requirements: (1) a terminal hydroxyl group provided by a primer or a recombination intermediate, (2) DNA polymerase, and (3) interactions with additional factors to help load the replisome. [146].
It is hence tempting to speculate that the LUCA relied on using RNA polymerase (particularly due to its innate ability to bond nucleotides within its active site) due to the pressures of using RNA as a sole genetic material during the transition from the RNA world [147], with DNA polymerase being a later invention. Intriguingly, comparative genomics has revealed that the main components of the replisome do not share homology between bacteria and archaea/eukaryotes, with a notable exception of sliding clamps; the primordial cell relied on a separate set of enzymes to replicate its RNA genome [148].
6. Recombination Dependent Replication
The viral origin hypothesis—enunciated by Forterre—states that HGT from mobile genetic elements and viruses has contributed to the evolution of the vast array of replication machinery in archaea and eukaryotes. It is worthwhile to investigate the mechanisms of the differing methods employed to overcome the primer requirement, as the ‘clues’ provided may enable us to better characterise the ancestral features of replication initiation.
6.1. Clue No.1: Lessons from Viral Models
Viruses served as invaluable models of the replisome—for example, Alberts proposed the ‘trombone’ model to explain the coordination of the leading and lagging strands [149,150]. Studies into the life cycle of T4 bacteriophage were the first to propose a connecting link between replication and recombination and initiated a research line into recombination processes, which were regarded as a rudimentary ‘cut-and-paste’ mechanism [151]. As early as 1980, Mosig (for a review of the author’s work and citations therein, see [152]) suggested that the replication of the bacteriophage occurs through homologous recombination. In the early stages, the replication is initiated from fixed origins; however, at a later stage of the process, the very 3′ end of the lagging strand cannot be replicated. This results in the recruitment of the DNA strand exchange protein called UvsX to the 3′ ssDNA, thereby resulting in the formation of the D-loop (Displacement Loop) through strand invasion. A D-loop can therefore be defined as an intermediate structure that is formed during processes involving homologous recombination, whereby a single strand invades the dsDNA molecule in a strand exchange event. A similar mechanism is employed as part of the natural life cycle of bacteriophage T4. Although in the early stages of the cycle, most origins are used—some origins utilise the 3′ ends of RNA displacement loops or R-loops (i.e., a three-stranded nucleic acid structure, which involves an RNA–DNA hybrid from a transcript, displacing a DNA strand—commonly occurring during transcription) to directly prime replication [152,153]. Thus, through the formation of DNA:RNA duplexes, these RNA sequences can be used to initiate DNA synthesis, bypassing the need for an additional RNA primer synthesis. The 3′ ssDNA ends are generated either as a natural part of the replication process or through end processing via the 5 to 3′ exonuclease activity of T4-encoded RNaseHs. The necessity of these DNA breaks for RDR initiation was confirmed using in vivo models of artificially created double-strand breaks (DSBs). Then, the UvsX protein promoted strand exchange (n.b., UvsX has also been noted to be involved in branch migration and complementary DNA reannealing) to form the D-loop (see Figure 6). Several authors have questioned the necessity of this two-way mode of replication, as a similar mechanism has been utilised in bacteria. Is there any functional advantage if de novo replication in T4 bacteriophage requires not only a D-loop formed but also terminal redundancy supplemented by homologous sequences from a second copy of the genome? McGlynn and colleagues [154] reason that RDR maximises phage replication—compared to canonical origin-dependent initiation mechanisms—and constitutes an ad hoc mechanism to overcome replicative blocks and ensure replication restart. This presents the origins of replication as strict control points that have been favoured through evolution to replace a potentially dysregulated RDR initiation mode of replication.
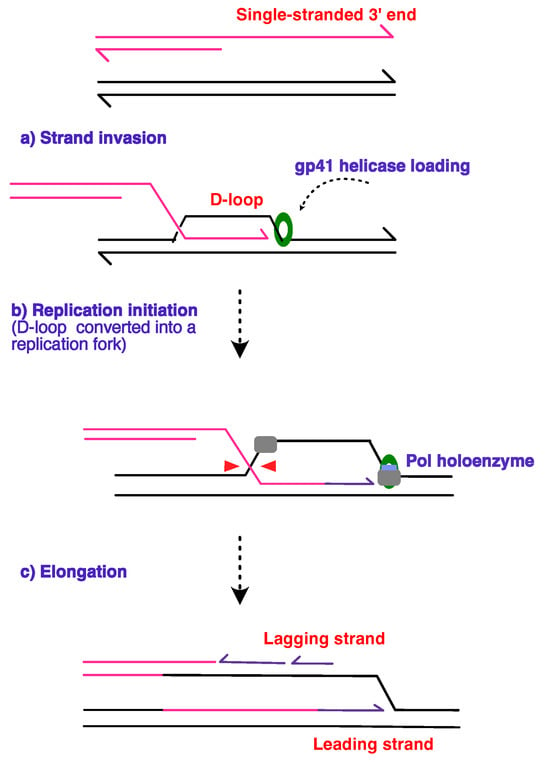
Figure 6.
Schematic diagram outlining the steps in the RDR process that occurs during the T4 bacteriophage lifecycle. The schematic depicts a model of D-loop formation through the (a) strand invasion mechanism proposed by Mosig [148], where the 3′ end of the DNA strand from the previous replication cycle is used to prime and initiate the next round of replication, thus the mechanism is described as self-regenerating. The subsequent cleavage (b) of the D-loop by the junction-cleaving nuclease or T4 gp41 establishes the directionality of the replication fork, followed by the loading of the replicative polymerase and the primer. The (pink) invading strand primes continuous replication (purple) on the leading strand in (c), while the discontinuous line denotes lagging strand synthesis, with the 3′ ends of the strands depicted as arrowheads. Figure adapted from [152].
The UvsX protein in bacteriophages that displays some sequence similarity to the bacterial RecA belongs to the RecA/Rad51/RadA superfamily of recombinases, found within the bacterial, eukaryotic, and archaeal domains, respectively. Bacterial RecA, which belongs to the Rad51-family, and archaeal RadA are all homologous to each other. Although it is tempting to speculate that the viral recombinase follows the same pattern due to some reports of weak homology of UvsX to RecA, structural analyses reveal that RecA has evolved through convergent evolution; UvsX and RecA/Rad51/RadA are orthologous [155].
The pressing problem in the RDR initiation research line is the missing gap between the initial D-loop formation and the molecular mechanisms leading up to full replisome assembly; in all three domains. However, in origin-independent replication in E. coli, DNA footprinting assays have revealed that PriA is not only able recognise the D-loop structure, but can also recruit the φX174-like primasome, leading to the formation of the replication fork [156,157]. PriA belongs to the 3′−5′ DExH helicases of the Superfamily 2 class. It becomes apparent that interactions between the helicase and the recombination intermediate may serve as a potential clue to the full elucidation of the replisome assembly mechanism; however, research into the interactions that occur between the recombination intermediate and the proteins that assist in the assembly of the replisome in the archaeal domain has been lacking. With the recent presentation of the archaeal domain as a novel platform for comparative molecular biology, archaea have been gaining increasing scientific interest; particularly with the discovery of several species that can replicate in an origin-independent way [158]. Archaea encode homologues to a number of eukaryotic replication proteins, but in addition, have a very flexible genome that allows for genetic manipulation, and a platform to investigate origin-independent mechanisms; implications of which can be extended to other life forms.
6.2. Clue No.2: Break-Induced DNA Replication in Eukaryotes
A form of RDR exists within the eukaryotic domain—termed BIR—that occurs during the G2 stage of the cell cycle, through homologous recombination (see Figure 7). The first evidence came from studies in S. cerevisiae when observing the telomere maintenance mechanisms within cells that lack telomerase [159]. Anand [160] emphasizes the lack of progress in understanding the conversion of D-loop structures into replisomes in BIR of budding yeast, as no homologues of the bacterial PriA have been discovered in eukaryotes. A potential lead is that a subunit of Pol δ—a PolB-like polymerase—has been shown to be essential in all BIR events. In higher eukaryotes, such as humans, HelQ helicase interacts with Pol δ to inhibit DNA synthesis, and in turn, promotes DNA repair pathways such as synthesis-dependent strand annealing [161]. One of the experimental methods employed was to induce artificial chromosomal DSBs using site-specific endonucleases, thereby stimulating strand invasion and initiation through BIR [162]. The simple model involves a 3′ end resection of the DSB, exposing a DNA strand that invades a homologous DNA molecule sequence to form a D-loop. The 3′ end acts as a primer in BIR to initiate synthesis through a migrating bubble, resulting in conservative inheritance. All pre-RC components of canonical origin-dependent replication were shown to act during BIR, in addition to recombination proteins such as Rad51, Rad52, Rad54, Rad55, and Rad57, which catalyse D-loop formation [163]. As with other helicases, it is still unknown through which interactions MCM is recruited to the D-loop.
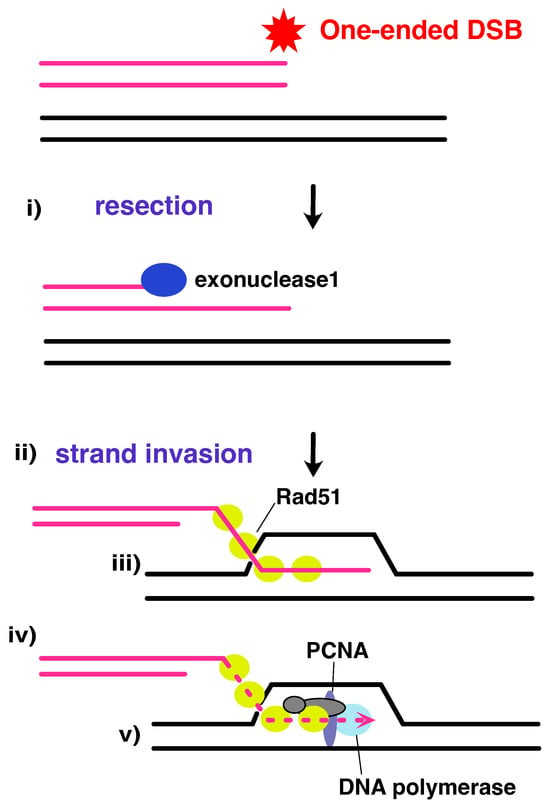
Figure 7.
Rad51-dependent BIR occurs via a bubble migration mechanism. Polα is implicated in the formation of the D-loop, and the replication factors that have been speculated to be involved are indicated. (i) During end resection, Exonuclease 1 catalyses 5′ to 3′ strand resection, leaving 3′ single-stranded DNA (ssDNA) ends initially coated by RPA. (ii) Rad51, with the help of mediator protein Rad52, displaces RPA, to coat the strand, and the (iii) Rad51 filament catalyses homology search and strand invasion, forming a D-loop structure. (iv) DNA synthesis followed by bubble migration catalysed by DNA polymerase, and (v) 3′ ssDNA being used as a direct primer, with the homologous DNA sequence used as a template for new strand synthesis. Figure adapted from [164].
6.3. Clue No.3: Origin-Independent Replication Initiation in Bacteria and Archaea
Kogoma and Lark [165] provided the first experimental evidence of an origin-independent replication process occurring in bacteria, expanding on their earlier paper that characterised E. coli replication, which continued through several rounds despite thymine deficiency. Replication initiation through tightly controlled actions of DnaA and oriC is the preferred pathway due to the added regulation step of specific-origin binding, and thus, the ability to self-regulate due to various imposed control mechanisms. However, the ‘cost’ of such a mechanism is repeated protein synthesis of all the replication components with every cycle—an energy-consuming process. It was found, however, that cells that undergo sustained replication in the absence of protein synthesis do so through the ‘stable DNA replication’ pathway. This condition can be induced through the activation of the DNA damage or SOS response pathway in E. coli; through either UV irradiation, DNA damage agents such as mitomycin, or thymine starvation. The mechanism of SDR that occurs as a result of this induction was hence termed ‘iSDR’ or induced stable DNA Replication, and is considered a special type of RDR [55] (see Figure 8).
Interestingly, E. coli with deletions for RNase HI (i.e., rnhA) have displayed another subcategory of SDR: constitutive SDR, or cSDR. As ∆rnhA E. coli were able to grow without DnaA and oriC, it was postulated that the increase in R-loop formation due to the deletion of RNase HI can promote replication. This avoids the use of an initiator protein that is sequence-specific, allowing replication to initiate at different sites across the genome.
In cSDR and iSDR, PriA has been shown to be essential [166]. Historically, replication initiation from R-loops did not gain much traction, owing to the lack of experimental methods to track R-loop formation in vivo. With the advent of DRIP (i.e., DNA:RNA immunoprecipitation using the S9.6 antibody, which binds to DNA:RNA hybrids), and DRIP-seq (high-throughput sequencing) techniques, it became possible to characterise the genome distribution of these structures. More importantly, the harmful biological implications of excessive R-loop accumulation have been implicated in human diseases like cancer, which stimulated a revival in research on the correlation between R-loops and genomic instability [167].
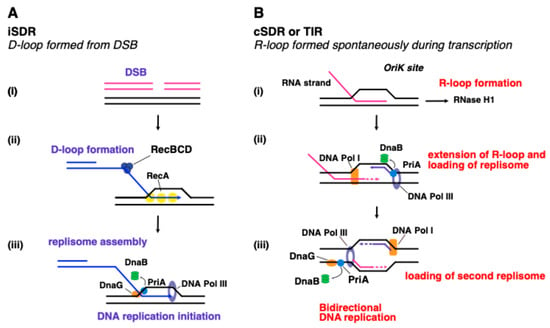
Figure 8.
The two pathways of stable DNA replication in E. coli that occur independently of DnaA and oriC. (A) iSDR mode of replication or ‘D-loop model—left pathway: iSDR differs from cSDR (right pathway) in that it occurs through D-loop formation, as opposed to R-loops, hence is referred to as the ‘D-loop model’. In the iSDR mode of replication following SOS induction, (i) the DSB generated at the oriM site is initially processed by RecBCD helicase, (ii,iii) with RecA recombinase catalysing the strand exchange reaction with the invading 3′ssDNA top strand, to result in a D-loop structure. This is followed by PriA-mediated replisome assembly; PriA recruits DnaB helicase; DnaG primase synthesises an RNA primer for the lagging strand, whereas the 3′ssDNA (invading strand) can be used directly to prime synthesis of the leading strand, catalysed by DNA Pol III. (B) cSDR mode of replication or ‘R-loop model—right pathway: cSDR is also referred to as transcription-induced replication (TIR), whereby an (i) RNA transcript invades the DNA duplex through a reaction called inverse strand exchange, thus resulting in an R-loop structure. The R-loop can result from stalled transcription—and is usually degraded by RNase H1 [168,169]. In ΔrnhA mutants, R-loops are stabilised and thus can be used as an intermediate for RDR. (ii) The 3′ssRNA end can be directly extended by DNA Pol I, forming a D-loop-like structure that acts as a substrate for (iii) PriA to bind and recruit DnaB helicase, together with DnaG primase, followed by the loading of the DNA Pol III to the resulting replisome. Conversely, the lagging strand extended by Pol I leads to the formation of another D-loop structure to repeat the PriA-dependent replisome formation process. Thus, the replication mechanism is bidirectional. Models originally proposed by [55] and adapted from [163].
There is a balancing act between the efficiency of DnaA-dependent initiation and SDR; the latter allows the bacterium to survive in adverse conditions, but occurs with low sequence specificity, and hence is inefficient for proper survival and growth in the normal environment [83]. It was therefore believed that origin-independent replication was only needed for ensuring the survival of the cell in harsh environments, at the expense of replication accuracy.
This paradigm was overturned when a paper in 2013 reported that Haloferax volcanii—a halophilic species found within the archaeal domain—is able to not only survive, but also display a 7.5% faster growth phenotype when all of its origins are deleted, compared to the wild-type strains [76]. There were several intriguing features. Firstly, replication profiles of genome copy numbers along the length of the chromosome revealed that this type of replication does not initiate from a fixed sequence; but rather, in a stochastic manner with initiation points dispersed all over the genome. Another observation was that when the RadA recombinase gene was put under a tryptophan-inducible promoter to regulate its levels, originless cells displayed an absolute requirement for this protein. Based on this body of evidence, alongside the previous known cases of a similar type of replication mechanism in E. coli [170], the authors suggested that an RDR mechanism must be involved, where RadA catalyses D-loop formation. The next line of questioning involved the replication machinery that is assembled during RDR; with MCM being a major player in the recruitment to the D-loop structure. The indispensability of RadA in archaeal RDR was confirmed this year [78], where it was shown to fluctuate according to the growth stage.
This phenomenon does not extend to archaeal cells lacking individual origins, which display a growth disadvantage. Given the known cases of sexual mating involving HGT in H. volcanii [171], the authors suggested that origins behave akin to selfish genetic elements, which prioritise the maintenance of their own ploidy. This could explain the discrepancy between the deletions of individual origins that have no growth advantage; however, the picture was only beginning to emerge. This discovery stimulated the birth of a new subfield—the study of the necessity and the nature of replication origins within the archaeal domain. In the more phylogenetically distant thermophilic archaeon—T. kodakarensis—the single origin can also be deleted and have no deleterious consequences on the phenotype [75]. Similarly, the results from the MFA technique were consistent with the hypothesis of dispersed sites of replication initiation during RDR. The picture becomes less clear when in a closely related species to H. volcanii—H. mediterranei—genuine origin deletion cannot be achieved because a dormant origin becomes activated [172]. What is then so special about the replication origins in H. volcanii? Finding an answer to this question may reveal new insights into the fundamental characteristics of replication origins that were previously unknown, having been ‘concealed’ during normal replication processes.
7. Conclusions: The Archaeal Domain as a Window into Our Evolutionary Past
The discovery of archaea as a separate domain has overturned the long-standing paradigm of the two-domain tree of life. Woese believed that the studies of protein synthesis at the time lacked an evolutionary underpinning, which was the reason for their lack of progress. His background in biophysics endowed him with a unique perspective: in a letter to Crick, he expressed how he intended to study the conservation of proteins and their variation amongst different domains of life [173]. Woese saw the potential in Sanger’s fingerprinting technique [174], and utilised it to sequence the small subunit of 16S rRNA, which appeared to have evolved from a common ancestor. From that, Woese and his postdoc, Fox, concluded that bacteria and archaebacteria (as Archaea were then called) constitute separate domains on the tree of life [63]—and redrew the evolutionary tree to show a tripartite division between eukarya, bacteria, and archaea (i.e., the ‘-bacteria’ suffix has been removed to highlight Archaea’s evolutionary distinction). Woese was highly criticised for the reductionist approach of attempting to rewrite the entire tree of life using a single molecule. To his defence, Zillig proposed the structural homology [175,176,177] between the RNAP molecules within the three domains, thus strengthening the proposal, and leading to the establishment of a new tripartite tree model [178]. At first glance, archaea share some obvious morphological similarities to bacteria, yet their genetic machinery highly resembles those commonly found within eukaryotes. Thus, archaea are often described as a ‘mosaic blend’ of eukaryotic and bacterial features.
Zillig’s work on RNAPs unveiled a previously unsuspected evolutionary connection between archaea and eukaryotes, prompting others to search for evolutionary links between other major enzymes such as DNA polymerases [175]. With no intermediates between eukaryotes and prokaryotes, we observe a formidable gap (some authors go as far as to call it a ‘quantum leap’ of eukaryotic organisational complexity) in evolutionary history. The moment that finally drew attention to archaea—specifically the halophiles—was that haloarchaeon Halobacterium halobium was found to be sensitive to the eukaryotic polymerase inhibitor aphidicolin [179]. Subsequently, it was confirmed that archaea and eukaryotes do indeed share B-family polymerases. One particularly intriguing finding was that PolD (which is insensitive to aphidicolin) is unique to the euryarchaeota group of archaea and is absent from eukaryotes [180].
Several hypotheses have emerged attempting to reconcile the missing link between archaea and eukarya in light of eukaryogenesis. Attempts to characterise the ancestral features of the last eukaryotic common ancestor (i.e., LECA, giving rise to all eukaryotic lineages), have led some to speculate on the archaeal origin of eukaryote, with the most commonly proposed scenarios involving an endosymbiotic event between an Asgard archaeon and alphaproteobacterium (refer to [181] for an in-depth review on eukaryogenesis theories). Many of these theories remained on the speculative side; however, the recent isolation and metagenomic analysis of the Asgard archaeaota superphylum (such as Lokiarchaeota) has revealed them to be the closest living relative of eukaryotes [182], thus strengthening the archaeal involvement in the evolution of the modern eukaryotic cell. Let us not forget about another missing piece—why has the genome evolved to consist of specific origins, yet retain alternative replication initiation mechanisms? Moreover, our understanding of the steps and enzymology of the full replisome assembly from recombination intermediates in archaea remains fragmentary. This is due to the small number of culturable model organisms that can replicate in an origin-independent manner.
The so-called ‘black hole’ of evolutionary biology persists; that is, the origin of the eukaryotic cell and the emergence of eukaryotic organisational complexity. The race is on to reconstitute the proto-eukaryote—and finding a genetically tractable species of Lokiarchaeaota to recapitulate the findings of originless haloarchaea might just be what gets us closer to the finishing line.
Author Contributions
Writing—original draft preparation, A.S. and T.A.; writing—review and editing, A.S. and T.A.; supervision, T.A.; funding acquisition, T.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Biotechnology and Biological Sciences Research Council, studentship [BB/M008770/1] to A.S., and by The Leverhulme Trust, grant number [RF-2023-286\2] to T.A.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analysed in this study. Data sharing is not applicable to this article.
Acknowledgments
We thank Laura Mitchell for her continuous technical support to the laboratory.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Gaillard, H.; García-Muse, T.; Aguilera, A. Replication Stress and Cancer. Nat. Rev. Cancer 2015, 15, 276–289. [Google Scholar] [CrossRef] [PubMed]
- Avery, O.T.; Macleod, C.M.; McCarthy, M. Studies on the Chemical Nature of the Substance Inducing Transformation of Pneumococcal Types. J. Exp. Med. 1944, 79, 137–158. [Google Scholar] [CrossRef]
- Cobb, M. Oswald Avery, DNA, and the Transformation of Biology. Curr. Biol. 2014, 24, R55–R60. [Google Scholar] [CrossRef] [PubMed]
- Stent, G.S. Prematurity and Uniqueness in Scientific Discovery. Sci. Am. 1972, 227, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Mirsky, A.E.; Pollister, A.W.; Chromosin, A. Desoxyribose Nucleoprotein Complex of the Cell Nucleus. J. Gen. Physiol. 1946, 30, 117–148. [Google Scholar] [CrossRef] [PubMed]
- Levene, P.A.; La Forge, F.B. On Chondrosamine. Proc. Natl. Acad. Sci. USA 1915, 1, 190–191. [Google Scholar] [CrossRef] [PubMed]
- Chargaff, E. Chemical Specificity of Nucleic Acids and Mechanism of Their Enzymatic Degradation. Experientia 1950, 6, 201–209. [Google Scholar] [CrossRef]
- Hershey, A.D.; Chase, M. Independent Functions of Viral Protein and Nucleic Acid in Growth of Bacteriophage. J. Gen. Physiol. 1952, 36, 39–56. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, H.V. How History Has Blended. Nature 1974, 249, 803–805. [Google Scholar] [CrossRef]
- Northrop, J. Growth and Phage Production of Lysogenic B. Megatherium. J. Gen. Physiol. 1951, 34, 715–735. [Google Scholar] [CrossRef]
- Swift, H. The Constancy of Desoxyribose Nucleic Acid in Plant Nuclei. Proc. Natl. Acad. Sci. USA 1950, 36, 643–654. [Google Scholar] [CrossRef]
- Watson, J.D.; Crick, F.H.C. Genetical Implications of the Structure of Deoxyribonucleic Acid. Nature 1953, 171, 964–967. [Google Scholar] [CrossRef]
- Franklin, R.; Gosling, R. Molecular Configuration in Sodium Thymonucleate. Nature 1953, 171, 740–741. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.; Stokes, A.; Wilson, H. Molecular Structure of Nucleic Acids: Molecular Structure of Deoxypentose Nucleic Acids. Nature 1953, 171, 738–740. [Google Scholar] [CrossRef] [PubMed]
- Gulland, J.M.; Jordan, D.O.; Taylor, H.F.W. Deoxypentose Nucleic Acids. Part II. Electrometric Titration of the Acidic and the Basic Groups of the Deoxypentose Nucleic Acid of Calf Thymus. J. Chem. Soc. Resumed 1947, 25, 1131–1141. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.D.; Crick, F.H.C. Molecular Structure of Nucleic Acids; a Structure for Deoxyribose Nucleic Acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef]
- Bloch, D.P. A Possible Mechanism for the Replication of the Helical Structure of Desoxyribonucleic Acid. Proc. Natl. Acad. Sci. USA 1955, 41, 1058–1064. [Google Scholar] [CrossRef]
- Taylor, J.H.; Woods, P.S.; Hughes, W.L. The Organization and Duplication Of Chromosomes as Revealed by Autoradiographic Studies Using Tritium-Labeled Thymidine. Proc. Natl. Acad. Sci. USA 1957, 43, 122–128. [Google Scholar] [CrossRef]
- Meselson, M.; Stahl, F.W. The Replication of DNA in Escherichia coli. Proc. Natl. Acad. Sci. USA 1958, 44, 671–682. [Google Scholar] [CrossRef]
- Holmes, F.L. The DNA Replication Problem, 1953–1958. Cell 1998, 23, 117–120. [Google Scholar] [CrossRef]
- Levinthal, C. The Mechanism of DNA Replication and Genetic Recombination in Phage. Proc. Natl. Acad. Sci. USA 1956, 42, 394–404. [Google Scholar] [CrossRef] [PubMed]
- Stent, G.S.; Jerne, N.K. The Distribution of Parental Phosphorus Atoms Among Bacteriophage Progeny. Proc. Natl. Acad. Sci. USA 1955, 41, 704–709. [Google Scholar] [CrossRef]
- Lehman, I.R.; Bessman, M.J.; Simms, E.S.; Kornberg, A. Enzymatic Synthesis of Deoxyribonucleic Acid. I. Preparation of Substrates and Partial Purification of an Enzyme from Escherichia coli. J. Biol. Chem. 1958, 233, 163–170. [Google Scholar] [CrossRef]
- Bessman, M.J.; Lehman, I.R.; Simms, E.S.; Kornberg, A. Enzymatic Synthesis of Deoxyribonucleic Acid. II. General Properties of the Reaction. J. Biol. Chem. 1958, 233, 171–177. [Google Scholar] [CrossRef]
- Lieberman, I.; Kornberg, A.; Simms, E.S. Enzymatic Syntheses of Pyrimidine and Purine Nucleotides. II.1 Orotidine-5′-phosphate Pyrophosphorylase and Decarboxylase. J. Am. Chem. Soc. 1954, 76, 2844–2845. [Google Scholar] [CrossRef]
- Cori, G.T.; Cori, C.F. Crystalline Muscle Phosphorylase. J. Biol. Chem. 1943, 151, 57–63. [Google Scholar] [CrossRef]
- Brutlag, D.; Kornberg, A. Enzymatic Synthesis of Deoxyribonucleic Acid. 36. A Proofreading Funtion for the 3′ Leads to 5′ Exonuclease Activity in the Deoxyribonucleic Acid Polymerases. J. Biol. Chem. 1972, 247, 241–248. [Google Scholar] [CrossRef]
- Delarue, M.; Poch, O.; Tordo, N.; Moras, D.; Argos, P. An Attempt to Unify the Structure of Polymerases. Protein Eng. Des. Sel. 1990, 3, 461–467. [Google Scholar] [CrossRef]
- Joyce, C.M.; Steitz, T.A. Function and Structure Relationships in DNA Polymerases. Annu. Rev. Biochem. 1994, 63, 777–822. [Google Scholar] [CrossRef]
- Bebenek, K.; Kunkel, T.A. Functions of DNA Polymerases. In Advances in Protein Chemistry; Elsevier: Amsterdam, The Netherlands, 2004; Volume 69, pp. 137–165. ISBN 978-0-12-034269-3. [Google Scholar]
- Braithwaite, D.K.; Ito, J. Compilation, Alignment, and Phylogenetic Relationships of DNA Polymerases. Nucleic Acids Res. 1993, 21, 787–802. [Google Scholar] [CrossRef]
- Beese, L.S.; Derbyshire, V.; Steitz, T.A. Structure of DNA Polymerase I Klenow Fragment Bound to Duplex DNA. Science 1993, 260, 352–355. [Google Scholar] [CrossRef] [PubMed]
- Morin, J.A.; Cao, F.J.; Lázaro, J.M.; Arias-Gonzalez, J.R.; Valpuesta, J.M.; Carrascosa, J.L.; Salas, M.; Ibarra, B. Active DNA Unwinding Dynamics during Processive DNA Replication. Proc. Natl. Acad. Sci. USA 2012, 109, 8115–8120. [Google Scholar] [CrossRef]
- Bérut, A.; Arakelyan, A.; Petrosyan, A.; Ciliberto, S.; Dillenschneider, R.; Lutz, E. Experimental Verification of Landauer’s Principle Linking Information and Thermodynamics. Nature 2012, 483, 187–189. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, J.; Nozawa, Y. Self-Reproducing System Can Behave as Maxwell’s Demon: Theoretical Illustration under Prebiotic Conditions. J. Theor. Biol. 1998, 194, 205–221. [Google Scholar] [CrossRef]
- Forterre, P.; Filée, J.; Myllykallio, H. Origin and Evolution of DNA and DNA Replication Machineries. In The Genetic Code and the Origin of Life; Springer: Boston, MA, USA, 2004; pp. 145–168. ISBN 978-0-306-47843-7. [Google Scholar]
- Rothwell, P.J.; Waksman, G. Structure and Mechanism of DNA Polymerases. In Advances in Protein Chemistry; Elsevier: Amsterdam, The Netherlands, 2005; Volume 71, pp. 401–440. ISBN 978-0-12-034271-6. [Google Scholar]
- Steitz, T.A. DNA- and RNA-Dependent DNA Polymerases. Curr. Opin. Struct. Biol. 1993, 3, 31–38. [Google Scholar] [CrossRef]
- Bębenek, A.; Ziuzia-Graczyk, I. Fidelity of DNA Replication—A Matter of Proofreading. Curr. Genet. 2018, 64, 985–996. [Google Scholar] [CrossRef]
- Ekundayo, B.; Bleichert, F. Origins of DNA Replication. PLoS Genet. 2019, 15, e1008320. [Google Scholar] [CrossRef]
- Jacob, F.; Monod, J. Genetic Regulatory Mechanisms in the Synthesis of Proteins. J. Mol. Biol. 1961, 3, 318–356. [Google Scholar] [CrossRef]
- Ryter, A.; Hirota, Y.; Jacob, F. DNA-Membrane Complex and Nuclear Segregation in Bacteria. Cold Spring Harb. Symp. Quant. Biol. 1968, 33, 669–676. [Google Scholar] [CrossRef]
- Morange, M. What History Tells Us XXXI. The Replicon Model: Between Molecular Biology and Molecular Cell Biology. J. Biosci. 2013, 38, 225–227. [Google Scholar] [CrossRef]
- Jacob, F.; Brenner, S.; Cuzin, F. On the Regulation of DNA Replication in Bacteria. Cold Spring Harb. Symp. Quant. Biol. 1963, 28, 329–348. [Google Scholar] [CrossRef]
- Huberman, J.A.; Riggs, A.D. On the Mechanism of DNA Replication in Mammalian Chromosomes. J. Mol. Biol. 1968, 32, 327–341. [Google Scholar] [CrossRef] [PubMed]
- Novick, R.P. Plasmid Incompatibility. Microbiol. Rev. 1987, 51, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Kohiyama, M.; Hiraga, S.; Matic, I.; Radman, M. Bacterial Sex: Playing Voyeurs 50 Years Later. Science 2003, 301, 802–803. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, S.; Hirota, Y. Cloning and Mapping of the Replication Origin of Escherichia Coli. Proc. Natl. Acad. Sci. USA 1977, 74, 5458–5462. [Google Scholar] [CrossRef]
- Mackiewicz, P.; Zakrzewska-Czerwińska, J.; Zawilak, A.; Dudek, M.; Cebrat, M. Where Does Bacterial Replication Start? Rules for Predicting the oriC Region. Nucleic Acids Res. 2004, 32, 3781–3791. [Google Scholar] [CrossRef]
- Chakraborty, T.; Yoshinaga, K.; Lother, H.; Messer, W. Purification of the E. Coli dnaA Gene Product. EMBO J. 1982, 1, 1545–1549. [Google Scholar] [CrossRef]
- Fujita, M.Q.; Yoshikawa, H. Structure of the dnaA Region of Pseudomonas putida: Conservation among Three Bacteria, Bacillus subtilis, Escherichia coli and P. putida. Mol. Gen. Genet. MGG 1989, 215, 381–387. [Google Scholar]
- Chan, C.S.; Tye, B.K. Autonomously Replicating Sequences in Saccharomyces Cerevisiae. Proc. Natl. Acad. Sci. USA 1980, 77, 6329–6333. [Google Scholar] [CrossRef]
- Broach, J.R.; Li, Y.-Y.; Feldman, J.; Jayaram, M.; Abraham, J.; Nasmyth, K.A.; Hicks, J.B. Localization and Sequence Analysis of Yeast Origins of DNA Replication. Cold Spring Harb. Symp. Quant. Biol. 1983, 47, 1165–1173. [Google Scholar] [CrossRef]
- Bell, S.P.; Stillman, B. ATP-Dependent Recognition of Eukaryotic Origins of DNA Replication by a Multiprotein Complex. Nature 1992, 357, 128–134. [Google Scholar] [CrossRef]
- Kogoma, T. Stable DNA Replication: Interplay between DNA Replication, Homologous Recombination, and Transcription. Microbiol. Mol. Biol. Rev. 1997, 61, 212–238. [Google Scholar] [PubMed]
- Gavin, K.A.; Hidaka, M.; Stillman, B. Conserved Initiator Proteins in Eukaryotes. Science 1995, 270, 1667–1671. [Google Scholar] [CrossRef]
- Taylor, J.H. Increase in DNA Replication Sites in Cells Held at the Beginning of S Phase. Chromosoma 1977, 62, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Hamlin, J.L.; Mesner, L.D.; Lar, O.; Torres, R.; Chodaparambil, S.V.; Wang, L. A Revisionist Replicon Model for Higher Eukaryotic Genomes. J. Cell. Biochem. 2008, 105, 321–329. [Google Scholar] [CrossRef]
- diCenzo, G.C.; Finan, T.M. The Divided Bacterial Genome: Structure, Function, and Evolution. Microbiol. Mol. Biol. Rev. 2017, 81, e00019-17. [Google Scholar] [CrossRef] [PubMed]
- Harrison, P.W.; Lower, R.P.J.; Kim, N.K.D.; Young, J.P.W. Introducing the Bacterial ‘Chromid’: Not a Chromosome, Not a Plasmid. Trends Microbiol. 2010, 18, 141–148. [Google Scholar] [CrossRef]
- Krawiec, S.; Riley, M. Organization of the Bacterial Chromosome. Microbiol. Rev. 1990, 54, 502–539. [Google Scholar] [CrossRef]
- Suwanto, A.; Kaplan, S. Physical and Genetic Mapping of the Rhodobacter sphaeroides 2.4.1 Genome: Presence of Two Unique Circular Chromosomes. J. Bacteriol. 1989, 171, 5850–5859. [Google Scholar] [CrossRef]
- Woese, C.R.; Fox, G.E. Phylogenetic Structure of the Prokaryotic Domain: The Primary Kingdoms. Proc. Natl. Acad. Sci. USA 1977, 74, 5088–5090. [Google Scholar] [CrossRef]
- Bult, C.J.; White, O.; Olsen, G.J.; Zhou, L.; Fleischmann, R.D.; Sutton, G.G.; Blake, J.A.; FitzGerald, L.M.; Clayton, R.A.; Gocayne, J.D.; et al. Complete Genome Sequence of the Methanogenic Archaeon, Methanococcus jannaschii. Science 1996, 273, 1058–1073. [Google Scholar] [CrossRef]
- Myllykallio, H.; Lopez, P.; López-García, P.; Heilig, R.; Saurin, W.; Zivanovic, Y.; Philippe, H.; Forterre, P. Bacterial Mode of Replication with Eukaryotic-Like Machinery in a Hyperthermophilic Archaeon. Science 2000, 288, 2212–2215. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, F.; Forterre, P.; Ishino, Y.; Myllykallio, H. In Vivo Interactions of Archaeal Cdc6/Orc1 and Minichromosome Maintenance Proteins with the Replication Origin. Proc. Natl. Acad. Sci. USA 2001, 98, 11152–11157. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, F.; Norais, C.; Forterre, P.; Myllykallio, H. Identification of Short ‘Eukaryotic’ Okazaki Fragments Synthesized from a Prokaryotic Replication Origin. EMBO Rep. 2003, 4, 154–158. [Google Scholar] [CrossRef]
- Robinson, N.P.; Dionne, I.; Lundgren, M.; Marsh, V.L.; Bernander, R.; Bell, S.D. Identification of Two Origins of Replication in the Single Chromosome of the Archaeon Sulfolobus solfataricus. Cell 2004, 116, 25–38. [Google Scholar] [CrossRef]
- Lundgren, M.; Andersson, A.; Chen, L.; Nilsson, P.; Bernander, R. Three Replication Origins in Sulfolobus Species: Synchronous Initiation of Chromosome Replication and Asynchronous Termination. Proc. Natl. Acad. Sci. USA 2004, 101, 7046–7051. [Google Scholar] [CrossRef] [PubMed]
- She, Q.; Peng, X.; Zillig, W.; Garrett, R.A. Gene Capture in Archaeal Chromosomes. Nature 2001, 409, 478. [Google Scholar] [CrossRef]
- Robinson, N.P.; Bell, S.D. Extrachromosomal Element Capture and the Evolution of Multiple Replication Origins in Archaeal Chromosomes. Proc. Natl. Acad. Sci. USA 2007, 104, 5806–5811. [Google Scholar] [CrossRef]
- Zhang, R.; Zhang, C.-T. Multiple Replication Origins of the Archaeon Halobacterium Species NRC-1. Biochem. Biophys. Res. Commun. 2003, 302, 728–734. [Google Scholar] [CrossRef]
- Wu, Z.; Liu, H.; Liu, J.; Liu, X.; Xiang, H. Diversity and Evolution of Multiple Orc/Cdc6-Adjacent Replication Origins in Haloarchaea. BMC Genomics 2012, 13, 478. [Google Scholar] [CrossRef]
- Hawkins, M.S. DNA Replication Origins in Haloferax volcanii. Ph.D. Thesis, University of Nottingham, Nottingham, UK, 2009. [Google Scholar]
- Gehring, A.M.; Astling, D.P.; Matsumi, R.; Burkhart, B.W.; Kelman, Z.; Reeve, J.N.; Jones, K.L.; Santangelo, T.J. Genome Replication in Thermococcus kodakarensis Independent of Cdc6 and an Origin of Replication. Front. Microbiol. 2017, 8, 2084. [Google Scholar] [CrossRef]
- Hawkins, M.; Malla, S.; Blythe, M.J.; Nieduszynski, C.A.; Allers, T. Accelerated Growth in the Absence of DNA Replication Origins. Nature 2013, 503, 544–547. [Google Scholar] [CrossRef] [PubMed]
- Liman, G.L.S.; Lennon, C.W.; Mandley, J.L.; Galyon, A.M.; Zatopek, K.M.; Gardner, A.F.; Santangelo, T.J. Intein Splicing Efficiency and RadA Levels Can Control the Mode of Archaeal DNA Replication. Sci. Adv. 2024, 10, eadp4995. [Google Scholar] [CrossRef]
- Mc Teer, L.; Moalic, Y.; Cueff-Gauchard, V.; Catchpole, R.; Hogrel, G.; Lu, Y.; Laurent, S.; Hemon, M.; Aubé, J.; Leroy, E.; et al. Cooperation between Two Modes for DNA Replication Initiation in the Archaeon Thermococcus barophilus. mBio 2024, 15, e03200-23. [Google Scholar] [CrossRef]
- Dulermo, R. Archaeal DNA Replication Initiation: Bridging LUCA’s Legacy and Modern Mechanisms. Front. Microbiol. 2025, 16, 1561973. [Google Scholar] [CrossRef] [PubMed]
- Kreuzer, K.N.; Brister, J.R. Initiation of Bacteriophage T4 DNA Replication and Replication Fork Dynamics: A Review in the Virology Journal Series on Bacteriophage T4 and Its Relatives. Virol. J. 2010, 7, 358. [Google Scholar] [CrossRef] [PubMed]
- Richter, S.; Hagemann, M.; Messer, W. Transcriptional Analysis and Mutation of a dnaA-Like Gene in Synechocystis sp. Strain PCC 6803. J. Bacteriol. 1998, 180, 4946–4949. [Google Scholar] [CrossRef]
- Ohbayashi, R.; Hirooka, S.; Onuma, R.; Kanesaki, Y.; Hirose, Y.; Kobayashi, Y.; Fujiwara, T.; Furusawa, C.; Miyagishima, S. Evolutionary Changes in DnaA-Dependent Chromosomal Replication in Cyanobacteria. Front. Microbiol. 2020, 11, 786. [Google Scholar] [CrossRef]
- Masai, H. Replicon Hypothesis Revisited. Biochem. Biophys. Res. Commun. 2022, 633, 77–80. [Google Scholar] [CrossRef]
- Dyall-Smith, M.L.; Pfeiffer, F.; Klee, K.; Palm, P.; Gross, K.; Schuster, S.C.; Rampp, M.; Oesterhelt, D. Haloquadratum walsbyi: Limited Diversity in a Global Pond. PLoS ONE 2011, 6, e20968. [Google Scholar] [CrossRef]
- Xu, Y.; Bremer, H. Chromosome Replication in Escherichia coli Induced by Oversupply of DnaA. Mol Gen Genet 1988, 211, 138–142. [Google Scholar] [CrossRef]
- Grimwade, J.E.; Ryan, V.T.; Leonard, A.C. IHF Redistributes Bound Initiator Protein, DnaA, on Supercoiled oriC of Escherichia coli. Mol. Microbiol. 2000, 35, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Rozgaja, T.A.; Grimwade, J.E.; Iqbal, M.; Czerwonka, C.; Vora, M.; Leonard, A.C. Two Oppositely Oriented Arrays of Low-Affinity Recognition Sites in oriC Guide Progressive Binding of DnaA during Escherichia coli Pre-RC Assembly: DnaA-oriC Interaction at Arrayed Sites. Mol. Microbiol. 2011, 82, 475–488. [Google Scholar] [CrossRef] [PubMed]
- Matsui, M.; Oka, A.; Takanami, M.; Yasuda, S.; Hirota, Y. Sites of dnaA Protein-Binding in the Replication Origin of the Escherichia coli K-12 Chromosome. J. Mol. Biol. 1985, 184, 529–533. [Google Scholar] [CrossRef]
- Erzberger, J.P. The Structure of Bacterial DnaA: Implications for General Mechanisms Underlying DNA Replication Initiation. EMBO J. 2002, 21, 4763–4773. [Google Scholar] [CrossRef]
- Erzberger, J.P.; Berger, J.M. Evolutionary Relationships and Structural Mechanisms of AAA+ Proteins. Annu. Rev. Biophys. Biomol. Struct. 2006, 35, 93–114. [Google Scholar] [CrossRef] [PubMed]
- Katayama, T.; Kasho, K.; Kawakami, H. The DnaA Cycle in Escherichia Coli: Activation, Function and Inactivation of the Initiator Protein. Front. Microbiol. 2017, 8, 2496. [Google Scholar] [CrossRef]
- Kowalski, D.; Eddy, M.J. The DNA Unwinding Element: A Novel, Cis-Acting Component That Facilitates Opening of the Escherichia coli Replication Origin. EMBO J. 1989, 8, 4335–4344. [Google Scholar] [CrossRef]
- Wegrzyn, K.; Konieczny, I. Toward an Understanding of the DNA Replication Initiation in Bacteria. Front. Microbiol. 2024, 14, 1328842. [Google Scholar] [CrossRef]
- Robinson, N.P.; Bell, S.D. Origins of DNA Replication in the Three Domains of Life. FEBS J. 2005, 272, 3757–3766. [Google Scholar] [CrossRef]
- Schwob, E. Flexibility and Governance in Eukaryotic DNA Replication. Curr. Opin. Microbiol. 2004, 7, 680–690. [Google Scholar] [CrossRef]
- Segurado, M.; Gómez, M.; Antequera, F. Increased Recombination Intermediates and Homologous Integration Hot Spots at DNA Replication Origins. Mol. Cell 2002, 10, 907–916. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, H.; Chetlangia, N.; Prasanth, S.G. Chromatin’s Influence on Pre-Replication Complex Assembly and Function. Biology 2024, 13, 152. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, D.M.; Takebayashi, S.-I.; Ryba, T.; Lu, J.; Pope, B.D.; Wilson, K.A.; Hiratani, I. Space and Time in the Nucleus: Developmental Control of Replication Timing and Chromosome Architecture. Cold Spring Harb. Symp. Quant. Biol. 2010, 75, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Labib, K. How Do Cdc7 and Cyclin-Dependent Kinases Trigger the Initiation of Chromosome Replication in Eukaryotic Cells? Genes Dev. 2010, 24, 1208–1219. [Google Scholar] [CrossRef]
- O’Donnell, M.; Langston, L.; Stillman, B. Principles and Concepts of DNA Replication in Bacteria, Archaea, and Eukarya. Cold Spring Harb. Perspect. Biol. 2013, 5, a010108. [Google Scholar] [CrossRef]
- Hartwell, L.H. Sequential Function of Gene Products Relative to DNA Synthesis in the Yeast Cell Cycle. J. Mol. Biol. 1976, 104, 803–817. [Google Scholar] [CrossRef]
- Hofmann, J.F.X.; Beach, D. Cdt 1 Is an Essential Target of the Cdc 1 O/Sct 1 Transcription Factor: Requirement for DNA Replication and Inhibition of Mitosis. EMBO J. 1994, 13, 425–434. [Google Scholar] [CrossRef]
- Nishitani, H.; Lygerou, Z.; Nishimoto, T.; Nurse, P. The Cdt1 Protein Is Required to License DNA for Replication in fission Yeast. Nature 2000, 404, 625–628. [Google Scholar] [CrossRef]
- Schmidt, J.M.; Yang, R.; Kumar, A.; Hunker, O.; Seebacher, J.; Bleichert, F. A Mechanism of Origin Licensing Control through Autoinhibition of S. cerevisiae ORC·DNA·Cdc6. Nat. Commun. 2022, 13, 1059. [Google Scholar] [CrossRef]
- Randell, J.C.W.; Bowers, J.L.; Rodríguez, H.K.; Bell, S.P. Sequential ATP Hydrolysis by Cdc6 and ORC Directs Loading of the Mcm2-7 Helicase. Mol. Cell 2006, 21, 29–39. [Google Scholar] [CrossRef]
- Costa, A.; Diffley, J.F.X. The Initiation of Eukaryotic DNA Replication. Annu. Rev. Biochem. 2022, 91, 107–131. [Google Scholar] [CrossRef] [PubMed]
- Ilves, I.; Petojevic, T.; Pesavento, J.J.; Botchan, M.R. Activation of the MCM2-7 Helicase by Association with Cdc45 and GINS Proteins. Mol. Cell 2010, 37, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Zegerman, P.; Diffley, J.F.X. Phosphorylation of Sld2 and Sld3 by Cyclin-Dependent Kinases Promotes DNA Replication in Budding Yeast. Nature 2007, 445, 281–285. [Google Scholar] [CrossRef]
- Ausiannikava, D.; Allers, T. Diversity of DNA Replication in the Archaea. Genes 2017, 8, 56. [Google Scholar] [CrossRef]
- Wu, Z.; Liu, J.; Yang, H.; Xiang, H. DNA Replication Origins in Archaea. Front. Microbiol. 2014, 5, 179. [Google Scholar] [CrossRef]
- Greci, M.D.; Bell, S.D. Archaeal DNA Replication. Annu. Rev. Microbiol. 2020, 74, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Lopez, P.; Philippe, H.; Myllykallio, H.; Forterre, P. Identification of Putative Chromosomal Origins of Replication in Archaea. Mol. Microbiol. 1999, 32, 883–886. [Google Scholar] [CrossRef]
- Liu, J.; Smith, C.L.; DeRyckere, D.; DeAngelis, K.; Martin, G.S.; Berger, J.M. Structure and Function of Cdc6/Cdc18: Implications for Origin Recognition and Checkpoint Control. Mol. Cell 2000, 6, 637–648. [Google Scholar] [CrossRef]
- Matsunaga, F.; Glatigny, A.; Mucchielli-Giorgi, M.-H.; Agier, N.; Delacroix, H.; Marisa, L.; Durosay, P.; Ishino, Y.; Aggerbeck, L.; Forterre, P. Genomewide and Biochemical Analyses of DNA-Binding Activity of Cdc6/Orc1 and Mcm Proteins in Pyrococcus sp. Nucleic Acids Res. 2007, 35, 3214–3222. [Google Scholar] [CrossRef]
- Matsunaga, F.; Takemura, K.; Akita, M.; Adachi, A.; Yamagami, T.; Ishino, Y. Localized Melting of Duplex DNA by Cdc6/Orc1 at the DNA Replication Origin in the Hyperthermophilic Archaeon Pyrococcus furiosus. Extremophiles 2010, 14, 21–31. [Google Scholar] [CrossRef]
- Gaudier, M.; Schuwirth, B.S.; Westcott, S.L.; Wigley, D.B. Structural Basis of DNA Replication Origin Recognition by an ORC Protein. Science 2007, 317, 1213–1216. [Google Scholar] [CrossRef] [PubMed]
- Grainge, I. Biochemical Analysis of Components of the Pre-Replication Complex of Archaeoglobus fulgidus. Nucleic Acids Res. 2003, 31, 4888–4898. [Google Scholar] [CrossRef] [PubMed]
- Samson, R.Y.; Xu, Y.; Gadelha, C.; Stone, T.A.; Faqiri, J.N.; Li, D.; Qin, N.; Pu, F.; Liang, Y.X.; She, Q.; et al. Specificity and Function of Archaeal DNA Replication Initiator Proteins. Cell Rep. 2013, 3, 485–496. [Google Scholar] [CrossRef]
- Dueber, E.L.C.; Corn, J.E.; Bell, S.D.; Berger, J.M. Replication Origin Recognition and Deformation by a Heterodimeric Archaeal Orc1 Complex. Science 2007, 317, 1210–1213. [Google Scholar] [CrossRef] [PubMed]
- Grainge, I.; Gaudier, M.; Schuwirth, B.S.; Westcott, S.L.; Sandall, J.; Atanassova, N.; Wigley, D.B. Biochemical Analysis of a DNA Replication Origin in the Archaeon Aeropyrum pernix. J. Mol. Biol. 2006, 363, 355–369. [Google Scholar] [CrossRef]
- Akita, M.; Adachi, A.; Takemura, K.; Yamagami, T.; Matsunaga, F.; Ishino, Y. Cdc6/Orc1 from Pyrococcus furiosus May Act as the Origin Recognition Protein and Mcm Helicase Recruiter. Genes Cells 2010, 15, 537–552. [Google Scholar] [CrossRef]
- Kelman, L.M.; O’Dell, W.B.; Kelman, Z. Unwinding 20 Years of the Archaeal Minichromosome Maintenance Helicase. J. Bacteriol. 2020, 202, e00729-19. [Google Scholar] [CrossRef]
- Kasiviswanathan, R.; Shin, J.-H.; Kelman, Z. Interactions between the Archaeal Cdc6 and MCM Proteins Modulate Their Biochemical Properties. Nucleic Acids Res. 2005, 33, 4940–4950. [Google Scholar] [CrossRef][Green Version]
- Samson, R.Y.; Abeyrathne, P.D.; Bell, S.D. Mechanism of Archaeal MCM Helicase Recruitment to DNA Replication Origins. Mol. Cell 2016, 61, 287–296. [Google Scholar] [CrossRef]
- Samson, R.Y.; Bell, S.D. MCM Loading—An Open-and-Shut Case? Mol. Cell 2013, 50, 457–458. [Google Scholar] [CrossRef]
- Mohammed Khalid, A.A.; Parisse, P.; Medagli, B.; Onesti, S.; Casalis, L. Atomic Force Microscopy Investigation of the Interactions between the MCM Helicase and DNA. Materials 2021, 14, 687. [Google Scholar] [CrossRef] [PubMed]
- Kelman, Z.; Lee, J.-K.; Hurwitz, J. The Single Minichromosome Maintenance Protein of Methanobacterium Thermoautotrophicum DeltaH Contains DNA Helicase Activity. Proc. Natl. Acad. Sci. USA 1999, 96, 14783–14788. [Google Scholar] [CrossRef]
- Haugland, G.T.; Shin, J.-H.; Birkeland, N.-K.; Kelman, Z. Stimulation of MCM Helicase Activity by a Cdc6 Protein in the Archaeon Thermoplasma acidophilum. Nucleic Acids Res. 2006, 34, 6337–6344. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Arnaiz, P.; Dattani, A.; Smith, V.; Allers, T. Haloferax volcanii—A Model Archaeon for Studying DNA Replication and Repair. Open Biol. 2020, 10, 200293. [Google Scholar] [CrossRef]
- Ishino, S.; Kelman, L.M.; Kelman, Z.; Ishino, Y. The Archaeal DNA Replication Machinery: Past, Present and Future. Genes Genet. Syst. 2013, 88, 315–319. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sauguet, L.; Raia, P.; Henneke, G.; Delarue, M. Shared Active Site Architecture between Archaeal PolD and Multi-Subunit RNA Polymerases Revealed by X-Ray Crystallography. Nat. Commun. 2016, 7, 12227. [Google Scholar] [CrossRef] [PubMed]
- Čuboňová, L.; Richardson, T.; Burkhart, B.W.; Kelman, Z.; Connolly, B.A.; Reeve, J.N.; Santangelo, T.J. Archaeal DNA Polymerase D but Not DNA Polymerase B Is Required for Genome Replication in Thermococcus kodakarensis. J. Bacteriol. 2013, 195, 2322–2328. [Google Scholar] [CrossRef]
- Oki, K.; Yamagami, T.; Nagata, M.; Mayanagi, K.; Shirai, T.; Adachi, N.; Numata, T.; Ishino, S.; Ishino, Y. DNA Polymerase D Temporarily Connects Primase to the CMG-like Helicase before Interacting with Proliferating Cell Nuclear Antigen. Nucleic Acids Res. 2021, 49, 4599–4612. [Google Scholar] [CrossRef]
- Oki, K.; Nagata, M.; Yamagami, T.; Numata, T.; Ishino, S.; Oyama, T.; Ishino, Y. Family D DNA Polymerase Interacts with GINS to Promote CMG-Helicase in the Archaeal Replisome. Nucleic Acids Res. 2022, 50, 3601–3615. [Google Scholar] [CrossRef]
- Martínez-Carranza, M.; Vialle, L.; Madru, C.; Cordier, F.; Tekpinar, A.D.; Haouz, A.; Legrand, P.; Le Meur, R.A.; England, P.; Dulermo, R.; et al. Communication between DNA Polymerases and Replication Protein A within the Archaeal Replisome. Nat. Commun. 2024, 15, 10926. [Google Scholar] [CrossRef]
- Cairns, J. The Bacterial Chromosome and Its Manner of Replication as Seen by Autoradiography. J. Mol. Biol. 1963, 6, 208–213. [Google Scholar] [CrossRef]
- Wu, C.A.; Zechner, E.L.; Marians, K.J. Coordinated Leading- and Lagging-Strand Synthesis at the Escherichia coli DNA Replication Fork. I. Multiple Effectors Act to Modulate Okazaki Fragment Size. J. Biol. Chem. 1992, 267, 4030–4044. [Google Scholar] [CrossRef] [PubMed]
- Cha, T.A.; Alberts, B.M. The Bacteriophage T4 DNA Replication Fork. J. Biol. Chem. 1989, 264, 12220–12225. [Google Scholar] [CrossRef]
- Nakai, H.; Richardson, C.C. Leading and Lagging Strand Synthesis at the Replication Fork of Bacteriophage T7. Distinct Properties of T7 Gene 4 Protein as a Helicase and Primase. J. Biol. Chem. 1988, 263, 9818–9830. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, K.; Okazaki, T.; Okazaki, R. Mechanism of DNA Chain Growth, II. Accumulation of Newly Synthesized Short Chains in E. coli Infected with Ligase-Defective T4 Phages. Proc. Natl. Acad. Sci. USA 1968, 60, 1356–1362. [Google Scholar] [CrossRef] [PubMed]
- Cronan, G.E.; Kouzminova, E.A.; Kuzminov, A. Near-Continuously Synthesized Leading Strands in Escherichia coli Are Broken by Ribonucleotide Excision. Proc. Natl. Acad. Sci. USA 2019, 116, 1251–1260. [Google Scholar] [CrossRef]
- Kelman, L.M.; Kelman, Z. Archaea: An Archetype for Replication Initiation Studies? Mol. Microbiol. 2003, 48, 605–615. [Google Scholar] [CrossRef]
- Shen, W.; Wang, Z.; Ning, K.; Cheng, F.; Engelhardt, J.F.; Yan, Z.; Qiu, J. Hairpin Transfer-Independent Parvovirus DNA Replication Produces Infectious Virus. J. Virol. 2021, 95, e01108-21. [Google Scholar] [CrossRef]
- Lujan, S.A.; Williams, J.S.; Kunkel, T.A. DNA Polymerases Divide the Labor of Genome Replication. Trends Cell Biol. 2016, 26, 640–654. [Google Scholar] [CrossRef]
- Iyer, L.M. Origin and Evolution of the Archaeo-Eukaryotic Primase Superfamily and Related Palm-Domain Proteins: Structural Insights and New Members. Nucleic Acids Res. 2005, 33, 3875–3896. [Google Scholar] [CrossRef]
- Forterre, P. Why Are There So Many Diverse Replication Machineries? J. Mol. Biol. 2013, 425, 4714–4726. [Google Scholar] [CrossRef]
- Georgescu, R.; Langston, L.; O’Donnell, M. A Proposal: Evolution of PCNA’s Role as a Marker of Newly Replicated DNA. DNA Repair 2015, 29, 4–15. [Google Scholar] [CrossRef]
- Leipe, D.D.; Aravind, L.; Koonin, E.V. Did DNA Replication Evolve Twice Independently? Nucleic Acids Res. 1999, 27, 3389–3401. [Google Scholar] [CrossRef]
- Sinha, N.K.; Morris, C.F.; Alberts, B.M. Efficient in Vitro Replication of Double-Stranded DNA Templates by a Purified T4 Bacteriophage Replication System. J. Biol. Chem. 1980, 255, 4290–4303. [Google Scholar] [CrossRef]
- Alberts, B.M.; Barry, J.; Bedinger, P.; Formosa, T.; Jongeneel, C.V.; Kreuzer, K.N. Studies on DNA Replication in the Bacteriophage T4 In Vitro System. Cold Spring Harb Symp Quant Biol 1983, 47, 655–668. [Google Scholar] [CrossRef]
- Kreuzer, K.N. Recombination-Dependent DNA Replication in Phage T4. Trends Biochem. Sci. 2000, 25, 165–173. [Google Scholar] [CrossRef]
- Mosig, G.; Gewin, J.; Luder, A.; Colowick, N.; Vo, D. Two Recombination-Dependent DNA Replication Pathways of Bacteriophage T4, and Their Roles in Mutagenesis and Horizontal Gene Transfer. Proc. Natl. Acad. Sci. USA 2001, 98, 8306–8311. [Google Scholar] [CrossRef]
- Belanger, K.G.; Kreuzer, K.N. Bacteriophage T4 Initiates Bidirectional DNA Replication through a Two-Step Process. Mol. Cell 1998, 2, 693–701. [Google Scholar] [CrossRef]
- Syeda, A.H.; Hawkins, M.; McGlynn, P. Recombination and Replication. Cold Spring Harb. Perspect. Biol. 2014, 6, a016550. [Google Scholar] [CrossRef]
- Yang, S.; VanLoock, M.S.; Yu, X.; Egelman, E.H. Comparison of Bacteriophage T4 UvsX and Human Rad51 Filaments Suggests That RecA-like Polymers May Have Evolved independently. J. Mol. Biol. 2001, 312, 999–1009. [Google Scholar] [CrossRef]
- Liu, J.; Marians, K.J. PriA-Directed Assembly of a Primosome on D Loop DNA. J. Biol. Chem. 1999, 274, 25033–25041. [Google Scholar] [CrossRef]
- Jones, J.M. The Phi X174-Type Primosome Promotes Replisome Assembly at the Site of Recombination in Bacteriophage Mu Transposition. EMBO J. 1997, 16, 6886–6895. [Google Scholar] [CrossRef]
- Ferreira-Cerca, S. (Ed.) Archaea: Methods and Protocols; Methods in Molecular Biology; Springer: New York, NY, USA, 2022; Volume 2522, ISBN 978-1-07-162444-9. [Google Scholar]
- Epum, E.A.; Haber, J.E. DNA Replication: The Recombination Connection. Trends Cell Biol. 2022, 32, 45–57. [Google Scholar] [CrossRef]
- Anand, R.P.; Lovett, S.T.; Haber, J.E. Break-Induced DNA Replication. Cold Spring Harb. Perspect. Biol. 2013, 5, a010397. [Google Scholar] [CrossRef]
- He, L.; Lever, R.; Cubbon, A.; Tehseen, M.; Jenkins, T.; Nottingham, A.O.; Horton, A.; Betts, H.; Fisher, M.; Hamdan, S.M.; et al. Interaction of Human HelQ with DNA Polymerase Delta Halts DNA Synthesis and Stimulates DNA Single-Strand Annealing. Nucleic Acids Res. 2023, 51, 1740–1749. [Google Scholar] [CrossRef]
- Malkova, A.; Ira, G. Break-Induced Replication: Functions and Molecular Mechanism. Curr. Opin. Genet. Dev. 2013, 23, 271–279. [Google Scholar] [CrossRef]
- Ravoitytė, B.; Wellinger, R. Non-Canonical Replication Initiation: You’re Fired! Genes 2017, 8, 54. [Google Scholar] [CrossRef]
- Rossi, M.J.; DiDomenico, S.F.; Patel, M.; Mazin, A.V. RAD52: Paradigm of Synthetic Lethality and New Developments. Front. Genet. 2021, 12, 780293. [Google Scholar] [CrossRef]
- Kogoma, T.; Lark, K.G. Characterization of the Replication of Escherichia Coli DNA in the Absence of Protein Synthesis: Stable DNA Replication. J. Mol. Biol. 1975, 94, 243–256. [Google Scholar] [CrossRef]
- Masai, H.; Asai, T.; Kubota, Y.; Arai, K.; Kogoma, T. Escherichia Coil PriA Protein Is Essential for Inducible and Constitutive Stable DNA Replication. EMBO J. 1994, 13, 5338–5345. [Google Scholar] [CrossRef]
- Crossley, M.P.; Bocek, M.; Cimprich, K.A. R-Loops as Cellular Regulators and Genomic Threats. Mol. Cell 2019, 73, 398–411. [Google Scholar] [CrossRef] [PubMed]
- Zaitsev, E.N.; Kowalczykowski, S.C. A Novel Pairing Process Promoted by Escherichia Coli RecA Protein: Inverse DNA and RNA Strand Exchange. Genes Dev. 2000, 14, 740–749. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, M.; Clikeman, J.A.; Bates, D.B.; Kogoma, T. RecA Protein-Dependent R-Loop Formation in Vitro. Genes Dev. 2000, 14, 360–365. [Google Scholar] [CrossRef]
- Kogoma, T. Recombination by Replication. Cell 1996, 85, 625–627. [Google Scholar] [CrossRef] [PubMed]
- Rosenshine, I.; Tchelet, R.; Mevarech, M. The Mechanism of DNA Transfer in the Mating System of an Archaebacterium. Science 1989, 245, 1387–1389. [Google Scholar] [CrossRef]
- Yang, H.; Wu, Z.; Liu, J.; Liu, X.; Wang, L.; Cai, S.; Xiang, H. Activation of a Dormant Replication Origin Is Essential for Haloferax Mediterranei Lacking the Primary Origins. Nat. Commun. 2015, 6, 8321. [Google Scholar] [CrossRef]
- Pace, N.R.; Sapp, J.; Goldenfeld, N. Phylogeny and beyond: Scientific, Historical, and Conceptual Significance of the First Tree of Life. Proc. Natl. Acad. Sci. USA 2012, 109, 1011–1018. [Google Scholar] [CrossRef]
- Sanger, F.; Brownlee, G.G.; Barrell, B.G. A Two-Dimensional Fractionation Procedure for Radioactive Nucleotides. J. Mol. Biol. 1965, 13, 373–398. [Google Scholar] [CrossRef]
- Albers, S.-V.; Forterre, P.; Prangishvili, D.; Schleper, C. The Legacy of Carl Woese and Wolfram Zillig: From Phylogeny to Landmark Discoveries. Nat. Rev. Microbiol. 2013, 11, 713–719. [Google Scholar] [CrossRef]
- Zillig, W.; Klenk, H.-P.; Palm, P.; Pühler, G.; Gropp, F.; Garrett, R.A.; Leffers, H. The Phylogenetic Relations of DNA-Dependent RNA Polymerases of Archaebacteria, Eukaryotes, and Eubacteria. Can. J. Microbiol. 1989, 35, 73–80. [Google Scholar] [CrossRef]
- Zillig, W. Comparative Biochemistry of Archaea and Bacteria. Curr. Opin. Genet. Dev. 1991, 1, 544–551. [Google Scholar] [CrossRef] [PubMed]
- Woese, C.R. Towards a Natural System of Organisms: Proposal for the Domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA 1990, 87, 4576–4579. [Google Scholar] [CrossRef]
- Forterre, P.; Elie, C.; Kohiyama, M. Aphidicolin Inhibits Growth and DNA Synthesis in Halophilic Arachaebacteria. J. Bacteriol. 1984, 159, 800–802. [Google Scholar] [CrossRef] [PubMed]
- Ishino, Y.; Komori, K.; Cann, I.K.O.; Koga, Y. A Novel DNA Polymerase Family Found in Archaea. J. Bacteriol. 1998, 180, 2232–2236. [Google Scholar] [CrossRef] [PubMed]
- Baum, B.; Spang, A. On the Origin of the Nucleus: A Hypothesis. Microbiol. Mol. Biol. Rev. 2023, 87, e00186-21. [Google Scholar] [CrossRef]
- Rinke, C.; Chuvochina, M.; Mussig, A.J.; Chaumeil, P.-A.; Davín, A.A.; Waite, D.W.; Whitman, W.B.; Parks, D.H.; Hugenholtz, P. A Standardized Archaeal Taxonomy for the Genome Taxonomy Database. Nat. Microbiol. 2021, 6, 946–959. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).