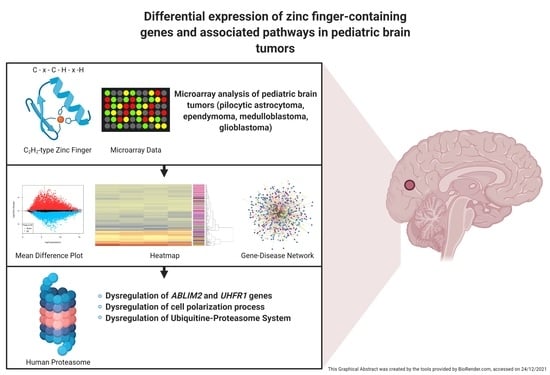
Central Role of C2H2-Type Zinc Finger-Containing Genes in Pediatric Brain Tumors
Abstract
:1. Introduction
1.1. Zinc Fingers
1.2. Role of Zinc Fingers in Brain Physiology and Disease
1.3. Pediatric Brain Tumors
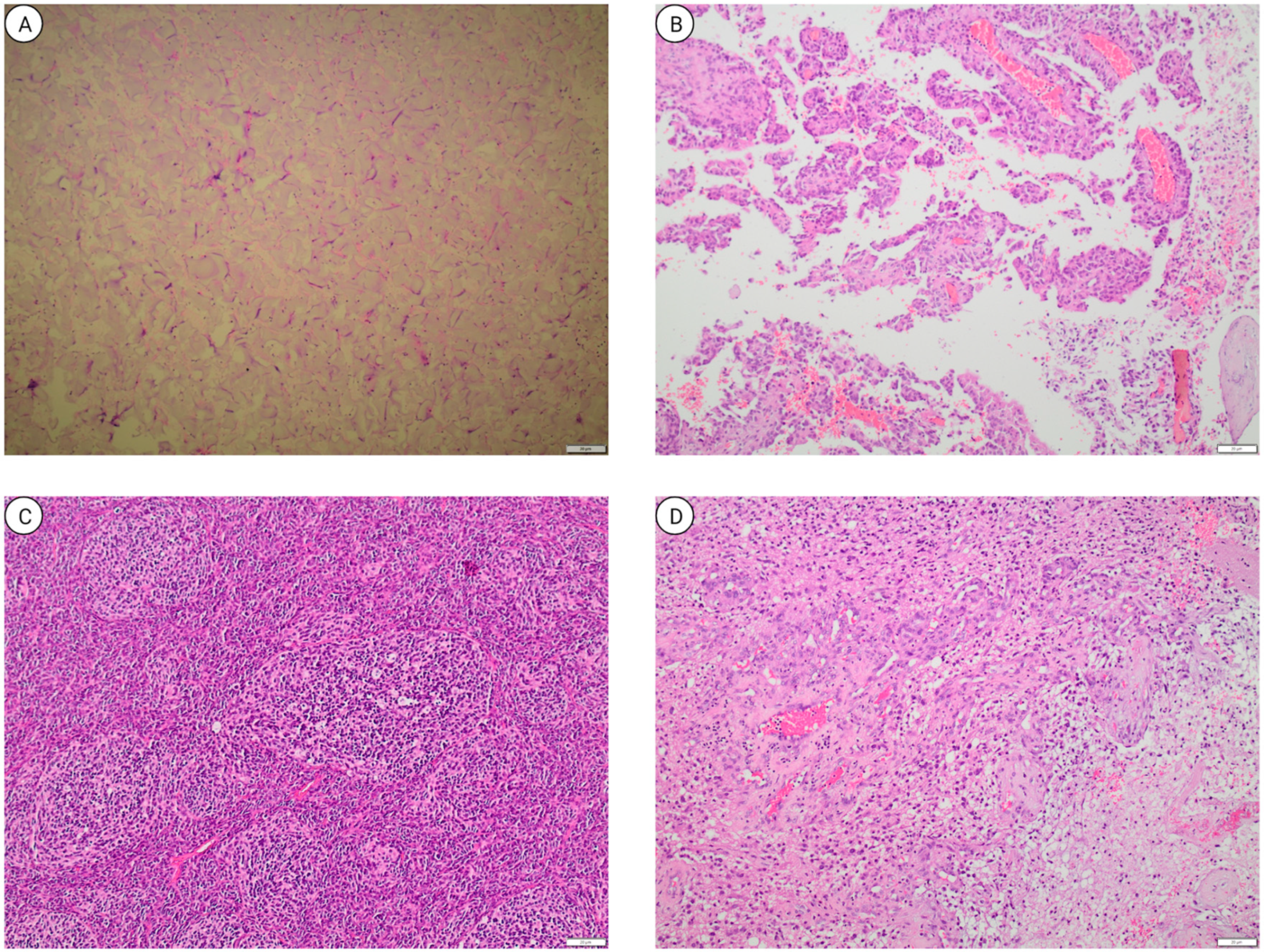
1.4. Pediatric Brain Tumor Pathology
2. Materials and Methods
2.1. Dataset Queries
2.2. Zinc Finger Information Acquisition
2.3. Microarray Analysis
2.3.1. Raw Data Quality Assessment and Normalization
2.3.2. Differential Gene Expression Analysis
2.3.3. Functional Enrichment Analysis and Gene-Disease Networks Construction
3. Results
3.1. Final Dataset
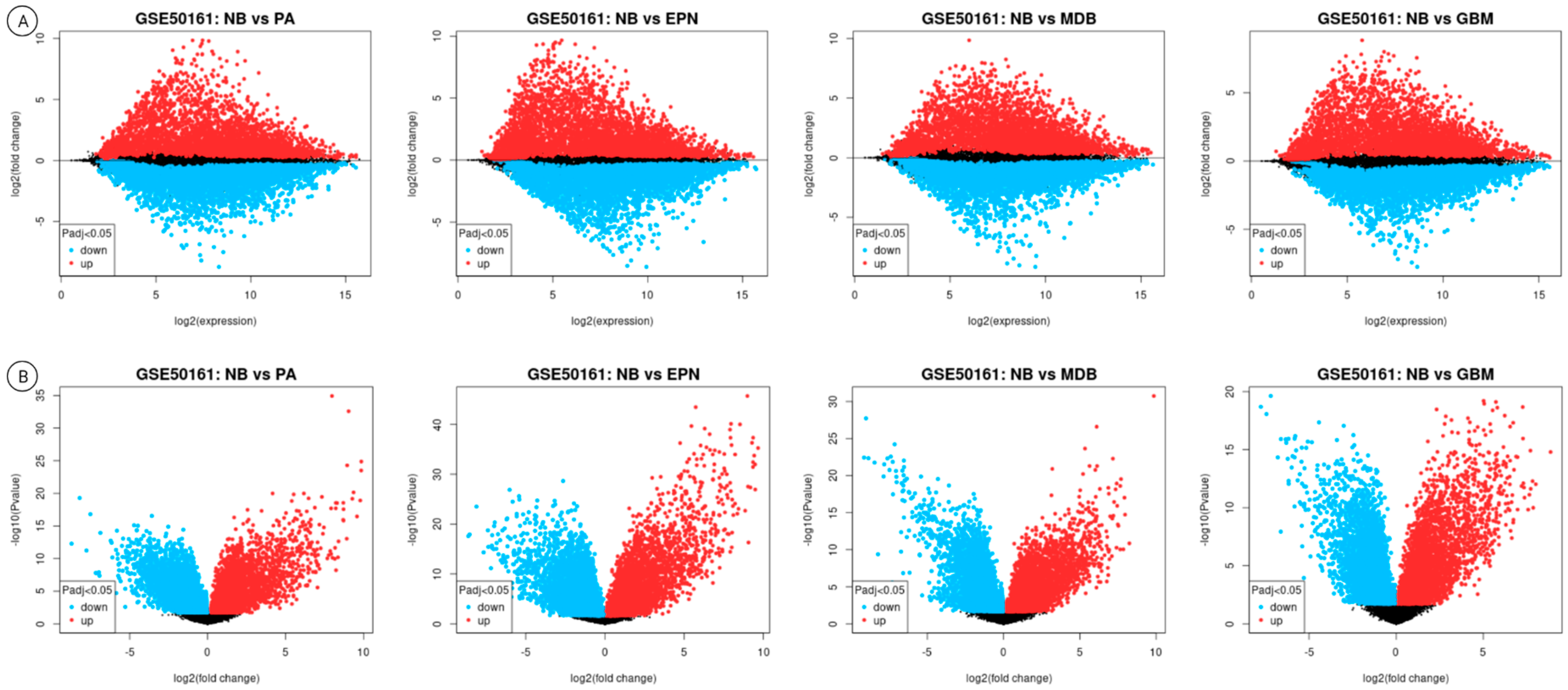
3.2. Differential Gene Expression Results
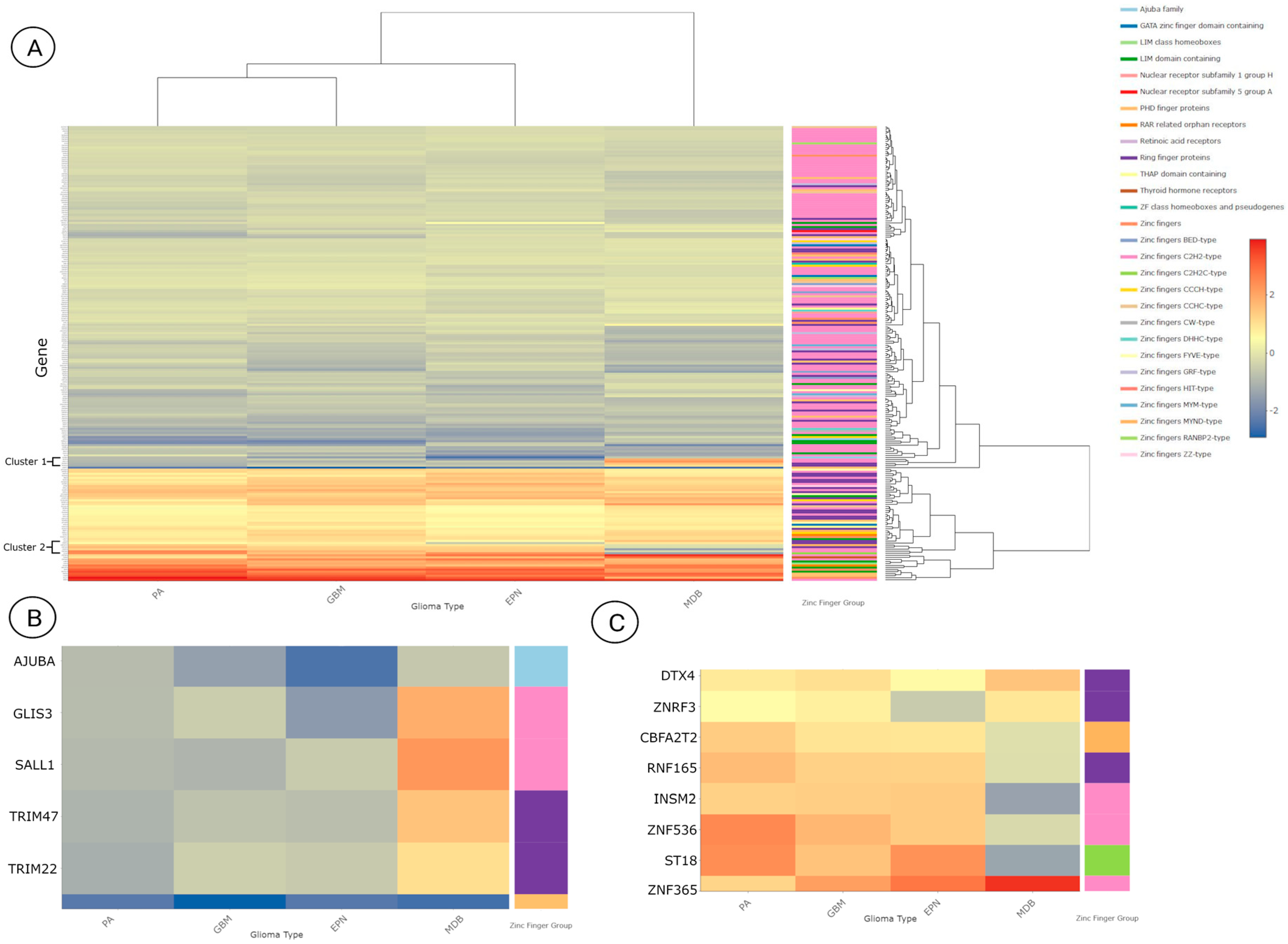
3.3. Differentially Expressed Zinc Finger-Containing Genes
3.4. Over-Representation Analysis
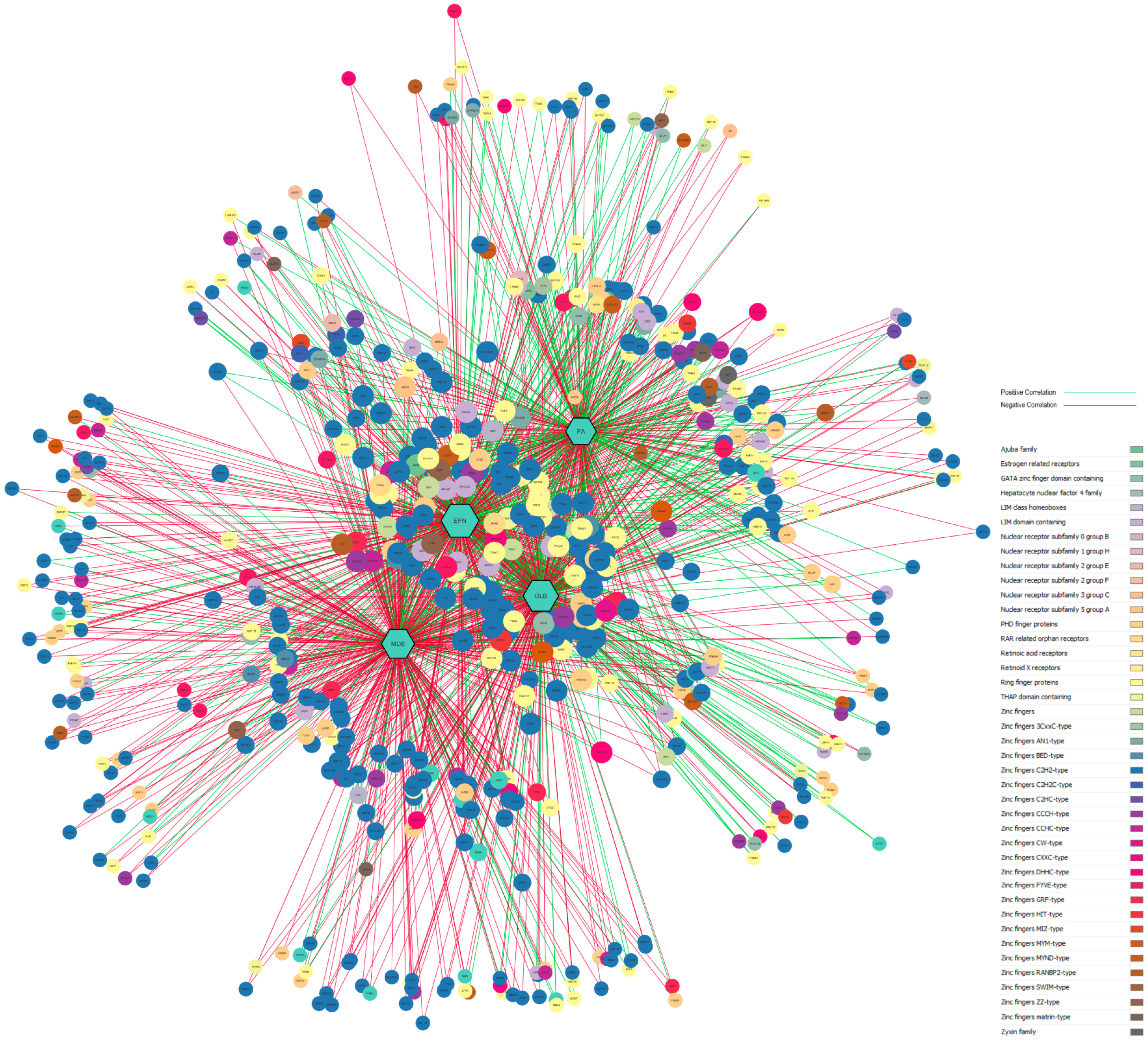
3.5. Gene-Disease Network Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Alliance | Alliance of Genome Resources |
| ATRT | Atypical Teratoid/Rhabdoid Tumors |
| CNS | Central Nervous System |
| DAG | Diacylglycerol |
| DIPGs | Diffuse Intrinsic Pontine Gliomas |
| DNET | Dysembryoplastic Neuroepithelial Tumors |
| EMT | Epithelial-Mesenchymal Transition |
| ENA | European Nucleotide Archive |
| EPNs | Ependymomas |
| FDR | False Discovery Rate |
| GBMs | Glioblastomas |
| GLI3 | GLI Family Zinc Finger 3 |
| GO | Gene Ontology |
| GSEA | Gene Set Enrichment Analysis |
| HGNC | HUGO Gene Nomenclature Committee |
| Log2FC | Log2 Fold Change |
| MD | Mean Difference |
| MDBs | Medulloblastomas |
| MyT1 | Myelin Transcription Factor 1 |
| NF-κB | Nuclear Factor kappa B |
| NZF-1 | Neural Zinc Finger Factor-1 |
| ORA | Over-Representation Analysis |
| PAs | Pilocytic Astrocytomas |
| PCA | Principal Component Analysis |
| PLP | Proteolipid Protein |
| PPIs | Protein–Protein Interactions |
| QC | Quality Control |
| RGPs | Radical Progenitor Cells |
| SHH | Sonic Hedgehog |
| ST18 | Suppression of Tumorigenicity 18 |
| TFs | Transcription Factors |
| UHRF1 | Ubiquitin-like with PHD and Ring Finger Domains 1 |
| UMAP | Uniform Manifold Approximation Projections |
| UniProtKB | UniProt KnowledgeBase |
| UPS | Ubiquitin-Proteasome System |
| UTR | 3′ Untranslated Region |
| ZFAND3 | AN1/A20 Zinc Finger Domain Containing Protein 3 |
| ZHX1 | Zinc Fingers and Homeoboxes Protein 1 |
| β-RAR | β-Retinoic Acid Receptor |
References
- Isernia, C.; Malgieri, G.; Russo, L.; D’Abrosca, G.; Baglivo, I.; Pedone, P.V.; Fattorusso, R. Zinc Fingers. Met. Ions. Life Sci. 2020, 20, 415–436. [Google Scholar] [CrossRef]
- Roy, S.; Dutta, S.; Khanna, K.; Singla, S.; Sundar, D. Prediction of DNA-binding specificity in zinc finger proteins. J. Biosci. 2012, 37, 483–491. [Google Scholar] [CrossRef]
- Persikov, A.V.; Wetzel, J.L.; Rowland, E.F.; Oakes, B.L.; Xu, D.J.; Singh, M.; Noyes, M.B. A systematic survey of the Cys2His2 zinc finger DNA-binding landscape. Nucleic. Acids. Res. 2015, 43, 1965–1984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mackay, J.P.; Crossley, M. Zinc fingers are sticking together. Trends Biochem. Sci. 1998, 23, 1–4. [Google Scholar] [CrossRef]
- Fu, M.; Blackshear, P.J. RNA-binding proteins in immune regulation: A focus on CCCH zinc finger proteins. Nat. Rev. Immunol. 2017, 17, 130–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, F.; Ilbert, M.; Varadan, R.; Cremers, C.M.; Hoyos, B.; Acin-Perez, R.; Vinogradov, V.; Cowburn, D.; Jakob, U.; Hammerling, U. Are Zinc-Finger Domains of Protein Kinase C Dynamic Structures That Unfold by Lipid or Redox Activation? Antioxid. Redox Signal. 2010, 14, 757–766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Priem, D.; Devos, M.; Druwé, S.; Martens, A.; Slowicka, K.; Ting, A.T.; Pasparakis, M.; Declercq, W.; Vandenabeele, P.; van Loo, G.; et al. A20 protects cells from TNF-induced apoptosis through linear ubiquitin-dependent and -independent mechanisms. Cell Death Dis. 2019, 10, 692. [Google Scholar] [CrossRef]
- Matera, A.G.; Hebert, M.D. The survival motor neurons protein uses its ZPR for nuclear localization. Nat. Cell Biol. 2001, 3, E93–E94. [Google Scholar] [CrossRef]
- Al-Naama, N.; Mackeh, R.; Kino, T. C2H2-Type Zinc Finger Proteins in Brain Development, Neurodevelopmental, and Other Neuropsychiatric Disorders: Systematic Literature-Based Analysis. Front. Neurol. 2020, 11, 32. [Google Scholar] [CrossRef] [Green Version]
- Nowick, K.; Gernat, T.; Almaas, E.; Stubbs, L. Differences in human and chimpanzee gene expression patterns define an evolving network of transcription factors in brain. Proc. Natl. Acad. Sci. USA 2009, 106, 22358. [Google Scholar] [CrossRef] [Green Version]
- Manukyan, A.; Kowalczyk, I.; Melhuish, T.A.; Lemiesz, A.; Wotton, D. Analysis of transcriptional activity by the Myt1 and Myt1l transcription factors. J. Cell. Biochem. 2018, 119, 4644–4655. [Google Scholar] [CrossRef]
- Besold, A.N.; Oluyadi, A.A.; Michel, S.L.J. Switching Metal Ion Coordination and DNA Recognition in a Tandem CCHHC-type Zinc Finger Peptide. Inorg. Chem. 2013, 52, 4721–4728. [Google Scholar] [CrossRef]
- Jiang, Y.; Yu, V.C.; Buchholz, F.; O’Connell, S.; Rhodes, S.J.; Candeloro, C.; Xia, Y.-R.; Lusis, A.J.; Rosenfeld, M.G. A Novel Family of Cys-Cys, His-Cys Zinc Finger Transcription Factors Expressed in Developing Nervous System and Pituitary Gland (*). J. Biol. Chem. 1996, 271, 10723–10730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Siqueira, M.F.; Behl, Y.; Alikhani, M.; Graves, D.T. The transcription factor ST18 regulates proapoptotic and proinflammatory gene expression in fibroblasts. FASEB J. 2008, 22, 3956–3967. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.G.; Armstrong, R.C.; Agoston, D.v.; Robinsky, A.; Wiese, C.; Nagle, J.; Hudson, L.D. Myelin transcription factor 1 (Myt1) of the oligodendrocyte lineage, along with a closely related CCHC zinc finger, is expressed in developing neurons in the mammalian central nervous system. J. Neurosci. Res. 1997, 50, 272–290. [Google Scholar] [CrossRef] [Green Version]
- Stevens, S.J.; van Ravenswaaij-Arts, C.M.; Janssen, J.W.; Klein Wassink-Ruiter, J.S.; van Essen, A.J.; Dijkhuizen, T.; van Rheenen, J.; Heuts-Vijgen, R.; Stegmann, A.P.; Smeets, E.E.; et al. MYT1L is a candidate gene for intellectual disability in patients with 2p25.3 (2pter) deletions. Am. J. Med. Genet. A 2011, 155, 2739–2745. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, A.; Oka, A.; Ito, M.; Tanaka, F.; Okoshi, Y.; Takashima, S. Myelin transcription factor 1 (MyT1) immunoreactivity in infants with periventricular leukomalacia. Dev. Brain Res. 2003, 140, 85–92. [Google Scholar] [CrossRef]
- Schuster, A.; Klein, E.; Neirinckx, V.; Knudsen, A.M.; Fabian, C.; Hau, A.-C.; Dieterle, M.; Oudin, A.; Nazarov, P.V.; Golebiewska, A.; et al. AN1-type zinc finger protein 3 (ZFAND3) is a transcriptional regulator that drives Glioblastoma invasion. Nat. Commun. 2020, 11, 6366. [Google Scholar] [CrossRef]
- Zhang, J.; Luo, J.; Jiang, H.; Xie, T.; Zheng, J.; Tian, Y.; Li, R.; Wang, B.; Lin, J.; Xu, A.; et al. The Tumor Suppressor Role of Zinc Finger Protein 671 (ZNF671) in Multiple Tumors Based on Cancer Single-Cell Sequencing. Front. Oncol. 2019, 9, 1214. [Google Scholar] [CrossRef] [Green Version]
- Kwon, R.-J.; Han, M.-E.; Kim, Y.-J.; Kim, Y.H.; Kim, J.-y.; Liu, L.; Heo, W.; Oh, S.-O. Roles of zinc-fingers and homeoboxes 1 during the proliferation, migration, and invasion of glioblastoma cells. Tumor Biol. 2017, 39, 1010428317694575. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Fletcher, M.; Gu, Z.; Wang, Q.; Costa, B.; Bertoni, A.; Man, K.-H.; Schlotter, M.; Felsberg, J.; Mangei, J.; et al. Glioblastoma epigenome profiling identifies SOX10 as a master regulator of molecular tumour subtype. Nat. Commun. 2020, 11, 6434. [Google Scholar] [CrossRef]
- Melhuish, T.A.; Kowalczyk, I.; Manukyan, A.; Zhang, Y.; Shah, A.; Abounader, R.; Wotton, D. Myt1 and Myt1l transcription factors limit proliferation in GBM cells by repressing YAP1 expression. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 983–995. [Google Scholar] [CrossRef]
- Hauser, P. Classification and Treatment of Pediatric Gliomas in the Molecular Era. Child 2021, 8, 739. [Google Scholar] [CrossRef]
- Pfister, S.M.; Capper, D.; Jones, D.T.W. Modern Principles of CNS Tumor Classification. In Brain Tumors in Children; Gajjar, A., Reaman, G.H., Racadio, J.M., Smith, F.O., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 117–129. [Google Scholar]
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Cancer, I.A.F.R.O. WHO Classification of Tumours of the Central Nervous System, 4th revised ed.; IARC Press: Lyon, France, 2016. [Google Scholar]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [Green Version]
- Udaka, Y.T.; Packer, R.J. Pediatric Brain Tumors. Neurol. Clin. 2018, 36, 533–556. [Google Scholar] [CrossRef]
- Toland, A.; McNulty, S.N.; Pekmezci, M.; Evenson, M.; Huntoon, K.; Pierson, C.R.; Boue, D.R.; Perry, A.; Dahiya, S. Pediatric meningioma: A clinicopathologic and molecular study with potential grading implications. Brain Pathol. 2020, 30, 1134–1143. [Google Scholar] [CrossRef]
- Schulz, M.; Afshar-Bakshloo, M.; Koch, A.; Capper, D.; Driever, P.H.; Tietze, A.; Grün, A.; Thomale, U.-W. Management of pineal region tumors in a pediatric case series. Neurosurg. Rev. 2021, 44, 1417–1427. [Google Scholar] [CrossRef] [PubMed]
- Tabash, M.A. Characteristics, survival and incidence rates and trends of pilocytic astrocytoma in children in the United States; SEER-based analysis. J. Neurol. Sci. 2019, 400, 148–152. [Google Scholar] [CrossRef]
- Oushy, S.; Perry, A.; Graffeo, C.S.; Raghunathan, A.; Carlstrom, L.P.; Daniels, D.J. Pediatric ganglioglioma of the brainstem and cervicomedullary junction: A retrospective cohort study. J. Neurosurg. Pediatr. 2019, 25, 30–36. [Google Scholar] [CrossRef]
- Singla, A.K.; Madan, R.; Gupta, K.; Goyal, S.; Kumar, N.; Sahoo, S.K.; Uppal, D.K.; Ahuja, C.K. Clinical behaviour and outcome in pediatric glioblastoma: Current scenario. Radiat. Oncol. J. 2021, 39, 72–77. [Google Scholar] [CrossRef]
- Chamdine, O.; Gajjar, A. Molecular characteristics of pediatric high-grade gliomas. CNS Oncol. 2014, 3, 433–443. [Google Scholar] [CrossRef]
- Perrone, M.G.; Ruggiero, A.; Centonze, A.; Carrieri, A.; Ferorelli, S.; Scilimati, A. Diffuse Intrinsic Pontine Glioma (DIPG): Breakthrough and Clinical Perspective. Curr. Med. Chem. 2021, 28, 3287–3317. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Gittleman, H.; Fulop, J.; Liu, M.; Blanda, R.; Kromer, C.; Wolinsky, Y.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2008–2012. Neuro-oncology 2015, 17 (Suppl. S4), iv1–iv62. [Google Scholar] [CrossRef]
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Burger, P.C.; Jouvet, A.; Scheithauer, B.W.; Kleihues, P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007, 114, 97–109. [Google Scholar] [CrossRef] [Green Version]
- Bayliss, J.; Mukherjee, P.; Lu, C.; Jain, S.U.; Chung, C.; Martinez, D.; Sabari, B.; Margol, A.S.; Panwalkar, P.; Parolia, A.; et al. Lowered H3K27me3 and DNA hypomethylation define poorly prognostic pediatric posterior fossa ependymomas. Sci. Transl. Med. 2016, 8, 366ra161. [Google Scholar] [CrossRef] [Green Version]
- Pajtler, K.W.; Witt, H.; Sill, M.; Jones, D.T.W.; Hovestadt, V.; Kratochwil, F.; Wani, K.; Tatevossian, R.; Punchihewa, C.; Johann, P.; et al. Molecular Classification of Ependymal Tumors across All CNS Compartments, Histopathological Grades, and Age Groups. Cancer Cell 2015, 27, 728–743. [Google Scholar] [CrossRef] [Green Version]
- Ostrom, Q.T.; Gittleman, H.; Truitt, G.; Boscia, A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011–2015. Neuro-oncology 2018, 20, iv1–iv86. [Google Scholar] [CrossRef] [Green Version]
- Harrison, P.W.; Ahamed, A.; Aslam, R.; Alako, B.T.F.; Burgin, J.; Buso, N.; Courtot, M.; Fan, J.; Gupta, D.; Haseeb, M.; et al. The European Nucleotide Archive in 2020. Nucleic Acids Res. 2021, 49, D82–D85. [Google Scholar] [CrossRef]
- Athar, A.; Füllgrabe, A.; George, N.; Iqbal, H.; Huerta, L.; Ali, A.; Snow, C.; Fonseca, N.A.; Petryszak, R.; Papatheodorou, I.; et al. ArrayExpress update–from bulk to single-cell expression data. Nucleic Acids Res. 2019, 47, D711–D715. [Google Scholar] [CrossRef] [PubMed]
- Tweedie, S.; Braschi, B.; Gray, K.; Jones, T.E.M.; Seal, R.L.; Yates, B.; Bruford, E.A. Genenames.org: The HGNC and VGNC resources in 2021. Nucleic Acids Res. 2021, 49, D939–D946. [Google Scholar] [CrossRef]
- Krishna, S.S.; Majumdar, I.; Grishin, N.V. Structural classification of zinc fingers: Survey and summary. Nucleic Acids Res. 2003, 31, 532–550. [Google Scholar] [CrossRef] [Green Version]
- Cassandri, M.; Smirnov, A.; Novelli, F.; Pitolli, C.; Agostini, M.; Malewicz, M.; Melino, G.; Raschellà, G. Zinc-finger proteins in health and disease. Cell Death Discov. 2017, 3, 17071. [Google Scholar] [CrossRef] [Green Version]
- Matys, V.; Kel-Margoulis, O.V.; Fricke, E.; Liebich, I.; Land, S.; Barre-Dirrie, A.; Reuter, I.; Chekmenev, D.; Krull, M.; Hornischer, K.; et al. TRANSFAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006, 34, D108–D110. [Google Scholar] [CrossRef] [Green Version]
- The UniProt, C. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef] [PubMed]
- Alliance of Genome Resources Portal: Unified model organism research platform. Nucleic Acids Res. 2020, 48, D650–D658. [CrossRef] [PubMed] [Green Version]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [CrossRef]
- Carbon, S.; Ireland, A.; Mungall, C.J.; Shu, S.; Marshall, B.; Lewis, S. AmiGO: Online access to ontology and annotation data. Bioinformatics 2009, 25, 288–289. [Google Scholar] [CrossRef]
- Huber, W.; Carey, V.J.; Gentleman, R.; Anders, S.; Carlson, M.; Carvalho, B.S.; Bravo, H.C.; Davis, S.; Gatto, L.; Girke, T.; et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 2015, 12, 115–121. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 1 November 2021).
- RStudio Team. RStudio: Integrated Development for R. RstudioInc. Available online: https://www.rstudio.com/ (accessed on 1 November 2021).
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef] [Green Version]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [Green Version]
- Blighe, K.; Lun, A. PCAtools: Everything Principal Components Analysis. 2021. Available online: https://bioconductor.org/packages/release/bioc/vignettes/PCAtools/inst/doc/PCAtools.html (accessed on 11 November 2021).
- McInnes, L.; Healy, J.; Melville, J. Umap: Uniform manifold approximation and projection for dimension reduction. arXiv 2018, arXiv:1802.03426. [Google Scholar]
- Murrell, P. R Graphics. WIREs Comput. Stat. 2009, 1, 216–220. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic. Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khatri, P.; Sirota, M.; Butte, A.J. Ten years of pathway analysis: Current approaches and outstanding challenges. PLoS Comput. Biol. 2012, 8, e1002375. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic. Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Koutrouli, M.; Karatzas, E.; Paez-Espino, D.; Pavlopoulos, G.A. A Guide to Conquer the Biological Network Era Using Graph Theory. Front. Bioeng. Biotechnol. 2020, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Goh, K.I.; Cusick, M.E.; Valle, D.; Childs, B.; Vidal, M.; Barabási, A.L. The human disease network. Proc. Natl. Acad. Sci. USA 2007, 104, 8685–8690. [Google Scholar] [CrossRef] [Green Version]
- García Del Valle, E.P.; Lagunes García, G.; Prieto Santamaría, L.; Zanin, M.; Menasalvas Ruiz, E.; Rodríguez-González, A. Disease networks and their contribution to disease understanding: A review of their evolution, techniques and data sources. J. Biomed. Inform. 2019, 94, 103206. [Google Scholar] [CrossRef]
- Heer, J.; Card, S.; Landay, J. Prefuse: A toolkit for interactive information visualization. Proceedings of ACM CHI 2005 Conference on Human Factors in Computing Systems, Portland, OR, USA, 2 April 2005; pp. 421–430. [Google Scholar]
- Kawai, S. An Algorithm for Drawing General Undirected Graphs Tomihisa Kamada and Satoru Kawai. Inf. Process. Lett. 1989, 31, 7–15. [Google Scholar]
- Harrington, B. Inkscape Project. Available online: https://inkscape.org/ (accessed on 10 November 2021).
- Griesinger, A.M.; Birks, D.K.; Donson, A.M.; Amani, V.; Hoffman, L.M.; Waziri, A.; Wang, M.; Handler, M.H.; Foreman, N.K. Characterization of distinct immunophenotypes across pediatric brain tumor types. J. Immunol. 2013, 191, 4880–4888. [Google Scholar] [CrossRef] [Green Version]
- Galili, T.; O’Callaghan, A.; Sidi, J.; Sievert, C. Heatmaply: An R package for creating interactive cluster heatmaps for online publishing. Bioinformatics 2018, 34, 1600–1602. [Google Scholar] [CrossRef] [PubMed]
- Chi, D.; Zhang, W.; Jia, Y.; Cong, D.; Hu, S. Spalt-Like Transcription Factor 1 (SALL1) Gene Expression Inhibits Cell Proliferation and Cell Migration of Human Glioma Cells Through the Wnt/β-Catenin Signaling Pathway. Med. Sci. Monit. Basic Res. 2019, 25, 128–138. [Google Scholar] [CrossRef]
- Ji, B.; Liu, L.; Guo, Y.; Ming, F.; Jiang, J.; Li, F.; Zhao, G.a.; Wen, J.; Li, N. Upregulated Tripartite Motif 47 Could Facilitate Glioma Cell Proliferation and Metastasis as a Tumorigenesis Promoter. Comput. Math. Methods Med. 2021, 2021, 5594973. [Google Scholar] [CrossRef]
- Ji, J.; Ding, K.; Luo, T.; Zhang, X.; Chen, A.; Zhang, D.; Li, G.; Thorsen, F.; Huang, B.; Li, X.; et al. TRIM22 activates NF-κB signaling in glioblastoma by accelerating the degradation of IκBα. Cell Death Differ. 2021, 28, 367–381. [Google Scholar] [CrossRef]
- Chen, J.; Ananthanarayanan, B.; Springer, K.S.; Wolf, K.J.; Sheyman, S.M.; Tran, V.D.; Kumar, S. Suppression of LIM Kinase 1 and LIM Kinase 2 Limits Glioblastoma Invasion. Cancer Res. 2020, 80, 69–78. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Liu, Y.; Xuan, C.; Liu, Y.; Shi, H.; Gao, Y. Identification of ubiquitination-related genes in human glioma as indicators of patient prognosis. PLoS ONE 2021, 16, e0250239. [Google Scholar] [CrossRef] [PubMed]
- Scholz, N.; Kurian, K.M.; Siebzehnrubl, F.A.; Licchesi, J.D.F. Targeting the Ubiquitin System in Glioblastoma. Front. Oncol. 2020, 10, 574011. [Google Scholar] [CrossRef]
- Nady, N.; Lemak, A.; Walker, J.R.; Avvakumov, G.V.; Kareta, M.S.; Achour, M.; Xue, S.; Duan, S.; Allali-Hassani, A.; Zuo, X.; et al. Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein. J. Biol. Chem. 2011, 286, 24300–24311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajakumara, E.; Wang, Z.; Ma, H.; Hu, L.; Chen, H.; Lin, Y.; Guo, R.; Wu, F.; Li, H.; Lan, F.; et al. PHD finger recognition of unmodified histone H3R2 links UHRF1 to regulation of euchromatic gene expression. Mol. Cell. 2011, 43, 275–284. [Google Scholar] [CrossRef] [Green Version]
- Xie, S.; Jakoncic, J.; Qian, C. UHRF1 double tudor domain and the adjacent PHD finger act together to recognize K9me3-containing histone H3 tail. J. Mol. Biol. 2012, 415, 318–328. [Google Scholar] [CrossRef]
- Jung, Y.-D.; Shim, J.-W.; Park, S.-J.; Choi, S.H.; Yang, K.; Heo, K.; Park, M.-T. Downregulation of UHRF1 promotes EMT via inducing CXCR4 in human cancer cells. Int. J. Oncol. 2015, 46, 1232–1242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sidhu, H.; Capalash, N. UHRF1: The key regulator of epigenetics and molecular target for cancer therapeutics. Tumor Biol. 2017, 39, 1010428317692205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashraf, W.; Ibrahim, A.; Alhosin, M.; Zaayter, L.; Ouararhni, K.; Papin, C.; Ahmad, T.; Hamiche, A.; Mély, Y.; Bronner, C.; et al. The epigenetic integrator UHRF1: On the road to become a universal biomarker for cancer. Oncotarget 2017, 8, 51946. [Google Scholar] [CrossRef] [Green Version]

| Type Name | Zinc Finger Structure | Number of Genes | Number of TFs | |
|---|---|---|---|---|
| TRANSFAC | UniProtKB AND Alliance AND GO | |||
| Zinc fingers C2H2-type (ZNF) | C-x-C-x-H-x-H | 748 | 676 | 166 |
| Ring finger proteins (RNF) | C-x-C-x-C-x-H-xxx-C-x-C-x-C-x-C | 305 | 25 | 48 |
| PHD finger proteins (PHF) | C-x-C-x-C-x-C-xxx-H-x-C-x-C-x-C | 90 | 40 | 53 |
| LIM domain containing | C-x-C-x-H-x-C-x-C-x-C-x-C-x-(C,H,D) | 53 | 6 | 7 |
| LIM domain subgroups | - | 22 | 10 | 11 |
| Nuclear hormone receptors (NR) | C-x-C-x-C-x-C-xxx-C-x-C-x-C-x-C | 49 | 46 | 40 |
| Zinc fingers CCCH-type (ZC3H) | C-x-C-x-C-x-H | 37 | 5 | 2 |
| Zinc fingers FYVE-type (ZFYVE) | C-x-C-x-C-x-C-xxx-C-x-C-x-C-x-C | 31 | 0 | 1 |
| Zinc fingers CCHC-type (ZCCHC) | C-x-C-x-H-x-C | 24 | 0 | 2 |
| Zinc fingers DHHC-type (ZDHHC) | C-x-C-x-H-x-C-xxx-C-x-C-x-H-x-C | 24 | O | 0 |
| Zinc fingers MYND-type (ZMYND) | C-x-C-x-C-x-C-xxx-C-x-C-x-H-x-C | 21 | 5 | 8 |
| Zinc fingers RANBP2-type (ZRANB) | C-x-C-x-C-x-C | 21 | 3 | 5 |
| Zinc fingers ZZ-type (ZZZ) | C-x-C-x-C-x-C | 18 | 4 | 3 |
| Zinc fingers C2HC-type (ZC2HC) | C-x-C-x-H-x-C | 16 | 7 | 8 |
| GATA zinc finger domain containing (GATAD) | C-x-C-x-C-x-C | 15 | 15 | 15 |
| ZF class homeoboxes and pseudogenes | C-x-C-x-H-x-H | 15 | 13 | 9 |
| THAP domain containing (THAP) | C-x-C-x-C-x-H | 12 | 11 | 5 |
| Zinc fingers CXXC-type (CXXC) | C-x-C-x-C-x-C-xxx-C-x-C-x-C-x-C | 12 | 7 | 7 |
| Zinc fingers SWIM-type (ZSWIM) | C-x-C-x-C-x-H | 10 | 0 | 0 |
| Zinc fingers AN1-type (ZFAND) | C-x-C-x-C-x-C-xxx-C-x-H-x-H-x-C | 8 | 1 | 1 |
| Zinc fingers 3CxxC-type (Z3CXXC) | C-x-C-x-H-x-C | 8 | 0 | 0 |
| Zinc fingers CW-type (ZCW) | C-x-C-x-C-x-C | 7 | 0 | 0 |
| Zinc fingers GRF-type (ZGRF) | C-x-C-x-C-x-C | 7 | 0 | 2 |
| Zinc fingers MIZ-type (ZMIZ) | C-x-C-x-H-x-C | 7 | 1 | 6 |
| Zinc fingers BED-type (ZBED) | C-x-C-x-H-x-H | 6 | 6 | 4 |
| Zinc fingers HIT-type (ZNHIT) | C-x-C-x-C-x-C-xxx-C-x-C-x-H-x-C | 6 | 0 | 0 |
| Zinc fingers MYM-type (ZMYM) | C-x-C-x-C-x-C | 6 | 1 | 0 |
| Zinc fingers matrin-type (ZMAT) | C-x-C-x-H-x-H | 5 | 0 | 0 |
| Zinc fingers C2H2C-type | C-x-C-x-H-x-H | 3 | 3 | 3 |
| Zinc fingers DBF-type (ZDBF) | C-x-C-x-H-x-H | 3 | 1 | 0 |
| Zinc fingers PARP-type | C-x-C-x-H-x-C | 2 | 0 | 1 |
| Zinc finger encoding genes that are not grouped into further subsets | - | 15 | 1 | 2 |
| Gene Set | Description | Adj. p-Value | FDR |
|---|---|---|---|
| Pilocytic Astrocytoma | |||
| Gene Ontology | |||
| GO:0000209 | Protein polyubiquitination | 5.55 × 10−16 | 4.72 × 10−13 |
| GO:0030522 | Intracellular receptor signaling pathway | 3.32 × 10−12 | 1.41 × 10−9 |
| GO:0051865 | Protein autoubiquitination | 4.02 × 10−10 | 1.03 × 10−7 |
| GO:0006513 | Protein monoubiquitination | 4.85 × 10−10 | 1.03 × 10−7 |
| GO:0009755 | Hormone-mediated signaling pathway | 7.89 × 10−9 | 1.34 × 10−6 |
| GO:0016569 | Covalent chromatin modification | 1.02 × 10−7 | 1.45 × 10−5 |
| GO:0048545 | Response to steroid hormone | 2.83 × 10−7 | 3.43 × 10−5 |
| GO:0009896 | Positive regulation of catabolic process | 8.52 × 10−6 | 8.27 × 10−4 |
| GO:0010498 | Proteasomal protein catabolic process | 8.75 × 10−6 | 8.27 × 10−4 |
| GO:0032606 | Type I interferon production | 1.33 × 10−5 | 1.13 × 10−3 |
| GO:0052192 | Movement in environment of other organism involved in symbiotic interaction | 1.73 × 10−5 | 1.27 × 10−3 |
| GO:0018205 | Peptidyl-lysine modification | 1.79 × 10−5 | 1.27 × 10−3 |
| KEGG | |||
| hsa04120 | Ubiquitin mediated proteolysis | 2.02 × 10−5 | 4.12 × 10−3 |
| hsa05202 | Transcriptional misregulation in cancer | 2.60 × 10−5 | 4.12 × 10−3 |
| Ependymoma | |||
| Gene Ontology | |||
| GO:0016569 | Covalent chromatin modification | 0 | 0 |
| GO:0000209 | Protein polyubiquitination | 0 | 0 |
| GO:0051865 | Protein autoubiquitination | 0 | 0 |
| GO:0030522 | Intracellular receptor signaling pathway | 8.90 × 10−13 | 1.89 × 10−10 |
| GO:0006513 | Protein monoubiquitination | 5.31 × 10−12 | 9.03 × 10−10 |
| GO:0018205 | Peptidyl-lysine modification | 1.17 × 10−10 | 1.66 × 10−8 |
| GO:0009755 | Hormone-mediated signaling pathway | 2.77 × 10−10 | 2.94 × 10−8 |
| GO:0043543 | Protein acylation | 2.77 × 10−10 | 2.94 × 10−8 |
| GO:0048545 | Response to steroid hormone | 1.85 × 10−7 | 1.75 × 10−5 |
| GO:0018198 | Peptidyl-cysteine modification | 1.23 × 10−6 | 1.04 × 10−4 |
| GO:0006352 | DNA-templated transcription, initiation | 1.47 × 10−6 | 1.14 × 10−4 |
| GO:0021953 | Central nervous system neuron differentiation | 1.83 × 10−6 | 1.26 × 10−4 |
| GO:0052192 | Movement in environment of other organism involved in symbiotic interaction | 1.98 × 10−6 | 1.26 × 10−4 |
| GO:0040029 | Regulation of gene expression, epigenetic | 2.07 × 10−6 | 1.26 × 10−4 |
| GO:0010498 | Proteasomal protein catabolic process | 7.75 × 10−6 | 4.39 × 10−4 |
| GO:0000726 | Non-recombinational repair | 3.18 × 10−5 | 1.56 × 10−3 |
| GO:0006302 | Double-strand break repair | 3.30 × 10−5 | 1.56 × 10−3 |
| GO:0008213 | Protein alkylation | 3.31 × 10−5 | 1.56 × 10−3 |
| GO:0009896 | Positive regulation of catabolic process | 1.04 × 10−4 | 4.64 × 10−3 |
| GO:0045444 | Fat cell differentiation | 2.14 × 10−4 | 9.08 × 10−3 |
| GO:0048732 | Gland development | 2.38 × 10−4 | 9.63 × 10−3 |
| KEGG | |||
| hsa05202 | Transcriptional misregulation in cancer | 6.68 × 10−7 | 2.12 × 10−4 |
| Medulloblastoma | |||
| Gene Ontology | |||
| GO:0016569 | Covalent chromatin modification | 0 | 0 |
| GO:0018205 | Peptidyl-lysine modification | 1.04 × 10−12 | 4.41 × 10−10 |
| GO:0043543 | Protein acylation | 6.04 × 10−11 | 1.71 × 10−8 |
| GO:0040029 | Regulation of gene expression, epigenetic | 3.03 × 10−10 | 6.43 × 10−8 |
| GO:0006513 | Protein monoubiquitination | 4.90 × 10−10 | 8.33 × 10−8 |
| GO:0000209 | Protein polyubiquitination | 1.43 × 10−9 | 2.02 × 10−7 |
| GO:0051865 | Protein autoubiquitination | 5.05 × 10−9 | 6.13 × 10−7 |
| GO:0009755 | Hormone-mediated signaling pathway | 1.25 × 10−6 | 1.33 × 10−4 |
| GO:0030522 | Intracellular receptor signaling pathway | 3.81 × 10−6 | 3.60 × 10−4 |
| GO:0021953 | Central nervous system neuron differentiation | 9.88 × 10−6 | 8.40 × 10−4 |
| GO:0071514 | Genetic imprinting | 1.26 × 10−5 | 9.70 × 10−4 |
| GO:0006397 | Mrna processing | 1.37 × 10−5 | 9.70 × 10−4 |
| GO:0048545 | Response to steroid hormone | 3.58 × 10−5 | 2.23 × 10−3 |
| GO:0098727 | Maintenance of cell number | 3.67 × 10−5 | 2.23 × 10−3 |
| GO:0008213 | Protein alkylation | 4.49 × 10−5 | 2.54 × 10−3 |
| GO:0006352 | DNA-templated transcription, initiation | 5.83 × 10−5 | 3.10 × 10−3 |
| GO:0045444 | Fat cell differentiation | 6.22 × 10−5 | 3.11 × 10−3 |
| GO:1903706 | Regulation of hemopoiesis | 9.34 × 10−5 | 4.41 × 10−3 |
| GO:1990823 | Response to leukemia inhibitory factor | 1.08 × 10−4 | 4.64 × 10−3 |
| GO:0033044 | Regulation of chromosome organization | 1.09 × 10−4 | 4.64 × 10−3 |
| GO:0010948 | Negative regulation of cell cycle process | 1.35 × 10−4 | 5.45 × 10−3 |
| GO:0032259 | Methylation | 1.84 × 10−4 | 7.10 × 10−3 |
| GO:0030099 | Myeloid cell differentiation | 2.06 × 10−4 | 7.62 × 10−3 |
| Glioblastoma | |||
| Gene Ontology | |||
| GO:0000209 | Protein polyubiquitination | 0 | 0 |
| GO:0051865 | Protein autoubiquitination | 4.44 × 10−16 | 1.89 × 10−13 |
| GO:0016569 | Covalent chromatin modification | 1.96 × 10−13 | 5.56 × 10−11 |
| GO:0030522 | Intracellular receptor signaling pathway | 6.42 × 10−13 | 1.36 × 10−10 |
| GO:0009755 | Hormone-mediated signaling pathway | 2.38 × 10−12 | 4.04 × 10−10 |
| GO:0006513 | Protein monoubiquitination | 4.77 × 10−11 | 6.75 × 10−9 |
| GO:0048545 | Response to steroid hormone | 6.85 × 10−9 | 8.32 × 10−7 |
| GO:0018205 | Peptidyl-lysine modification | 8.24 × 10−8 | 8.75 × 10−6 |
| GO:0045444 | Fat cell differentiation | 4.51 × 10−6 | 3.92 × 10−4 |
| GO:0006352 | DNA-templated transcription, initiation | 4.77 × 10−6 | 3.92 × 10−4 |
| GO:0040029 | Regulation of gene expression, epigenetic | 5.08 × 10−6 | 3.92 × 10−4 |
| GO:0043543 | Protein acylation | 7.13 × 10−6 | 5.05 × 10−4 |
| GO:0008213 | Protein alkylation | 5.57 × 10−5 | 3.38 × 10−3 |
| GO:0021953 | Central nervous system neuron differentiation | 5.57 × 10−5 | 3.38 × 10−3 |
| GO:0052192 | Movement in environment of other organism involved in symbiotic interaction | 6.78 × 10−5 | 3.84 × 10−3 |
| GO:0032259 | Methylation | 7.92 × 10−5 | 4.21 × 10−3 |
| GO:1903706 | Regulation of hemopoiesis | 1.25 × 10−4 | 6.23 × 10−3 |
| KEGG | |||
| hsa05202 | Transcriptional misregulation in cancer | 1.68 × 10−7 | 5.32 × 10−5 |
| hsa04120 | Ubiquitin mediated proteolysis | 2.63 × 10−5 | 4.16 × 10−3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanakoglou, D.S.; Pampalou, A.; Malakou, L.S.; Lakiotaki, E.; Loupis, T.; Vrachnos, D.M.; Glekas, P.D.; Klonou, A.; Giannopoulou, A.-I.; Carpenter, M.; et al. Central Role of C2H2-Type Zinc Finger-Containing Genes in Pediatric Brain Tumors. DNA 2022, 2, 1-21. https://doi.org/10.3390/dna2010001
Kanakoglou DS, Pampalou A, Malakou LS, Lakiotaki E, Loupis T, Vrachnos DM, Glekas PD, Klonou A, Giannopoulou A-I, Carpenter M, et al. Central Role of C2H2-Type Zinc Finger-Containing Genes in Pediatric Brain Tumors. DNA. 2022; 2(1):1-21. https://doi.org/10.3390/dna2010001
Chicago/Turabian StyleKanakoglou, Dimitrios S., Andromachi Pampalou, Lina S. Malakou, Eleftheria Lakiotaki, Theodoros Loupis, Dimitrios M. Vrachnos, Panayiotis D. Glekas, Alexia Klonou, Angeliki-Ioanna Giannopoulou, Madison Carpenter, and et al. 2022. "Central Role of C2H2-Type Zinc Finger-Containing Genes in Pediatric Brain Tumors" DNA 2, no. 1: 1-21. https://doi.org/10.3390/dna2010001
APA StyleKanakoglou, D. S., Pampalou, A., Malakou, L. S., Lakiotaki, E., Loupis, T., Vrachnos, D. M., Glekas, P. D., Klonou, A., Giannopoulou, A.-I., Carpenter, M., Korkolopoulou, P., & Piperi, C. (2022). Central Role of C2H2-Type Zinc Finger-Containing Genes in Pediatric Brain Tumors. DNA, 2(1), 1-21. https://doi.org/10.3390/dna2010001