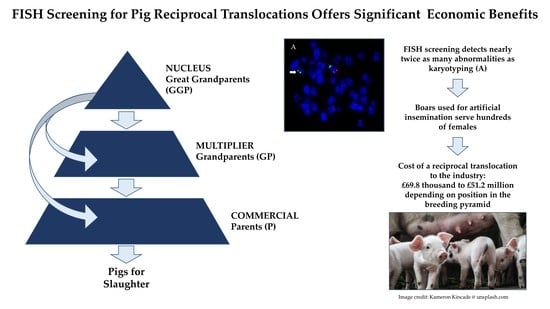
Incidence, Reproductive Outcome, and Economic Impact of Reciprocal Translocations in the Domestic Pig
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Preparation of Chromosome Metaphases and Karyotyping
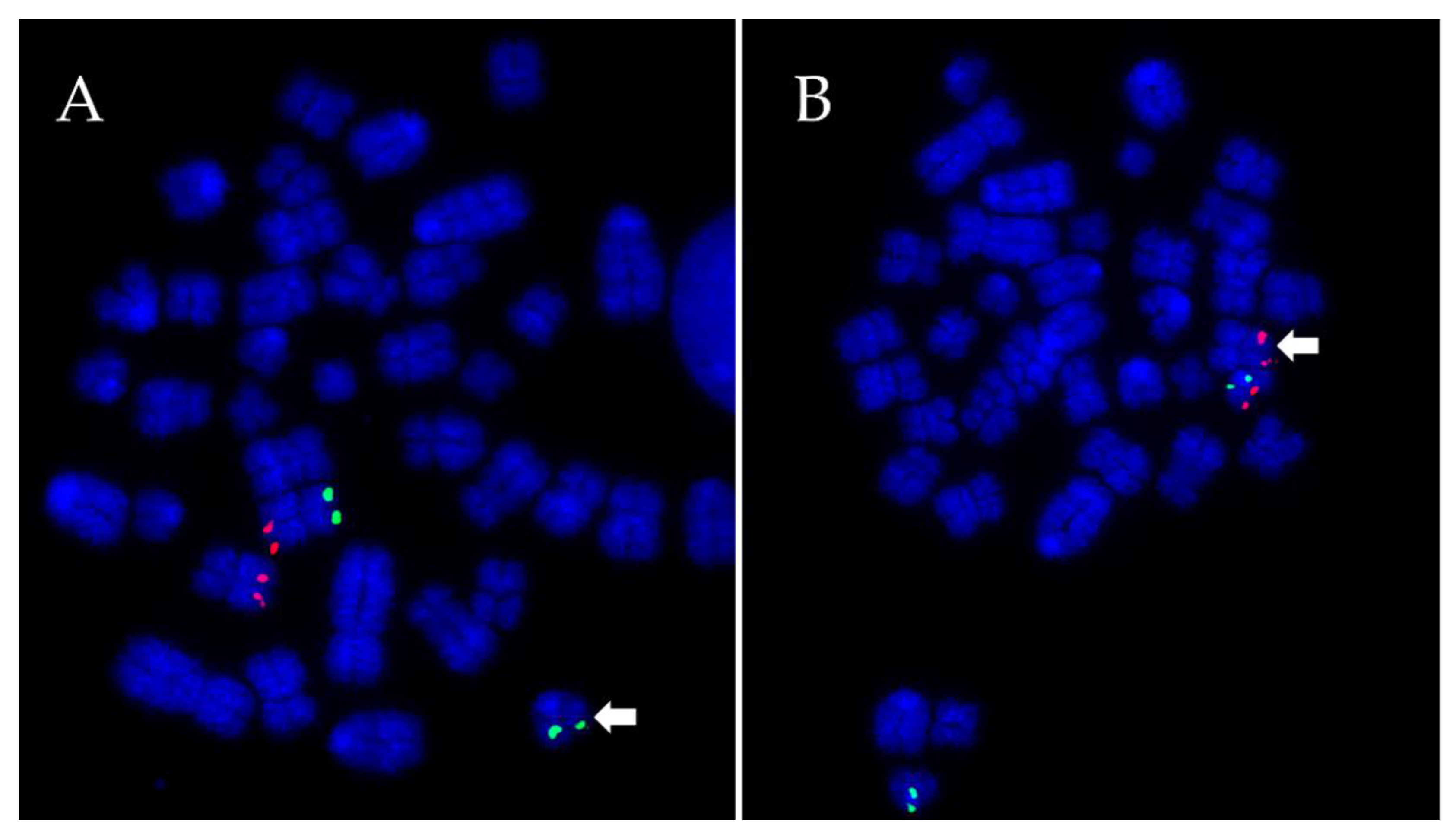
2.3. Fluorescence In Situ Hybridisation (FISH)
2.4. Microscopy and Image Analysis
2.5. Economic Impact Estimation Model
2.6. Data Analysis
3. Results
3.1. Prevalence and Incidence of Chromosomal Errors and RTs
3.2. The Economic Costs of RTs
- A commercial herd adopting a farrow to wean business model;
- A commercial herd adopting a farrow to finish business model;
- A multiplier herd;
- A nucleus herd producing terminal boars for use at stud;
- A nucleus herd, producing dam line sows.
3.3. Mitigating Losses by Proactive RT Screening
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. FAOSTAT. 2019. Available online: http://www.fao.org/faostat/en/#home (accessed on 26 December 2019).
- Knox, R. Artificial Insemination in Pigs Today. Theriogenology 2016, 85, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, A.; Sanz, E.; De Mercado, E.; Gómez, E.; Martín, M.; Carrascosa, C.; Gomez-Fidalgo, E.; Villagomez, D.; Sanchez-Sanchez, R. Reproductive Consequences of a Reciprocal Chromosomal Translocation in Two Duroc Boars used to Provide Semen for Artificial Insemination. Theriogenology 2010, 74, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Broekhuijse, M.L.W.J.; Šoštarić, E.; Feitsma, H.; Gadella, B.M. Application of computer-assisted semen analysis to explain variations in pig fertility1. J. Anim. Sci. 2012, 90, 779–789. [Google Scholar] [CrossRef] [PubMed]
- Kamphuis, C.; Duenk, P.; Veerkamp, R.F.; Visser, B.; Singh, G.; Nigsch, A.; De Mol, R.M.; Broekhuijse, M.L.W.J. Machine Learning to Further Improve the Decision which Boar Ejaculates to Process into Artificial Insemination Doses. Theriogenology 2020, 144, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Jennings, R.L.; Griffin, D.K.; O’Connor, R.E. A New Approach for Accurate Detection of Chromosome Rearrangements that Affect Fertility in Cattle. Animals 2020, 10, 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Connor, R.; Kiazim, L.; Rathje, C.; Jennings, R.; Griffin, D. Rapid Multi-Hybridisation FISH Screening for Balanced Porcine Reciprocal Translocations Suggests a Much Higher Abnormality Rate Than Previously Appreciated. Cells 2021, 10, 250. [Google Scholar] [CrossRef] [PubMed]
- Shams, F.; D’Souza, D.; Ezaz, T. Balanced Chromosomal Rearrangements Associated with Hypoprolificacy in Australian Boars (Sus scrofa domesticus). Cells 2021, 10, 2000. [Google Scholar] [CrossRef] [PubMed]
- Ducos, A.; Berland, H.-M.; Bonnet, N.; Calgaro, A.; Billoux, S.; Mary, N.; Garnier-Bonnet, A.; Darré, R.; Pinton, A. Chromosomal Control of Pig Populations in France: 2002–2006 Survey. Genet. Sel. Evol. 2007, 39, 583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez-Sánchez, R.; Gómez-Fidalgo, E.; Pérez-Garnelo, S.; Martín-Lluch, M.; De la Cruz-Vigo, P. Prevalence of Chromosomal Aberrations in Breeding Pigs in Spain. Reprod. Domest. Anim. 2019, 54, 98–101. [Google Scholar] [CrossRef] [PubMed]
- Danielak-Czech, B.; Kozubska-Sobocińska, A.; Smołucha, G.; Babicz, M. Breeding and Economic Aspects of Cytogenetic Screening Studies of Pigs Qualified for Reproduction. Animals 2020, 10, 1200. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Gil, J.E.; Estrada, E. Artificial Insemination in Boar Reproduction. In Boar Reproduction; Bonet, S., Casas, I., Holt, W.V., Yeste, M., Eds.; Springer: New York City, NY, USA, 2013; pp. 589–607. [Google Scholar]
- Grahofer, A.; Letko, A.; Häfliger, I.M.; Jagannathan, V.; Ducos, A.; Richard, O.; Peter, V.; Nathues, H.; Drögemüller, C. Chromo-somal Imbalance in Pigs Showing a Syndromic Form of Cleft Palate. BMC Genom. 2019, 20, 31068123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Visscher, P.; Pong-Wong, R.; Whittemore, C.; Haley, C. Impact of Biotechnology on (Cross)Breeding Programmes in Pigs. Livest. Prod. Sci. 2000, 65, 57–70. [Google Scholar] [CrossRef]
- O’Connor, R.; Fonseka, G.; Frodsham, R.; Archibald, A.; Lawrie, M.; Walling, G.A.; Griffin, D.K. Isolation of Subtelomeric Sequences of Porcine Chromosomes for Translocation Screening Reveals Errors in the Pig Genome Assembly. Anim. Genet. 2017, 48, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Agriculture and Horticulture Development Board (AHDB). Available online: https://ahdb.org.uk/ (accessed on 30 August 2021).
- Young, B.; Dewey, C.E.; Friendship, R.M. Management Factors Associated with Farrowing Rate in Commercial Sow Herds in On-tario. Can. Vet. J. 2010, 51, 185–189. [Google Scholar] [PubMed]
- Pierozan, C.; Callegari, M.; Dias, C.; de Souza, K.; Gasa, J.; da Silva, C. Herd-Level Factors Associated with Piglet Weight at Weaning, Kilograms of Piglets Weaned per Sow per Year and Sow Feed Conversion. Animals 2020, 14, 1283–1292. [Google Scholar] [CrossRef] [PubMed]
- Aherne, F.; Kirkwood, R. Factors Affecting Litter Size. Pig Articles from The Pig Site 2001. Available online: https://www.thepigsite.com/articles/factors-affecting-litter-size (accessed on 30 August 2021).
- McGlone, J.; Pond, W.G. Pig Production: Biological Principles and Applications; Delmar Learning, Inc.: Clifton Park, NY, USA, 2003. [Google Scholar]
- Freyer, G. Maximum Number of Total Born Piglets in a Parity and Individual Ranges in Litter Size Expressed as Specific Charac-teristics of Sows. J. Anim. Sci. Technol. 2018, 60, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Nowak, B.; Mucha, A.; Moska, M.; Kruszyński, W. Reproduction Indicators Related to Litter Size and Reproduction Cycle Length Among Sows of Breeds Considered Maternal and Paternal Components Kept on Medium-Size Farms. Animals 2020, 10, 1164. [Google Scholar] [CrossRef] [PubMed]
- Mainau, E.; Temple, D.; Manteca, X. Pre-Weaning Mortality in Piglets; Farm Animal Welfare Education Centre: Barcelona, Spain, 2015. [Google Scholar]
- Gebhardt, J.T.; Tokach, M.D.; Dritz, S.S.; DeRouchey, J.M.; Woodworth, J.C.; Goodband, R.D.; Henry, S.C. Postweaning Mortality in Commercial Swine Production II: Review of Infectious Contributing Factors. Transl. Anim. Sci. 2020, 4, 485–506. [Google Scholar] [CrossRef] [PubMed]
- BCC Research. Food and Beverages Market. Research Report; BCC Research: Wellesley, MA, USA, 2019. [Google Scholar]
- Dourmad, J.; Ryschawy, J.; Trousson, T.; Bonneau, M.; Gonzalez, J.; Houwers, H.; Hviid, M.; Zimmer, C.; Nguyen, T.L.T.; Morgensen, L. Evaluating Environmental Impacts of Contrasting Pig Farming Systems with Life Cycle Assessment. Animals 2014, 8, 2027–2037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Parameter | Definition | Evidence | Key Figures |
|---|---|---|---|
| Boar productivity | How many sperm doses and piglets are produced by a boar | interviews | 1872 semen doses per year 33.1 semen doses per week |
| Number of matings | Total matings by a single boar per year, each mating using multiple semen doses | interviews | 2.1 AI (artificial insemination) doses per mating 820.1 mattings per year |
| Boar prevalence | The maximum likely proportion of nucleus farm sows inseminated with a specific boar | interviews | 10% |
| Farrowing rate | The proportion of sows that are served that go on to farrow | [17] | 85–90% |
| Farrowing per year | The number of farrowings per sow per year | interviews [18] | 2.2 to 2.39 per sow |
| Total born alive | Total number of live piglets in a litter | interviews [19,20,21,22] | 8 to 16 piglets |
| Mortality | Piglets dying before reaching reproductive age or slaughter weight (as appropriate) | [20,23,24] | 5 to 35% pre-weaning 4 to 8% post-weaning |
| Dam selection rate | Proportion of dams that are selected to become parents of the next generation | interviews | 70% |
| Sire selection rate | Proportion of Great Grandparent (GGP) level boars that are selected to become sires to commercial pigs | interviews | 1.54 per litter |
| Availability rate | Proportion of females cycling and available for breeding | interviews | 90% |
| Corrective action time | Time interval between the first mating of an RT (reciprocal translocation) carrier boar and the discovery of a reduction in litter sizes for this animal | interviews | 16.6 weeks for first litters ≥19.6 weeks before sufficient data is gathered |
| Market values | Estimated monetary value of a commercial pig in British pounds (GBP) | [16] | £39.83 ± 0.81 per weaner pig £1.5 ± 0.02 per Kg of meat 88.79 ± 0.42 Kg average carcass weight |
| Chromosomal Error | Total Number of Cases | De Novo Cases |
|---|---|---|
| t(1;2) | 138 | 1 |
| t(1;3) | 1 | 1 |
| t(1;13) | 1 | 1 |
| t(1;17) | 1 | 1 |
| t(2;14) | 1 | 1 |
| t(3;7) | 1 | 1 |
| t(3;9) | 3 | 1 |
| t(3;17) | 1 | 1 |
| t(4;5) | 15 | 2 |
| t(5;6) | 1 | 1 |
| t(7;10) | 21 | 2 |
| t(9;10) | 1 | 1 |
| t(9;12) | 1 | 1 |
| t(9;13) | 2 | 2 |
| t(9;18) | 25 | 1 |
| t(10;15) | 1 | 1 |
| t(16;17) | 1 | 1 |
| XX/XY chimeric | 5 | 5 |
| other, complex | 2 | 2 |
| Key Scenario | Scenario Description |
|---|---|
| Commercial herd (farrow to wean) | A commercial level farm specializing in selling weaned pigs of about 7 kg in weight, purchasing AI semen doses from a stud farm. The RT is affecting one of the boars supplying the AI doses. |
| Commercial herd (farrow to finish) | This scenario mimics the previous one, but instead of selling piglets, this farm raises them to slaughter weight. |
| Multiplier herd | A multiplier herd is made up of purebred animals producing crossbred sows for commercial production. An RT affects a boar used to create these commercial sows. The calculation considers the cascade effect on the next generation of commercial level pigs. |
| Nucleus herd (terminal line) | A nucleus herd producing sires, where one of the boars is affected by an RT. This calculation considers the effects on the male’s offspring, and then on the generation of commercial pigs deriving from it. |
| Nucleus herd (dam line) | A nucleus herd aiming to produce dam line sires, where one of the boars is affected by an RT. This calculation considers the effects of the female and male offspring on the next generation of multiplier pigs and, further to that, commercial level pigs. |
| Key Scenario | Average Economic Impact (GBP) |
|---|---|
| Commercial herd (farrow to wean) | £69,802 ± 1413 |
| Commercial herd (farrow to finish) | £222,199 ± 3252 |
| Multiplier herd | £1,400,833 ± 28,351 |
| Nucleus herd (terminal line) | £4,442,945 ± £89,919 |
| Nucleus herd (dam line) | £51,215,378 ± £1,036,525 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lewis, N.M.; Rathje, C.C.; Canedo-Ribeiro, C.; Bosman, L.M.; Kiazim, L.G.; Jennings, R.L.; O’Connor, R.E.; Silvestri, G.; Griffin, D.K. Incidence, Reproductive Outcome, and Economic Impact of Reciprocal Translocations in the Domestic Pig. DNA 2021, 1, 68-76. https://doi.org/10.3390/dna1020007
Lewis NM, Rathje CC, Canedo-Ribeiro C, Bosman LM, Kiazim LG, Jennings RL, O’Connor RE, Silvestri G, Griffin DK. Incidence, Reproductive Outcome, and Economic Impact of Reciprocal Translocations in the Domestic Pig. DNA. 2021; 1(2):68-76. https://doi.org/10.3390/dna1020007
Chicago/Turabian StyleLewis, Nicole M., Claudia C. Rathje, Carla Canedo-Ribeiro, Lisa M. Bosman, Lucas G. Kiazim, Rebecca L. Jennings, Rebecca E. O’Connor, Giuseppe Silvestri, and Darren K. Griffin. 2021. "Incidence, Reproductive Outcome, and Economic Impact of Reciprocal Translocations in the Domestic Pig" DNA 1, no. 2: 68-76. https://doi.org/10.3390/dna1020007
APA StyleLewis, N. M., Rathje, C. C., Canedo-Ribeiro, C., Bosman, L. M., Kiazim, L. G., Jennings, R. L., O’Connor, R. E., Silvestri, G., & Griffin, D. K. (2021). Incidence, Reproductive Outcome, and Economic Impact of Reciprocal Translocations in the Domestic Pig. DNA, 1(2), 68-76. https://doi.org/10.3390/dna1020007