Demethylation of Non-CpG Sites in DNA Is Initiated by TET2 5-Methylcytosine Dioxygenase
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Purification of TET2 Dioxygenase
2.2.1. Purification of hTET2 Catalytic Domain
2.2.2. Purification of GST-Tagged hTET2 Catalytic Domain
2.2.3. Purification of Full-Length Myc-Tagged hTET2
2.3. 5mC Oxidation by TET2 Dioxygenase
2.4. MS/MS Parameters for Nucleosides
2.5. Liquid Chromatographic Conditions for Nucleosides under Different MS/MS Modes
3. Results and Discussion
3.1. Detection of Cytosine Derivatives Produced by Oxidative Demethylation of 5mCpG and 5mCpA Sites
3.2. Quantification of Cytosine Derivatives Produced by TET2-Mediated Oxidation of 5mCpH Sites; Effect of +1 Base
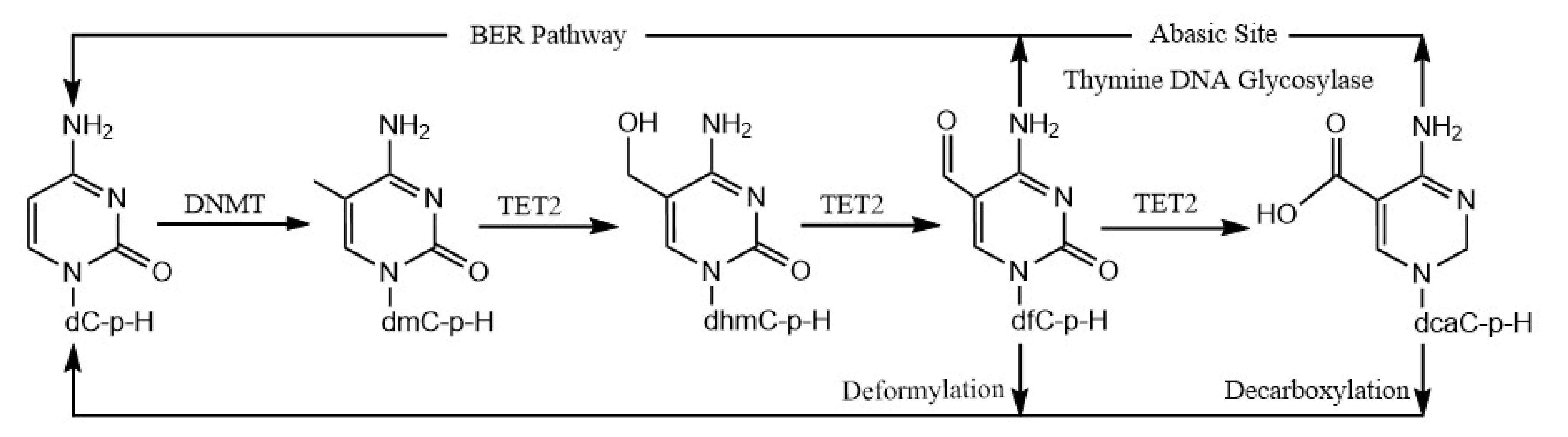
3.3. TET2 Can Initiate Demethylation of 5mCpH Sites
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
Correction Statement
Abbreviations
References
- Schultz, M.D.; He, Y.; Whitaker, J.W.; Hariharan, M.; Mukamel, E.; Leung, D.; Rajagopal, N.; Nery, J.R.; Urich, M.A.; Chen, H.; et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature 2015, 523, 212–216. [Google Scholar] [CrossRef] [PubMed]
- Bonasio, R.; Tu, S.; Reinberg, D. Molecular Signals of Epigenetic States. Science 2010, 330, 612–616. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Jacobsen, S.E.; Reik, W. Epigenetic Reprogramming in Plant and Animal Development. Science 2010, 330, 622–627. [Google Scholar] [CrossRef] [PubMed]
- Smith, Z.D.; Meissner, A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef]
- Jabbari, K.; Bernardi, G. Cytosine methylation and CpG, TpG (CpA) and TpA frequencies. Gene 2004, 333, 143–149. [Google Scholar] [CrossRef]
- Portela, A.; Esteller, M. Epigenetic modifications and human disease. Nat. Biotechnol. 2010, 28, 1057–1068. [Google Scholar] [CrossRef]
- Song, C.-X.; He, C. Balance of DNA methylation and demethylation in cancer development. Genome Biol. 2012, 13, 173. [Google Scholar] [CrossRef]
- Lister, R.; Pelizzola, M.; Dowen, R.H.; Hawkins, R.D.; Hon, G.; Tonti-Filippini, J.; Nery, J.R.; Lee, L.; Ye, Z.; Ngo, Q.-M.; et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009, 462, 315–322. [Google Scholar] [CrossRef]
- Laurent, L.; Wong, E.; Li, G.; Huynh, T.; Tsirigos, A.; Ong, C.T.; Low, H.M.; Sung, W.-K.; Rigoutsos, I.; Loring, J.; et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010, 20, 320–331. [Google Scholar] [CrossRef]
- Lister, R.; Pelizzola, M.; Kida, Y.; Hawkins, R.D.; Nery, J.R.; Hon, G.; Antosiewicz-Bourget, J.; O’Malley, R.; Castanon, R.; Klugman, S.; et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 2011, 471, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Ziller, M.J.; Müller, F.; Liao, J.; Zhang, Y.; Gu, H.; Bock, C.; Boyle, P.; Epstein, C.B.; Bernstein, B.E.; Lengauer, T.; et al. Genomic Distribution and Inter-Sample Variation of Non-CpG Methylation across Human Cell Types. PLoS Genet. 2011, 7, e1002389. [Google Scholar] [CrossRef] [PubMed]
- Ravichandran, M.; Jurkowska, R.Z.; Jurkowski, T.P. Target specificity of mammalian DNA methylation and demethylation machinery. Org. Biomol. Chem. 2017, 16, 1419–1435. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.U.; Su, Y.; Shin, J.H.; Shin, J.; Li, H.; Xie, B.; Zhong, C.; Hu, S.; Le, T.; Fan, G.; et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat. Neurosci. 2013, 17, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Patil, V.; Ward, R.; Hesson, L.B. The evidence for functional non-CpG methylation in mammalian cells. Epigenetics 2014, 9, 823–828. [Google Scholar] [CrossRef]
- Xiao, W.; Liu, X.; Niu, X.; Li, C.; Guo, Y.; Tan, J.; Xiong, W.; Fan, L.; Li, Y. The frequency of CpG and non-CpG methylation of Notch3 gene promoter determines its expression levels in breast cancer cells. Exp. Cell Res. 2019, 386, 111743. [Google Scholar] [CrossRef]
- Li, C.; Xiong, W.; Liu, X.; Xiao, W.; Guo, Y.; Tan, J.; Li, Y. Hypomethylation at non-CpG/CpG sites in the promoter of HIF-1α gene combined with enhanced H3K9Ac modification contribute to maintain higher HIF-1α expression in breast cancer. Oncogenesis 2019, 8, 26. [Google Scholar] [CrossRef] [PubMed]
- Kinde, B.; Gabel, H.W.; Gilbert, C.S.; Griffith, E.; Greenberg, M.E. Reading the unique DNA methylation landscape of the brain: Non-CpG methylation, hydroxymethylation, and MeCP2. Proc. Natl. Acad. Sci. 2015, 112, 6800–6806. [Google Scholar] [CrossRef]
- Sun, Z.; Dai, N.; Borgaro, J.G.; Quimby, A.; Sun, D.; Correa, I.; Zheng, Y.; Zhu, Z.; Guan, S. A Sensitive Approach to Map Genome-wide 5-Hydroxymethylcytosine and 5-Formylcytosine at Single-Base Resolution. Mol. Cell 2015, 57, 750–761. [Google Scholar] [CrossRef]
- Bestor, T.H. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000, 9, 2395–2402. [Google Scholar] [CrossRef] [PubMed]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef] [PubMed]
- Schübeler, D. Function and information content of DNA methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Shen, L.; Dai, Q.; Wu, S.C.; Collins, L.B.; Swenberg, J.A.; He, C.; Zhang, Y. Tet Proteins Can Convert 5-Methylcytosine to 5-Formylcytosine and 5-Carboxylcytosine. Science 2011, 333, 1300–1303. [Google Scholar] [CrossRef]
- He, Y.-F.; Li, B.-Z.; Li, Z.; Liu, P.; Wang, Y.; Tang, Q.; Ding, J.; Jia, Y.; Chen, Z.; Li, L.; et al. Tet-Mediated Formation of 5-Carboxylcytosine and Its Excision by TDG in Mammalian DNA. Science 2011, 333, 1303–1307. [Google Scholar] [CrossRef] [PubMed]
- Ponnaluri, V.C.; Maciejewski, J.P.; Mukherji, M. A mechanistic overview of TET-mediated 5-methylcytosine oxidation. Biochem. Biophys. Res. Commun. 2013, 436, 115–120. [Google Scholar] [CrossRef]
- Ramsahoye, B.H.; Biniszkiewicz, D.; Lyko, F.; Clark, V.; Bird, A.; Jaenisch, R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl. Acad. Sci. USA 2000, 97, 5237–5242. [Google Scholar] [CrossRef]
- Gowher, H.; Jeltsch, A. Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: The enzyme modifies DNA in a non-processive manner and also methylates non-CpA sites. J. Mol. Biol. 2001, 309, 1201–1208. [Google Scholar] [CrossRef]
- Shirane, K.; Toh, H.; Kobayashi, H.; Miura, F.; Chiba, H.; Ito, T.; Kono, T.; Sasaki, H. Mouse Oocyte Methylomes at Base Resolution Reveal Genome-Wide Accumulation of Non-CpG Methylation and Role of DNA Methyltransferases. PLoS Genet. 2013, 9, e1003439. [Google Scholar] [CrossRef]
- Barres, R.; Osler, M.E.; Yan, J.; Rune, A.; Fritz, T.; Caidahl, K.; Krook, A.; Zierath, J.R. Non-CpG Methylation of the PGC-1α Promoter through DNMT3B Controls Mitochondrial Density. Cell Metab. 2009, 10, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Ichiyanagi, T.; Ichiyanagi, K.; Miyake, M.; Sasaki, H. Accumulation and loss of asymmetric non-CpG methylation during male germ-cell development. Nucleic Acids Res. 2012, 41, 738–745. [Google Scholar] [CrossRef] [PubMed]
- Arand, J.; Spieler, D.; Karius, T.; Branco, M.; Meilinger, D.; Meissner, A.; Jenuwein, T.; Xu, G.; Leonhardt, H.; Wolf, V.; et al. In Vivo Control of CpG and Non-CpG DNA Methylation by DNA Methyltransferases. PLoS Genet. 2012, 8, e1002750. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Park, S.-J.; Nakai, K. Differential landscape of non-CpG methylation in embryonic stem cells and neurons caused by DNMT3s. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Fuso, A.; Ferraguti, G.; Grandoni, F.; Ruggeri, R.; Scarpa, S.; Strom, R.; Lucarelli, M. Early demethylation of non-CpG, CpC-rich, elements in the myogenin 5’-flanking region. Cell Cycle 2010, 9, 3965–3976. [Google Scholar] [CrossRef] [PubMed]
- Tognini, P.; Napoli, D.; Pizzorusso, T. Dynamic DNA methylation in the brain: A new epigenetic mark for experience-dependent plasticity. Front. Cell. Neurosci. 2015, 9, 331. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Li, Z.; Cheng, J.; Rao, Q.; Gong, W.; Liu, M.; Shi, Y.G.; Zhu, J.; Wang, P.; Xu, Y. Crystal Structure of TET2-DNA Complex: Insight into TET-Mediated 5mC Oxidation. Cell 2013, 155, 1545–1555. [Google Scholar] [CrossRef] [PubMed]
- Pais, J.E.; Dai, N.; Tamanaha, E.; Vaisvila, R.; Fomenkov, A.I.; Bitinaite, J.; Sun, Z.; Guan, S.; Correa, I.; Noren, C.J.; et al. Biochemical characterization of a Naegleria TET-like oxygenase and its application in single molecule sequencing of 5-methylcytosine. Proc. Natl. Acad. Sci. USA 2015, 112, 4316–4321. [Google Scholar] [CrossRef] [PubMed]
- Papin, C.; Ibrahim, A.; Le Gras, S.; Velt, A.; Stoll, I.; Jost, B.; Menoni, H.; Bronner, C.; Dimitrov, S.; Hamiche, A. Combinatorial DNA methylation codes at repetitive elements. Genome Res. 2017, 27, 934–946. [Google Scholar] [CrossRef]
- Feng, Y.; Xie, N.; Tao, W.; Ding, J.; You, X.; Ma, C.; Zhang, X.; Yi, C.; Zhou, X.; Yuan, B.; et al. Transformation of 5-Carboxylcytosine to Cytosine Through C–C Bond Cleavage in Human Cells Constitutes a Novel Pathway for DNA De-methylation. CCS Chem. 2020, 2, 994–1008. [Google Scholar] [CrossRef]
- Iwan, K.; Rahimoff, R.; Kirchner, A.; Spada, F.; Schröder, A.S.; Kosmatchev, O.; Ferizaj, S.; Steinbacher, J.; Parsa, E.; Müller, M.; et al. 5-Formylcytosine to cytosine conversion by C–C bond cleavage in vivo. Nat. Chem. Biol. 2017, 14, 72–78. [Google Scholar] [CrossRef]
- Maiti, A.; Drohat, A.C. Thymine DNA Glycosylase Can Rapidly Excise 5-Formylcytosine and 5-Carboxylcytosine: Potential Implications for Active Demethylation of CpG Sites. J. Biol. Chem. 2011, 286, 35334–35338. [Google Scholar] [CrossRef]
- DeNizio, J.E.; Dow, B.J.; Serrano, J.C.; Ghanty, U.; Drohat, A.C.; Kohli, R.M. TET-TDG Active DNA Demethylation at CpG and Non-CpG Sites. J. Mol. Biol. 2021, 433, 166877. [Google Scholar] [CrossRef]
- Bhattacharya, C.; Dey, A.; Ayon, N.J.; Gutheil, W.G.; Mukherji, I. Efficient Purification and LC-MS/MS-based Assay Development for Ten-Eleven Translocation-2 5-Methylcytosine Dioxygenase. J. Vis. Exp. 2018, 140, e57798. [Google Scholar] [CrossRef] [PubMed]
- Dey, A.S.; Ayon, N.J.; Bhattacharya, C.; Gutheil, W.G.; Mukherji, M. Positive/negative ion-switching-based LC–MS/MS method for quantification of cytosine derivatives produced by the TET-family 5-methylcytosine dioxygenases. Biol. Methods Protoc. 2020, 5, bpaa019. [Google Scholar] [CrossRef]
- Guan, Y.; Greenberg, E.F.; Hasipek, M.; Chen, S.; Liu, X.; Kerr, C.M.; Gackowski, D.; Zarakowska, E.; Radivoyevitch, T.; Gu, X.; et al. Context dependent effects of ascorbic acid treatment in TET2 mutant myeloid neoplasia. Commun. Biol. 2020, 3, 1–13. [Google Scholar] [CrossRef]
- Hong, S.; Cheng, X. DNA Base Flipping: A General Mechanism for Writing, Reading, and Erasing DNA Modifications. Adv. Exp. Med. Biol. 2016, 945, 321–341. [Google Scholar] [CrossRef]
- Handa, V.; Jeltsch, A. Profound Flanking Sequence Preference of Dnmt3a and Dnmt3b Mammalian DNA Methyltransferases Shape the Human Epigenome. J. Mol. Biol. 2005, 348, 1103–1112. [Google Scholar] [CrossRef] [PubMed]
- Adam, S.; Anteneh, H.; Hornisch, M.; Wagner, V.; Lu, J.; Radde, N.E.; Bashtrykov, P.; Song, J.; Jeltsch, A. DNA sequence-dependent activity and base flipping mechanisms of DNMT1 regulate genome-wide DNA methylation. Nat. Commun. 2020, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
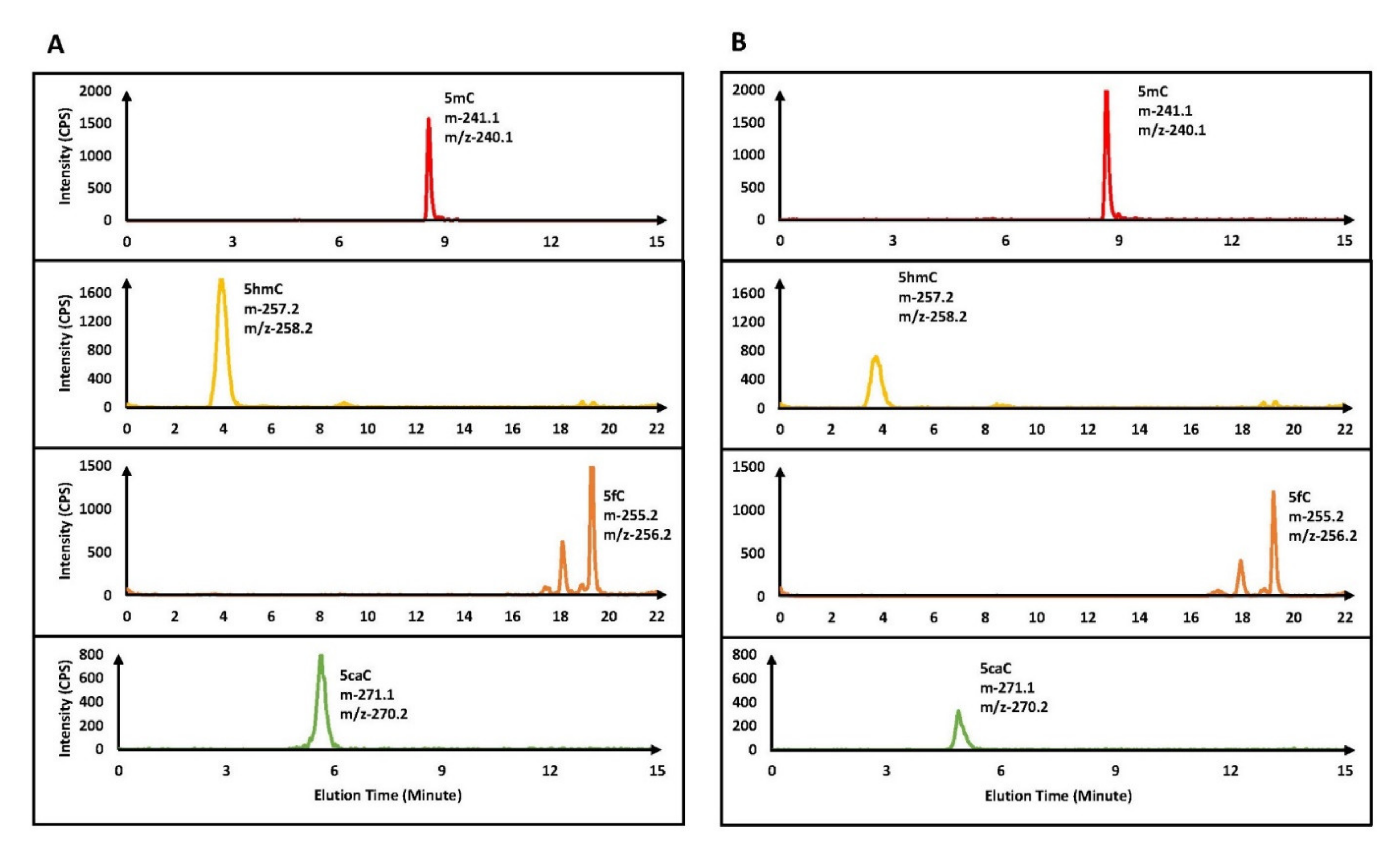
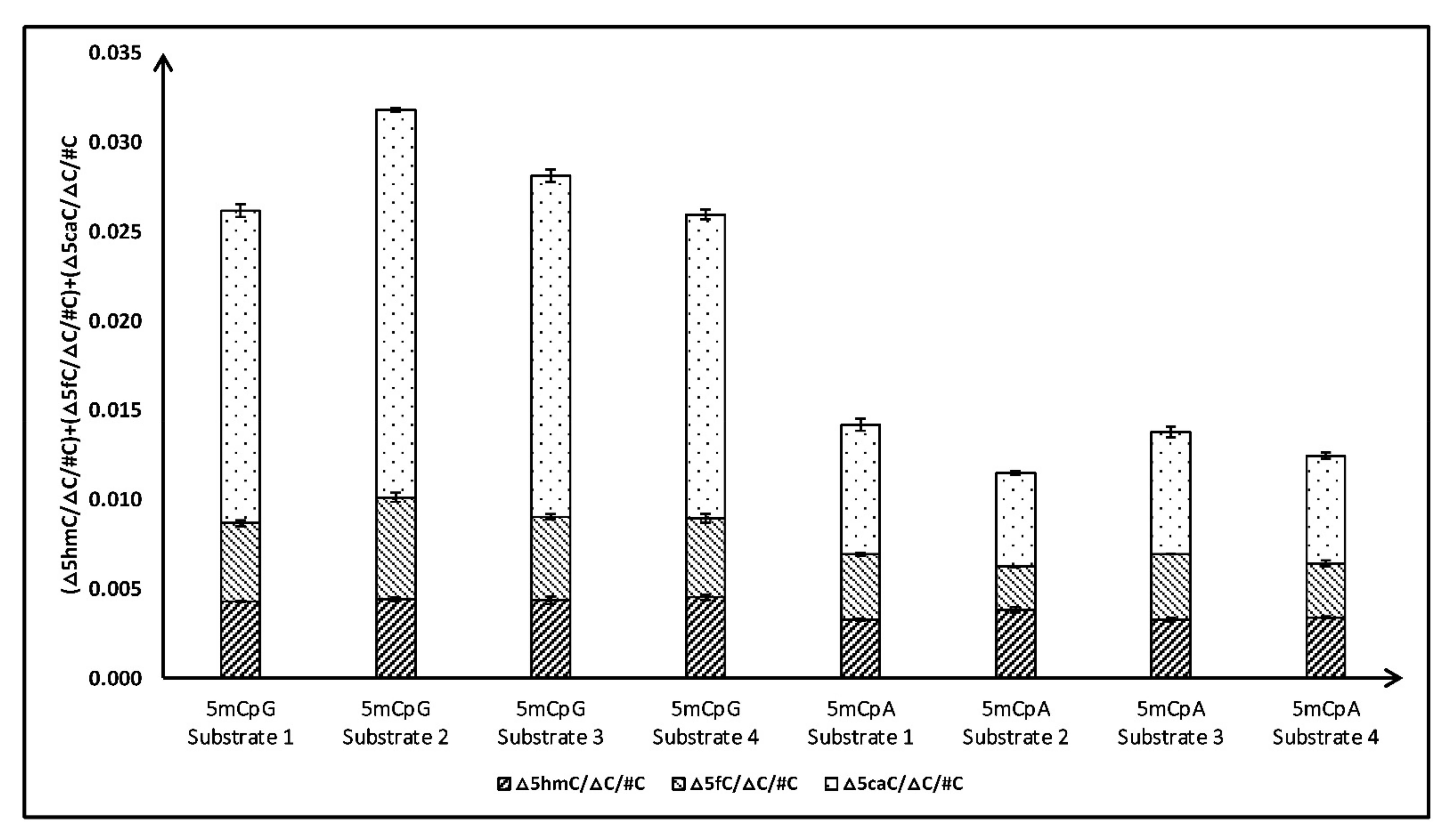

| 1A | |
|---|---|
| DNA Substrates | Sequence |
| 5mCpG Substrate 1 | Forward Strand 5′-GCGCCGGTCGTA/Me-C/GGCCGCTCCCGC-3′ Reverse Strand 5′-GCGGGAGCGGCCGTACGACCGGCGC-3′ |
| 5mCpA Substrate 1 | Forward Strand 5′-GCGCCGGTCGTA/Me-C/AGCCGCTCCCGC-3′ Reverse Strand 5′-GCGGGAGCGGCTGTACGACCGGCGC-3′ |
| 1B | |
| DNA Substrates | Sequence |
| 5mCpG Substrate 2 | Forward Strand 5′-GCGCCGGTCCCG/Me-C/GGGCGCTCCCGC-3′ Reverse Strand 5′-GCGGGAGCGCCCGCGGGACCGGCGC-3′ |
| 5mCpG Substrate 3 | Forward Strand 5′-GCGCCGGTCGTT/Me-C/GTCCGCTCCCGC-3′ Reverse Strand 5′-GCGGGAGCGGACGAACGACCGGCGC-3 |
| 5mCpG Substrate 4 | Forward Strand 5′-GCGCCGGTCAAC/Me-C/GACCGCTCCCGC-3′ Reverse Strand 5′-GCGGGAGCGGTCGGTTGACCGGCGC-3′ |
| 5mCpA Substrate 2 | Forward Strand 5′-GCGCCGGTCTAA/Me-C/AAACGCTCCCGC-3′ Reverse Strand 5′-GCGGGAGCGTTTGTTAGACCGGCGC-3′ |
| 5mCpA Substrate 3 | Forward Strand 5′-GCGCCGGTCGTT/Me-C/ATCCGCTCCCGC-3′ Reverse Strand 5′-GCGGGAGCGGATGAACGACCGGCGC-3′ |
| 5mCpA Substrate 4 | Forward Strand 5′-GCGCCGGTCGGA/Me-C/AACCGCTCCCGC-3′ Reverse Strand 5′-GCGGGAGCGGTTGTCCGACCGGCGC-3′ |
| 1C | |
| DNA Substrates | Sequence |
| 5mCpC Substrate | Forward Strand 5′-GCGCCGGTCCTG/Me-C/CCCCGCTCCCGC-3′ Reverse Strand 5′-GCGGGAGCGGGGGCAGGACCGGCGC-3′ |
| 5mCpT Substrate | Forward Strand 5′-GCGCCGGTCCTG/Me-C/TCCCGCTCCCGC-3′ Reverse Strand 5′-GCGGGAGCGGGAGCAGGACCGGCGC-3 |
| 2A | |||
|---|---|---|---|
| DNA Substrate | DNA Sequence | ||
| 5mCpG | Forward Strand 5′-GCGCCGGTCCCG/Me-C/GGGCGCTCCCGC-3′ Reverse Strand 5′-GCGGGAGCGTTTGTTAGACCGGCGC-3 | ||
| Enzyme | Full Length human TET2 | ||
| dC Area (Counts) ± Standard Error | 5hmC Area (Counts) ± Standard Error | 5fC Area (Counts) ± Standard Error | 5caC Area (Counts) ± Standard Error |
| (2.00 × 107) ± (6.11 × 105) | (3.11 × 104) ± (2.17 × 103) | (2.25 × 104) ± (1.29 × 103) | (6.12 × 103) ± (3.71 × 102) |
| Enzyme | GST Tagged human TET2 | ||
| dC Area (Counts) | 5hmC Area (Counts) | 5fC Area (Counts) | 5caC Area (Counts) |
| (6.63 × 107) ± (3.33 × 105) | (3.17 × 105) ± (2.11 × 104) | (2.50 × 105) ± (2.67 × 103) | (7.37 × 104) ± (1.89 × 103) |
| 2B | |||
| DNA Substrate | DNA Sequence | ||
| 5mCpA | Forward Strand 5′-GCGCCGGTCCTG/Me-C/ACCCGCTCCCGC-3′ Reverse Strand 5′-GCGGGAGCGTTTGTTAGACCGGCGC-3 | ||
| Enzyme | Full Length human TET2 | ||
| dC Area (Counts) ± Standard Error | 5hmC Area (Counts) ± Standard Error | 5fC Area (Counts) ± Standard Error | 5caC Area (Counts) ± Standard Error |
| (1.56 × 107) ± (6.43 × 105) | (1.91 × 104) ± (1.17 × 103) | (1.67 × 104) ± (1.00 × 103) | (2.89 × 103) ± (4.88 × 102) |
| Enzyme | GST Tagged human TET2 | ||
| dC Area (Counts) | 5hmC Area (Counts) | 5fC Area (Counts) | 5caC Area (Counts) |
| (7.73 × 107) ± (3.33 × 105) | (2.17 × 105) ± (1.18 × 104) | (1.41 × 105) ± (3.67 × 103) | (2.37 × 104) ± (1.65 × 103) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dey, A.S.; Bhattacharya, C.; Guan, Y.; Jha, B.K.; Mukherji, M. Demethylation of Non-CpG Sites in DNA Is Initiated by TET2 5-Methylcytosine Dioxygenase. DNA 2021, 1, 26-36. https://doi.org/10.3390/dna1010004
Dey AS, Bhattacharya C, Guan Y, Jha BK, Mukherji M. Demethylation of Non-CpG Sites in DNA Is Initiated by TET2 5-Methylcytosine Dioxygenase. DNA. 2021; 1(1):26-36. https://doi.org/10.3390/dna1010004
Chicago/Turabian StyleDey, Aninda Sundar, Chayan Bhattacharya, Yihong Guan, Babal Kant Jha, and Mridul Mukherji. 2021. "Demethylation of Non-CpG Sites in DNA Is Initiated by TET2 5-Methylcytosine Dioxygenase" DNA 1, no. 1: 26-36. https://doi.org/10.3390/dna1010004
APA StyleDey, A. S., Bhattacharya, C., Guan, Y., Jha, B. K., & Mukherji, M. (2021). Demethylation of Non-CpG Sites in DNA Is Initiated by TET2 5-Methylcytosine Dioxygenase. DNA, 1(1), 26-36. https://doi.org/10.3390/dna1010004








