Ligase A and RNase HI Participate in Completing Replication on the Chromosome in Escherichia coli
Abstract
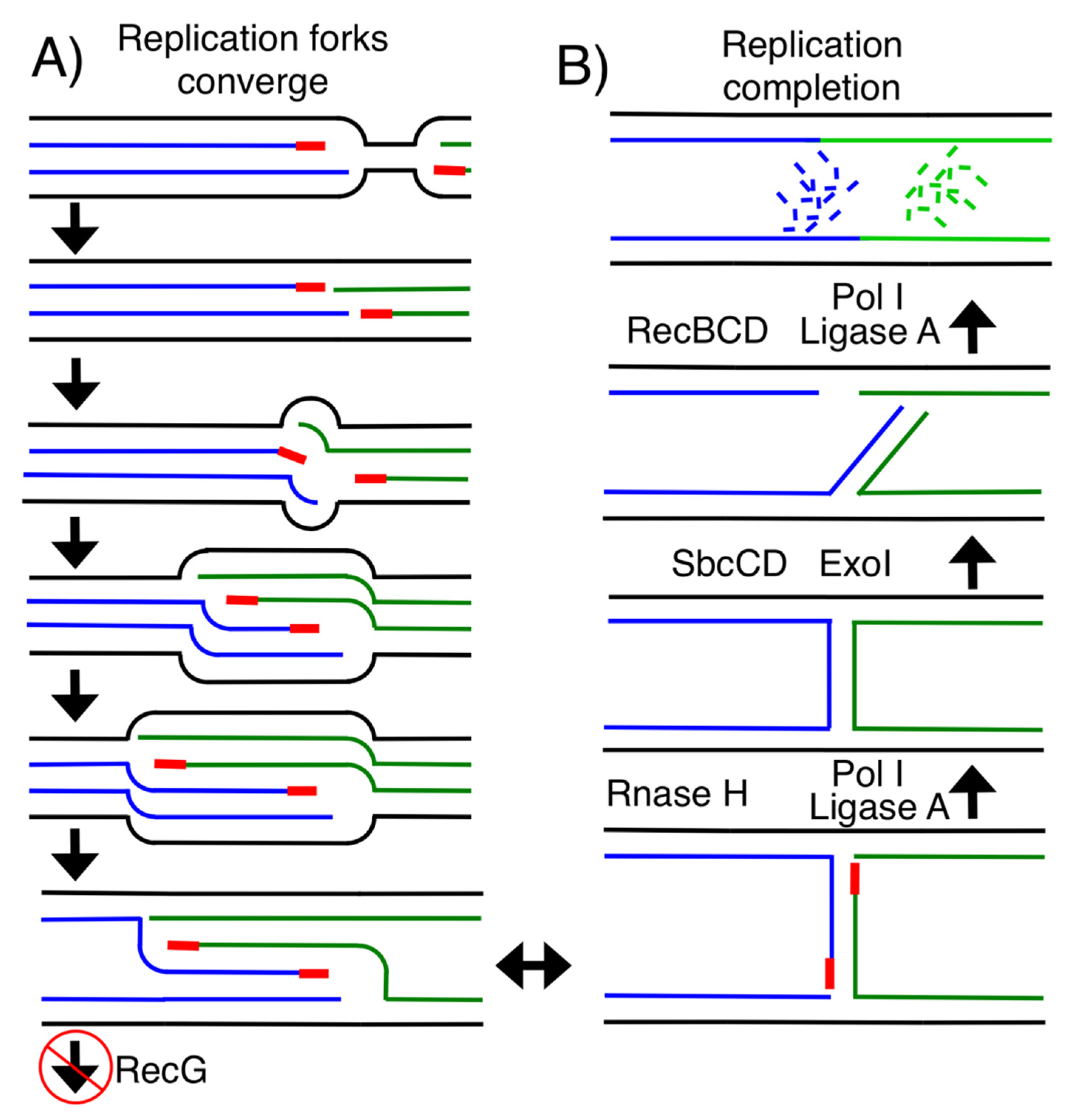
1. Introduction
2. Materials and Methods
2.1. Bacteria
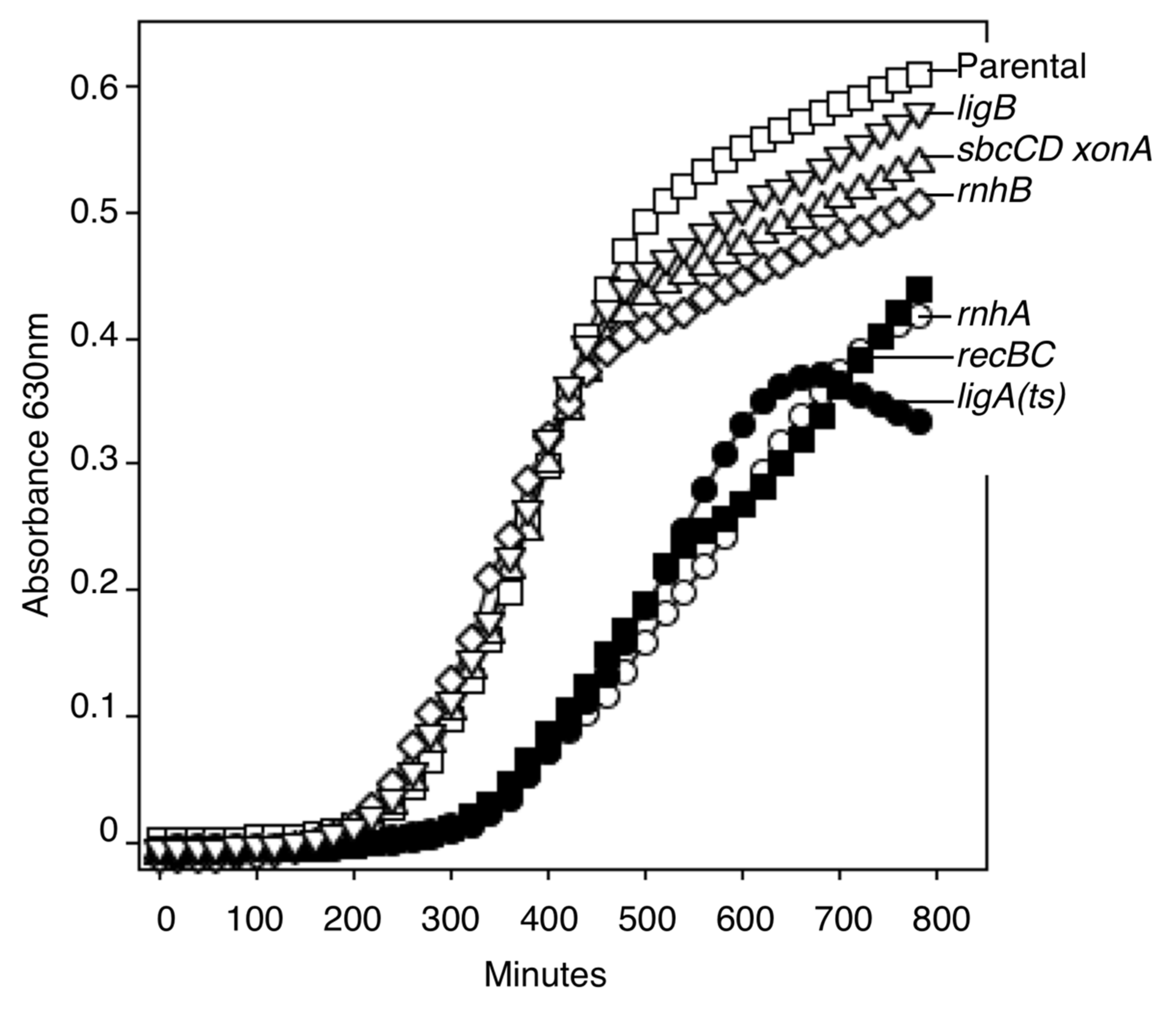
2.2. Growth Rates
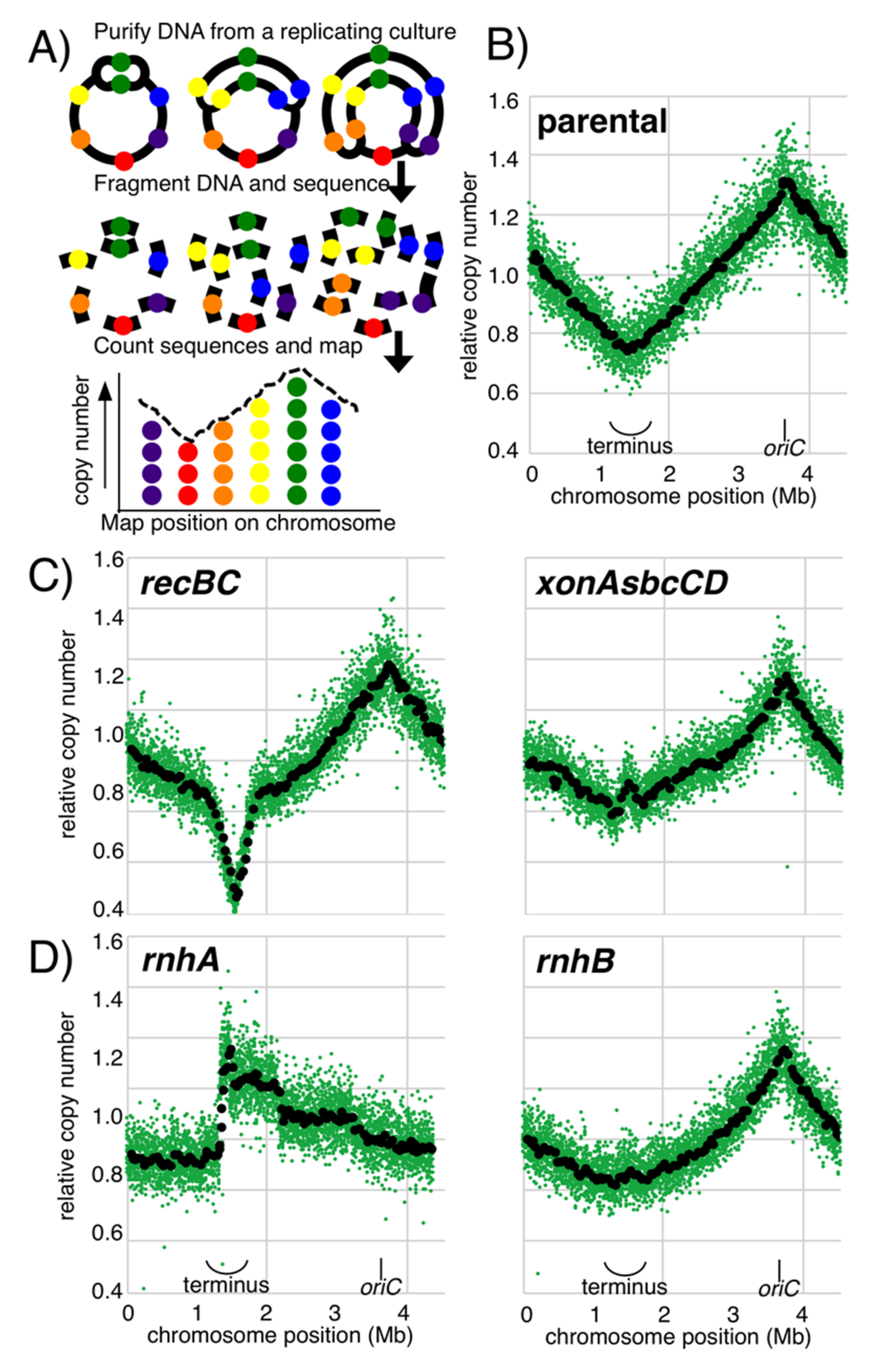
2.3. Replication Profiling
3. Results and Discussion
3.1. RNase HI, but Not RNase HII, Participates in Completing Replication on the Chromosome
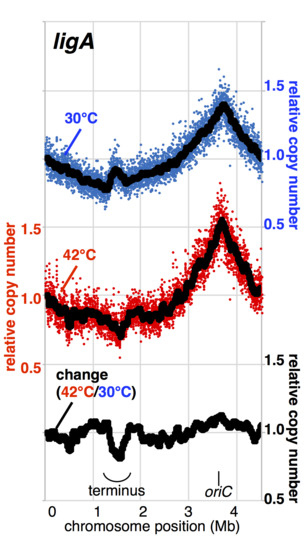
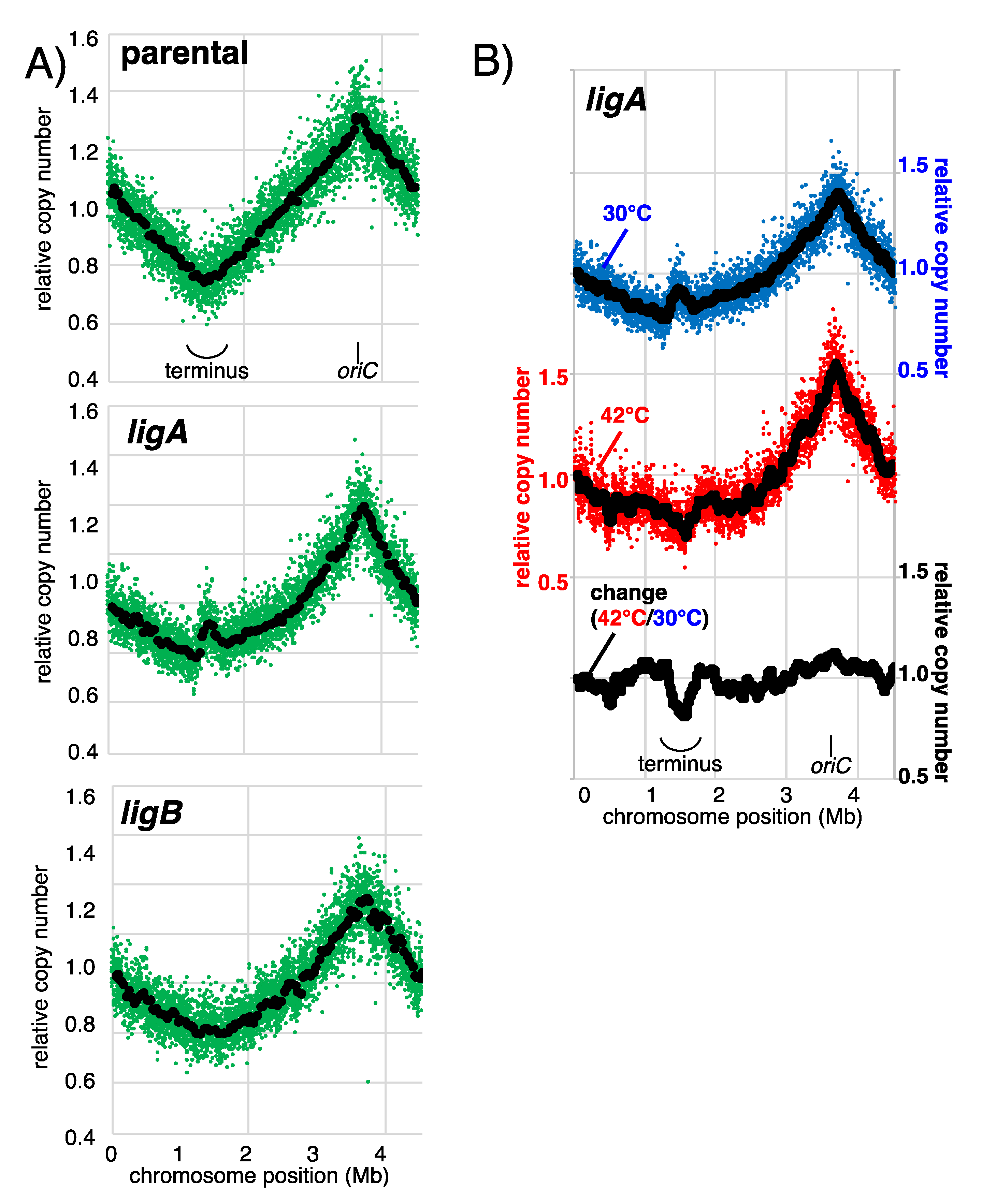
3.2. Ligase A, but Not Ligase B, Participates in Completing Replication on the Chromosome
3.3. Efficient Removal of Okazaki Primers and Joining of DNA Ends Is Important to Accurately Complete Replication on the Chromosome
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Costa, A.; Hood, I.V.; Berger, J.M. Mechanisms for initiating cellular DNA replication. Annu. Rev. Biochem. 2013, 82, 25–54. [Google Scholar] [CrossRef]
- O’Donnell, M.; Langston, L.; Stillman, B. Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb. Perspect. Biol. 2013, 5, a010108. [Google Scholar] [CrossRef] [PubMed]
- Dimude, J.U.; Stein, M.; Andrzejewska, E.E.; Khalifa, M.S.; Gajdosova, A.; Retkute, R.; Skovgaard, O.; Rudolph, C.J. Origins Left, Right, and Centre: Increasing the Number of Initiation Sites in the Escherichia coli Chromosome. Genes 2018, 9, 376. [Google Scholar] [CrossRef] [PubMed]
- Heichinger, C.; Penkett, C.J.; Bahler, J.; Nurse, P. Genome-wide characterization of fission yeast DNA replication origins. EMBO J. 2006, 25, 5171–5179. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.Y.; Nurse, P. Establishing the program of origin firing during S phase in fission Yeast. Cell 2009, 136, 852–864. [Google Scholar] [CrossRef]
- Wang, X.; Lesterlin, C.; Reyes-Lamothe, R.; Ball, G.; Sherratt, D.J. Replication and segregation of an Escherichia coli chromosome with two replication origins. Proc. Natl. Acad. Sci. USA 2011, 108, E243–E250. [Google Scholar] [CrossRef]
- Ivanova, D.; Taylor, T.; Smith, S.L.; Dimude, J.U.; Upton, A.L.; Mehrjouy, M.M.; Skovgaard, O.; Sherratt, D.J.; Retkute, R.; Rudolph, C.J. Shaping the landscape of the Escherichia coli chromosome: Replication-transcription encounters in cells with an ectopic replication origin. Nucleic Acids Res. 2015, 43, 7865–7877. [Google Scholar] [CrossRef]
- Hiasa, H.; Marians, K.J. Tus prevents overreplication of oriC plasmid DNA. J. Biol. Chem. 1994, 269, 26959–26968. [Google Scholar] [CrossRef]
- Rudolph, C.J.; Upton, A.L.; Harris, L.; Lloyd, R.G. Pathological replication in cells lacking RecG DNA translocase. Mol. Microbiol. 2009, 73, 352–366. [Google Scholar] [CrossRef]
- Dimude, J.U.; Stockum, A.; Midgley-Smith, S.L.; Upton, A.L.; Foster, H.A.; Khan, A.; Saunders, N.J.; Retkute, R.; Rudolph, C.J. The Consequences of Replicating in the Wrong Orientation: Bacterial Chromosome Duplication without an Active Replication Origin. MBio 2015, 6, e01294-15. [Google Scholar] [CrossRef]
- Midgley-Smith, S.L.; Dimude, J.U.; Taylor, T.; Forrester, N.M.; Upton, A.L.; Lloyd, R.G.; Rudolph, C.J. Chromosomal over-replication in Escherichia coli recG cells is triggered by replication fork fusion and amplified if replichore symmetry is disturbed. Nucleic Acids Res. 2018, 46, 7701–7715. [Google Scholar] [CrossRef]
- Midgley-Smith, S.L.; Dimude, J.U.; Rudolph, C.J. A role for 3′ exonucleases at the final stages of chromosome duplication in Escherichia coli. Nucleic Acids Res. 2019, 47, 1847–1860. [Google Scholar] [CrossRef] [PubMed]
- de Massy, B.; Fayet, O.; Kogoma, T. Multiple origin usage for DNA replication in sdrA(rnh) mutants of Escherichia coli K-12. Initiation in the absence of oriC. J. Mol. Biol. 1984, 178, 227–236. [Google Scholar] [CrossRef]
- de Massy, B.; Patte, J.; Louarn, J.M.; Bouche, J.P. oriX: A new replication origin in E. coli. Cell 1984, 36, 221–227. [Google Scholar] [CrossRef]
- Asai, T.; Kogoma, T. D-loops and R-loops: Alternative mechanisms for the initiation of chromosome replication in Escherichia coli. J. Bacteriol. 1994, 176, 1807–1812. [Google Scholar] [CrossRef] [PubMed]
- Wendel, B.M.; Courcelle, C.T.; Courcelle, J. Completion of DNA replication in Escherichia coli. Proc. Natl. Acad. Sci. USA 2014, 111, 16454–16459. [Google Scholar] [CrossRef] [PubMed]
- Wendel, B.M.; Cole, J.M.; Courcelle, C.T.; Courcelle, J. SbcC-SbcD and ExoI process convergent forks to complete chromosome replication. Proc. Natl. Acad. Sci. USA 2018, 115, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Hamperl, S.; Cimprich, K.A. The contribution of co-transcriptional RNA:DNA hybrid structures to DNA damage and genome instability. DNA Repair 2014, 19, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, V.; Barroso, S.I.; Garcia-Rubio, M.L.; Tumini, E.; Herrera-Moyano, E.; Aguilera, A. BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature 2014, 511, 362–365. [Google Scholar] [CrossRef]
- Donnianni, R.A.; Symington, L.S. Break-induced replication occurs by conservative DNA synthesis. Proc. Natl. Acad. Sci. USA 2013, 110, 13475–13480. [Google Scholar] [CrossRef] [PubMed]
- Magee, T.R.; Asai, T.; Malka, D.; Kogoma, T. DNA damage-inducible origins of DNA replication in Escherichia coli. EMBO J. 1992, 11, 4219–4225. [Google Scholar] [CrossRef]
- Brochu, J.; Vlachos-Breton, É.; Sutherland, S.; Martel, M.; Drolet, M. Topoisomerases I and III inhibit R-loop formation to prevent unregulated replication in the chromosomal Ter region of Escherichia coli. PLoS Genet. 2018, 14, e1007668. [Google Scholar] [CrossRef]
- Hill, T.M. Arrest of bacterial DNA replication. Annu. Rev. Microbiol. 1992, 46, 603–633. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Hidaka, M.; Horiuchi, T. Evidence of a ter specific binding protein essential for the termination reaction of DNA replication in Escherichia coli. EMBO J. 1989, 8, 2435–2441. [Google Scholar] [CrossRef]
- Roecklein, B.; Pelletier, A.; Kuempel, P. The tus gene of Escherichia coli: Autoregulation, analysis of flanking sequences and identification of a complementary system in Salmonella typhimurium. Res. Microbiol. 1991, 142, 169–175. [Google Scholar] [CrossRef]
- Duggin, I.G.; Wake, R.G.; Bell, S.D.; Hill, T.M. The replication fork trap and termination of chromosome replication. Mol. Microbiol. 2008, 70, 1323–1333. [Google Scholar] [CrossRef] [PubMed]
- Duggin, I.G.; Bell, S.D. Termination structures in the Escherichia coli chromosome replication fork trap. J. Mol. Biol. 2009, 387, 532–539. [Google Scholar] [CrossRef]
- Rudolph, C.J.; Upton, A.L.; Stockum, A.; Nieduszynski, C.A.; Lloyd, R.G. Avoiding chromosome pathology when replication forks collide. Nature 2013, 500, 608–611. [Google Scholar] [CrossRef]
- McGlynn, P.; Lloyd, R.G. RecG helicase activity at three- and four-strand DNA structures. Nucleic Acids Res. 1999, 27, 3049–3056. [Google Scholar]
- Hamilton, N.A.; Wendel, B.M.; Weber, E.A.; Courcelle, C.T.; Courcelle, J. RecBCD, SbcCD and ExoI process a substrate created by convergent replisomes to complete DNA replication. Mol. Microbiol. 2019, 111, 1638–1651. [Google Scholar] [CrossRef] [PubMed]
- Connelly, J.C.; de Leau, E.S.; Okely, E.A.; Leach, D.R. Overexpression, purification, and characterization of the SbcCD protein from Escherichia coli. J. Biol. Chem. 1997, 272, 19819–19826. [Google Scholar] [CrossRef]
- Cromie, G.A.; Millar, C.B.; Schmidt, K.H.; Leach, D.R. Palindromes as substrates for multiple pathways of recombination in Escherichia coli. Genetics 2000, 154, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Courcelle, J.; Wendel, B.M.; Livingstone, D.D.; Courcelle, C.T. RecBCD is required to complete chromosomal replication: Implications for double-strand break frequencies and repair mechanisms. DNA Repair 2015, 32, 86–95. [Google Scholar] [CrossRef][Green Version]
- Taylor, A.; Smith, G.R. Unwinding and rewinding of DNA by the RecBC enzyme. Cell 1980, 22, 447–457. [Google Scholar] [CrossRef]
- Taylor, A.F.; Schultz, D.W.; Ponticelli, A.S.; Smith, G.R. RecBC enzyme nicking at Chi sites during DNA unwinding: Location and orientation-dependence of the cutting. Cell 1985, 41, 153–163. [Google Scholar] [CrossRef]
- Taylor, A.F.; Smith, G.R. Substrate specificity of the DNA unwinding activity of the RecBC enzyme of Escherichia coli. J. Mol. Biol. 1985, 185, 431–443. [Google Scholar] [CrossRef]
- Amundsen, S.K.; Taylor, A.F.; Chaudhury, A.M.; Smith, G.R. recD: The gene for an essential third subunit of exonuclease V. Proc. Natl. Acad. Sci. USA 1986, 83, 5558–5562. [Google Scholar] [CrossRef]
- Taylor, A.F.; Smith, G.R. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature 2003, 423, 889–893. [Google Scholar] [CrossRef]
- Ponticelli, A.S.; Schultz, D.W.; Taylor, A.F.; Smith, G.R. Chi-dependent DNA strand cleavage by RecBC enzyme. Cell 1985, 41, 145–151. [Google Scholar] [CrossRef]
- Amundsen, S.K.; Taylor, A.F.; Smith, G.R. The RecD subunit of the Escherichia coli RecBCD enzyme inhibits RecA loading, homologous recombination, and DNA repair. Proc. Natl. Acad. Sci. USA 2000, 97, 7399–7404. [Google Scholar] [CrossRef]
- Capaldo, F.N.; Ramsey, G.; Barbour, S.D. Analysis of the growth of recombination-deficient strains of Escherichia coli K-12. J. Bacteriol. 1974, 118, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Carl, P.L.; Bloom, L.; Crouch, R.J. Isolation and mapping of a mutation in Escherichia coli with altered levels of ribonuclease H. J. Bacteriol. 1980, 144, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Itaya, M. Isolation and characterization of a second RNase H (RNase HII) of Escherichia coli K-12 encoded by the rnhB gene. Proc. Natl. Acad. Sci. USA 1990, 87, 8587–8591. [Google Scholar] [CrossRef]
- Miller, H.I.; Riggs, A.D.; Gill, G.N. Ribonuclease H (hybrid) in Escherichia coli. Identification and characterization. J. Biol. Chem. 1973, 248, 2621–2624. [Google Scholar] [CrossRef]
- Tannous, E.; Kanaya, E.; Kanaya, S. Role of RNase H1 in DNA repair: Removal of single ribonucleotide misincorporated into DNA in collaboration with RNase H2. Sci. Rep. 2015, 5, 9969. [Google Scholar] [CrossRef] [PubMed]
- Haruki, M.; Tsunaka, Y.; Morikawa, M.; Kanaya, S. Cleavage of a DNA-RNA-DNA/DNA chimeric substrate containing a single ribonucleotide at the DNA-RNA junction with prokaryotic RNases HII. FEBS Lett. 2002, 531, 204–208. [Google Scholar] [CrossRef]
- Ohtani, N.; Haruki, M.; Morikawa, M.; Crouch, R.J.; Itaya, M.; Kanaya, S. Identification of the genes encoding Mn2+-dependent RNase HII and Mg2+-dependent RNase HIII from Bacillus subtilis: Classification of RNases H into three families. Biochemistry 1999, 38, 605–618. [Google Scholar] [CrossRef]
- Ogawa, T.; Okazaki, T. Function of RNase H in DNA replication revealed by RNase H defective mutants of Escherichia coli. Mol. Genet. Genom. 1984, 193, 231–237. [Google Scholar] [CrossRef]
- Kogoma, T.; Subia, N.L.; von Meyenburg, K. Function of ribonuclease H in initiation of DNA replication in Escherichia coli K-12. Mol. Genet. Genom. 1985, 200, 103–109. [Google Scholar] [CrossRef]
- Ogawa, T.; Pickett, G.G.; Kogoma, T.; Kornberg, A. RNase H confers specificity in the dnaA-dependent initiation of replication at the unique origin of the Escherichia coli chromosome in vivo and in vitro. Proc. Natl. Acad. Sci. USA 1984, 81, 1040–1044. [Google Scholar] [CrossRef]
- Horiuchi, T.; Maki, H.; Sekiguchi, M. RNase H-defective mutants of Escherichia coli: A possible discriminatory role of RNase H in initiation of DNA replication. Mol. Genet. Genom. 1984, 195, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Maduike, N.Z.; Tehranchi, A.K.; Wang, J.D.; Kreuzer, K.N. Replication of the Escherichia coli chromosome in RNase HI-deficient cells: Multiple initiation regions and fork dynamics. Mol. Microbiol. 2014, 91, 39–56. [Google Scholar] [CrossRef] [PubMed]
- Vaisman, A.; McDonald, J.P.; Huston, D.; Kuban, W.; Liu, L.; Van Houten, B.; Woodgate, R. Removal of misincorporated ribonucleotides from prokaryotic genomes: An unexpected role for nucleotide excision repair. PLoS Genet. 2013, 9, e1003878. [Google Scholar] [CrossRef]
- Gellert, M. Formation of covalent circles of lambda DNA by E. coli extracts. Proc. Natl. Acad. Sci. USA 1967, 57, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, S.B.; Little, J.W.; Oshinsky, C.K.; Gellert, M. Enzymatic joining of DNA strands: A novel reaction of diphosphopyridine nucleotide. Proc. Natl. Acad. Sci. USA 1967, 57, 1841–1848. [Google Scholar] [CrossRef]
- Olivera, B.M.; Lehman, I.R. Diphosphopyridine nucleotide: A cofactor for the polynucleotide-joining enzyme from Escherichia coli. Proc. Natl. Acad. Sci. USA 1967, 57, 1700–1704. [Google Scholar] [CrossRef]
- Olivera, B.M.; Lehman, I.R. Linkage of polynucleotides through phosphodiester bonds by an enzyme from Escherichia coli. Proc. Natl. Acad. Sci. USA 1967, 57, 1426–1433. [Google Scholar] [CrossRef]
- Pauling, C.; Hamm, L. Properties of a temperature-sensitive radiation-sensitive mutant of Escherichia coli. Proc. Natl. Acad. Sci. USA 1968, 60, 1495–1502. [Google Scholar] [CrossRef] [PubMed]
- Modrich, P.; Lehman, I.R. Enzymatic characterization of a mutant of Escherichia coli with an altered DNA ligase. Proc. Natl. Acad. Sci. USA 1971, 68, 1002–1005. [Google Scholar] [CrossRef]
- Konrad, E.B.; Modrich, P.; Lehman, I.R. Genetic and enzymatic characterization of a conditional lethal mutant of Escherichia coli K12 with a temperature-sensitive DNA ligase. J. Mol. Biol. 1973, 77, 519–529. [Google Scholar] [CrossRef]
- Gottesman, M.M.; Hicks, M.L.; Gellert, M. Genetics and function of DNA ligase in Escherichia coli. J. Mol. Biol. 1973, 77, 531–547. [Google Scholar] [CrossRef]
- Pauling, C.; Hamm, L. Properties of a temperature-sensitive, radiation-sensitive mutant of Escherichia coli. II. DNA replication. Proc. Natl. Acad. Sci. USA 1969, 64, 1195–1202. [Google Scholar] [CrossRef]
- Dean, C.; Pauling, C. Properties of a deoxyribonucleic acid ligase mutant of Escherichia coli: X-ray sensitivity. J. Bacteriol. 1970, 102, 588–589. [Google Scholar] [CrossRef] [PubMed]
- Verly, W.G.; Gossard, F.; Crine, P. In vitro repair of apurinic sites in DNA. Proc. Natl. Acad. Sci. USA 1974, 71, 2273–2275. [Google Scholar] [CrossRef] [PubMed]
- Robertson, A.B.; Matson, S.W. Reconstitution of the very short patch repair pathway from Escherichia coli. J. Biol. Chem. 2012, 287, 32953–32966. [Google Scholar] [CrossRef] [PubMed]
- Lahue, R.S.; Au, K.G.; Modrich, P. DNA mismatch correction in a defined system. Science 1989, 245, 160–164. [Google Scholar] [CrossRef]
- Sriskanda, V.; Shuman, S. A second NAD+-dependent DNA ligase (LigB) in Escherichia coli. Nucleic Acids Res. 2001, 29, 4930–4934. [Google Scholar] [CrossRef]
- Bodine, T.J.; Evangelista, M.A.; Chang, H.T.; Ayoub, C.A.; Samuel, B.S.; Sucgang, R.; Zechiedrich, L. Escherichia coli DNA ligase B may mitigate damage from oxidative stress. PLoS ONE 2017, 12, e0180800. [Google Scholar] [CrossRef]
- Mellon, I.; Hanawalt, P.C. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature 1989, 342, 95–98. [Google Scholar] [CrossRef]
- Dermody, J.J.; Robinson, G.T.; Sternglanz, R. Conditional-lethal deoxyribonucleic acid ligase mutant of Escherichia coli. J. Bacteriol. 1979, 139, 701–704. [Google Scholar] [CrossRef]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Ellis, H.M.; Lee, E.C.; Jenkins, N.A.; Copeland, N.G.; Court, D.L. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 2000, 97, 5978–5983. [Google Scholar] [CrossRef]
- Barrick, J.E.; Colburn, G.; Deatherage, D.E.; Traverse, C.C.; Strand, M.D.; Borges, J.J.; Knoester, D.B.; Reba, A.; Meyer, A.G. Identifying structural variation in haploid microbial genomes from short-read resequencing data using breseq. BMC Genom. 2014, 15, 1039. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinform. 2010, 32, 11.7.1–11.7.14. [Google Scholar] [CrossRef]
- Hill, T.M.; Tecklenburg, M.L.; Pelletier, A.J.; Kuempel, P.L. tus, the trans-acting gene required for termination of DNA replication in Escherichia coli, encodes a DNA-binding protein. Proc. Natl. Acad. Sci. USA 1989, 86, 1593–1597. [Google Scholar] [CrossRef]
- Fuller, R.S.; Kaguni, J.M.; Kornberg, A. Enzymatic replication of the origin of the Escherichia coli chromosome. Proc. Natl. Acad. Sci. USA 1981, 78, 7370–7374. [Google Scholar] [CrossRef]
- Kogoma, T.; Lark, K.G. DNA replication in Escherihia coli: Replication in absence of protein synthesis after replication inhibition. J. Mol. Biol. 1970, 52, 143–164. [Google Scholar] [CrossRef]
- Guzman, E.C.; Jimenez-Sanchez, A.; Orr, E.; Pritchard, R.H. Heat stress in the presence of low RNA polymerase activity increases chromosome copy number of Escherichia coli. Mol. Genet. Genom. 1988, 212, 203–206. [Google Scholar] [CrossRef]
- Kogoma, T.; Skarstad, K.; Boye, E.; von Meyenburg, K.; Steen, H.B. RecA protein acts at the initiation of stable DNA replication in rnh mutants of Escherichia coli K-12. J. Bacteriol. 1985, 163, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Louarn, J.; Bouché, J.P.; Patte, J.; Louarn, J.M. Genetic inactivation of topoisomerase I suppresses a defect in initiation of chromosome replication in Escherichia coli. Mol. Genet. Genom. 1984, 195, 170–174. [Google Scholar] [CrossRef]
- Atlung, T. Allele-specific suppression of dnaA(Ts) mutations by rpoB mutations in Escherichia coli. Mol. Genet. Genom. 1984, 197, 125–128. [Google Scholar] [CrossRef]
- Torrey, T.A.; Atlung, T.; Kogoma, T. dnaA suppressor (dasF) mutants of Escherichia coli are stable DNA replication (sdrA/rnh) mutants. Mol. Genet. Genom. 1984, 196, 350–355. [Google Scholar] [CrossRef]
- Lobner-Olesen, A.; Atlung, T.; Rasmussen, K.V. Stability and replication control of Escherichia coli minichromosomes. J. Bacteriol. 1987, 169, 2835–2842. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, T.; Okazaki, T. Concurrent transcription from the gid and mioC promoters activates replication of an Escherichia coli minichromosome. Mol. Genet. Genom. 1991, 230, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Bates, D.B.; Boye, E.; Asai, T.; Kogoma, T. The absence of effect of gid or mioC transcription on the initiation of chromosomal replication in Escherichia coli. Proc. Natl. Acad. Sci. USA 1997, 94, 12497–12502. [Google Scholar] [CrossRef] [PubMed]
- Itoh, T.; Tomizawa, J. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc. Natl. Acad. Sci. USA 1980, 77, 2450–2454. [Google Scholar] [CrossRef]
- Naito, S.; Kitani, T.; Ogawa, T.; Okazaki, T.; Uchida, H. Escherichia coli mutants suppressing replication-defective mutations of the ColE1 plasmid. Proc. Natl. Acad. Sci. USA 1984, 81, 550–554. [Google Scholar] [CrossRef] [PubMed]
- Dimude, J.U.; Midgley-Smith, S.L.; Stein, M.; Rudolph, C.J. Replication Termination: Containing Fork Fusion-Mediated Pathologies in Escherichia coli. Genes 2016, 7, 40. [Google Scholar] [CrossRef]
- Konrad, E.B.; Lehman, I.R. A conditional lethal mutant of Escherichia coli K12 defective in the 5′ leads to 3′ exonuclease associated with DNA polymerase I. Proc. Natl. Acad. Sci. USA 1974, 71, 2048–2051. [Google Scholar] [CrossRef]
| Strain | Relevant Genotype | Source or Construction |
|---|---|---|
| GR501 | ligA251(ts) | [70] |
| JW0204 | rnhA::kan | [71] |
| JW0178 | rnhB::kan | [71] |
| JW3622 | ligB::kan | [71] |
| DY329 | W3110 Del(lacU169) nadA::Tn10, gal490, Lambda cI857, Del(cro-bioA) | [72] |
| CL1180 | DY329 (nupC-yfeA intergenic region)::cat | primers 5′ GTTACGGGTTGTACAAGCGGAAAGAGATTGCG TCTTGTCGATGAGACGTTGATCGGCAC, 5′ TCCTTTTCGACGATTCTCGCTGAGCAGTCGGGT TTTACTGCTTTCGAATTTCTGCCATTC to amplify cat, transformed into recombineering strain DY392. cat inserts within nupC-yfeA intergenic region. |
| CL1834 | GR501 (nupC-yfeA intergenic region)::cat | P1 transduction of (nupC-yfeA intergenic region)::cat from CL1180 into GR501 |
| SR108 | Λ-thyA deoC IN(rrnD-rrnE) | [69] |
| CL1056 | SR108 Δ(recC ptr recB recD)::cam | [17] |
| CL2357 | SR108 xonA::Cat300 sbcCD::Gm | [16] |
| CL3362 | SR108 rnhA::kan | P1 transduction of rnhA::kan from JW0204 into SR108 |
| CL3360 | SR108 rnhB::kan | P1 transduction of rnhB::kan from JW0178 into SR108 |
| CL3912 | SR108 ligA251(ts) (nupC-yfeA intergenic region)::cat | P1 transduction of ligA251(ts) (nupC-yfeA intergenic region)::cat from CL1834 into SR108 |
| CL3909 | SR108 ligB753::kan | P1 transduction of ligB753::kan from JW3622 into SR108 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wendel, B.M.; Hernandez, A.J.; Courcelle, C.T.; Courcelle, J. Ligase A and RNase HI Participate in Completing Replication on the Chromosome in Escherichia coli. DNA 2021, 1, 13-25. https://doi.org/10.3390/dna1010003
Wendel BM, Hernandez AJ, Courcelle CT, Courcelle J. Ligase A and RNase HI Participate in Completing Replication on the Chromosome in Escherichia coli. DNA. 2021; 1(1):13-25. https://doi.org/10.3390/dna1010003
Chicago/Turabian StyleWendel, Brian M., Adrian J. Hernandez, Charmain T. Courcelle, and Justin Courcelle. 2021. "Ligase A and RNase HI Participate in Completing Replication on the Chromosome in Escherichia coli" DNA 1, no. 1: 13-25. https://doi.org/10.3390/dna1010003
APA StyleWendel, B. M., Hernandez, A. J., Courcelle, C. T., & Courcelle, J. (2021). Ligase A and RNase HI Participate in Completing Replication on the Chromosome in Escherichia coli. DNA, 1(1), 13-25. https://doi.org/10.3390/dna1010003