Regulation of Antioxidant Expression in the Liver Tissue of Obese Rats Treated with Coriander Seed Ethanolic Extract: In Silico and In Vivo Studies
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Ethanolic Extraction of Coriander Seeds (Coriandrum sativum L.)
2.3. In Silico Study Procedures
2.3.1. Ligand Preparation
2.3.2. Protein Preparation
2.3.3. Molecular Docking
2.3.4. Drug-Likeness, Pharmacokinetics, and Toxicity Prediction
2.4. In Vivo Study Procedures
2.4.1. Tissue Homogenate Preparation and Total Protein Analysis
2.4.2. Nrf2 and FOXO3 Protein Expression Using Enzyme-Linked Immunosorbent Assay (ELISA)
2.4.3. Nrf2 Protein Expression Using Western Blot
2.4.4. MnSOD and GPx Relative mRNA Expression Analysis
2.4.5. Total SOD, MnSOD, and GPx Specific Enzymatic Activity Analysis
2.4.6. GSH Level Analysis
2.4.7. Statistical Analysis
3. Results
3.1. In Silico Study
3.1.1. Molecular Docking
3.1.2. Drug-Likeness, Pharmacokinetics, and Toxicity Prediction
3.2. In Vivo Study
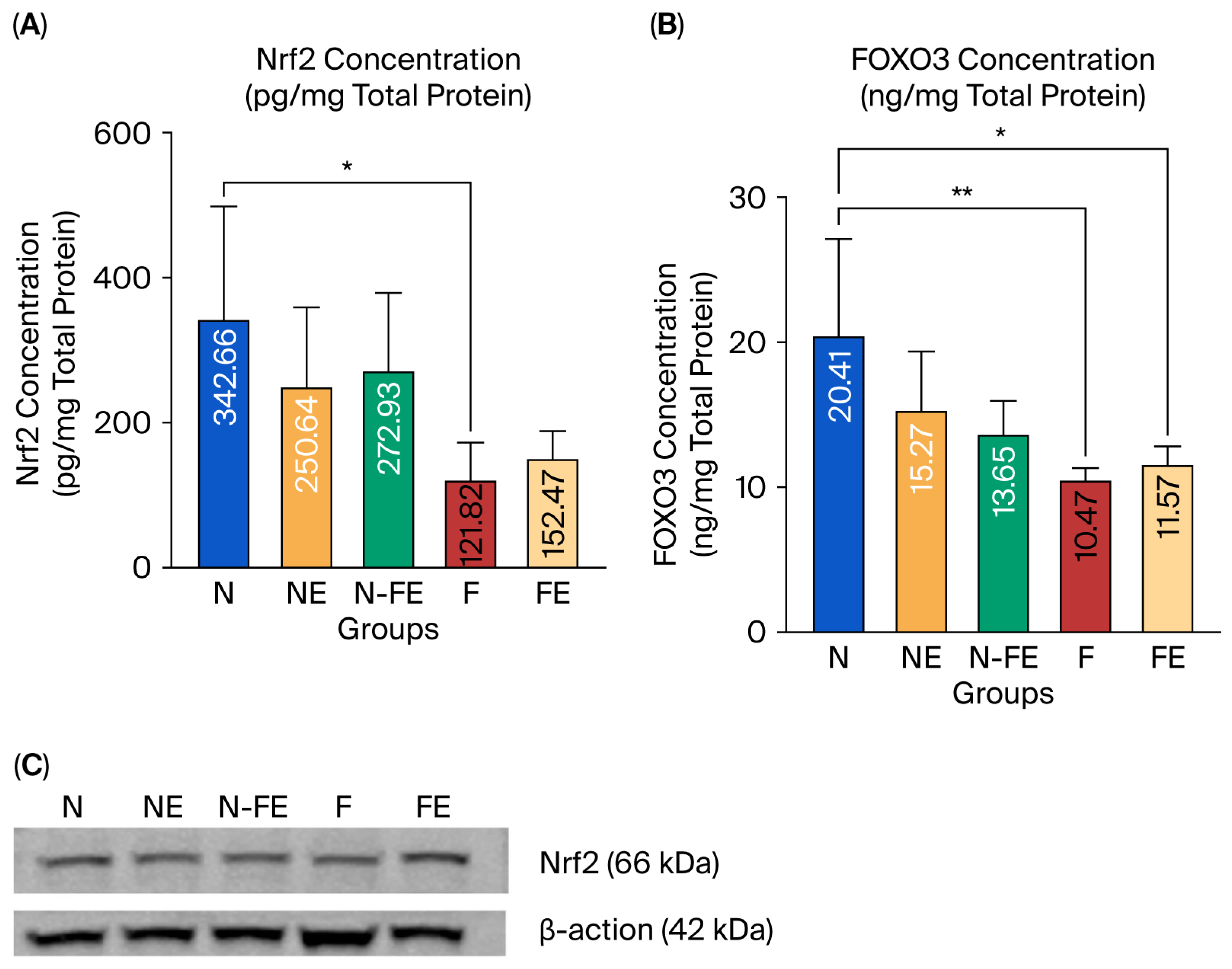
3.2.1. The Efficacy of Coriander Seed Ethanolic Extract on Hepatic Nrf2 and FOXO3 Protein Expression
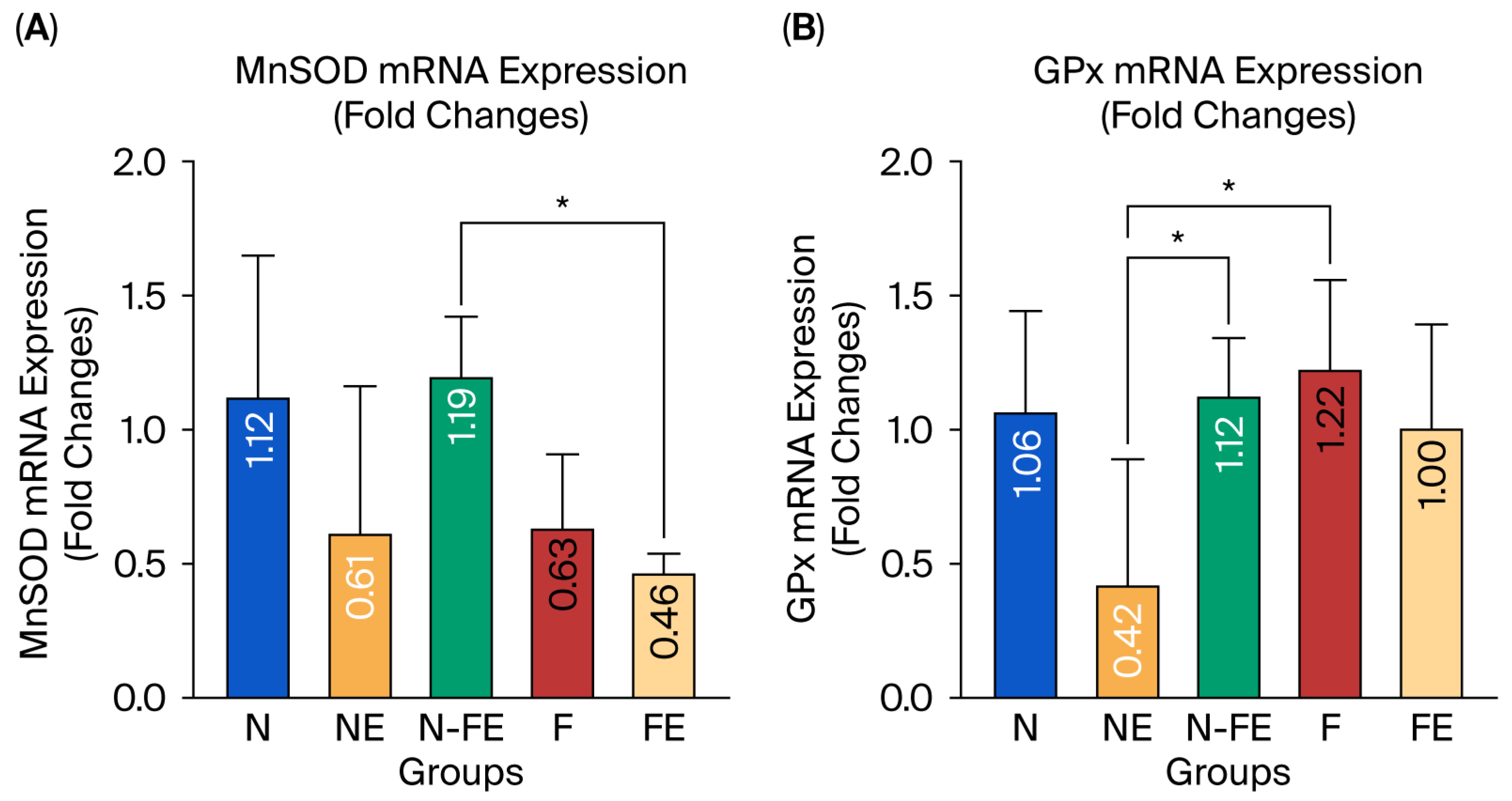
3.2.2. The Efficacy of Coriander Seed Ethanolic Extract on Hepatic MnSOD and GPx mRNA Expression
3.2.3. The Efficacy of Coriander Seed Ethanolic Extract on Hepatic Antioxidant Activity
3.2.4. Correlation Between the Transcription Factors (Nrf2 and FOXO3) and Their Downstream Effectors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 3′ UTR | 3′ untranslated region |
| BSA | Bovine serum albumin |
| CAT | Catalase |
| CBP | CREB-binding protein |
| FOXO3 | Forkhead Box O3 |
| GC-MS | Gas Chromatography–Mass Spectrophotometry |
| GPx | Glutathione peroxidase |
| GSH | Reduced glutathione |
| GSSG | Glutathione disulfide |
| H2O2 | Hydrogen peroxide |
| HDAC3 | histone deacetylase 3 |
| Keap1 | Kelch-like ECH-associated Protein 1 |
| Ki | Inhibition constant |
| LC-MS | Liquid Chromatography–Mass Spectrophotometry |
| MafK | MAF bZIP transcription factor K |
| MDA | Malondialdehyde |
| MnSOD | Manganese superoxide dismutase |
| NADPH | Nicotinamide adenine dinucleotide phosphate |
| NAFLD | Non-alcoholic fatty liver disease |
| NFκB | Nuclear factor-κB |
| Nrf2 | Nuclear factor erythroid 2-related factor 2 |
| PVDF | Polyvinylidene difluoride |
| ROS | Radical oxygen species |
| SDS-PAGE | Sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| SIRT | Sirtuin |
| TBST | Tris-buffered saline tween 0.1% |
| T-SOD | Total superoxide dismutase |
References
- Xia, Y.; Zhai, X.; Qiu, Y.; Lu, X.; Jiao, Y. The Nrf2 in Obesity: A Friend or Foe? Antioxidants 2022, 11, 2067. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Obesity and Overweight. Available online: https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight (accessed on 8 April 2025).
- Shabrina, E.; Briawan, D.; Ekayanti, I.; Riyadina, W. Changes in sugar, salt, and fat intake among obese adults: Cohort study. J. Gizi Diet. (Indones. Indones. J. Nutr. Diet.) 2022, 10, 109–118. [Google Scholar] [CrossRef]
- Jin, H.; Zhang, L.; He, J.; Wu, M.; Jia, L.; Guo, J. Role of FOXO3a Transcription Factor in the Regulation of Liver Oxidative Injury. Antioxidants 2022, 11, 2478. [Google Scholar] [CrossRef]
- Divella, R.; Mazzocca, A.; Daniele, A.; Sabbà, C.; Paradiso, A. Obesity, Nonalcoholic Fatty Liver Disease and Adipocytokines Network in Promotion of Cancer. Int. J. Biol. Sci. 2019, 15, 610–616. [Google Scholar] [CrossRef]
- Galicia-Moreno, M.; Lucano-Landeros, S.; Monroy-Ramirez, H.C.; Silva-Gomez, J.; Gutierrez-Cuevas, J.; Santos, A.; Armendariz-Borunda, J. Roles of Nrf2 in Liver Diseases: Molecular, Pharmacological, and Epigenetic Aspects. Antioxidants 2020, 9, 980. [Google Scholar] [CrossRef]
- Li, L.; Fu, J.; Sun, J.; Liu, D.; Chen, C.; Wang, H.; Hou, Y.; Xu, Y.; Pi, J. Is Nrf2-ARE a potential target in NAFLD mitigation? Curr. Opin. Toxicol. 2019, 13, 35–44. [Google Scholar] [CrossRef]
- Song, M.Y.; Lee, D.Y.; Chun, K.S.; Kim, E.-H. The Role of NRF2/KEAP1 Signaling Pathway in Cancer Metabolism. Int. J. Mol. Sci. 2021, 22, 4376. [Google Scholar] [CrossRef]
- Dobson, M.; Ramakrishnan, G.; Ma, S.; Kaplun, L.; Balan, V.; Fridman, R.; Tzivion, G. Bimodal regulation of FoxO3 by AKT and 14-3-3. Biochim. Biophys. Acta BBA Mol. Cell Res. 2011, 1813, 1453–1464. [Google Scholar] [CrossRef]
- Obsilova, V.; Vecer, J.; Herman, P.; Pabianova, A.; Sulc, M.; Teisinger, J.; Boura, E.; Obsil, T. 14-3-3 Protein Interacts with Nuclear Localization Sequence of Forkhead Transcription Factor FoxO4. Biochemistry 2005, 44, 11608–11617. [Google Scholar] [CrossRef]
- Khalili, F.; Vaisi-Raygani, A.; Shakiba, E.; Kohsari, M.; Dehbani, M.; Naseri, R.; Asadi, S.; Rahimi, Z.; Rahimi, M.; Rahimi, Z. Oxidative stress parameters and keap 1 variants in T2DM: Association with T2DM, diabetic neuropathy, diabetic retinopathy, and obesity. J. Clin. Lab. Anal. 2022, 36, e24163. [Google Scholar] [CrossRef]
- Yahya, M.A.; Alshammari, G.M.; Osman, M.A.; Al-Harbi, L.N.; Yagoub, A.E.A.; AlSedairy, S.A. Isoliquiritigenin attenuates high-fat diet-induced intestinal damage by suppressing inflammation and oxidative stress and through activating Nrf2. J. Funct. Foods 2022, 92, 105058. [Google Scholar] [CrossRef]
- Mathivanan, S.; Chunchagatta Lakshman, P.K.; Singh, M.; Giridharan, S.; Sathish, K.; Hurakadli, M.A.; Bharatham, K.; Kamariah, N. Structure of a 14-3-3ε:FOXO3apS253 Phosphopeptide Complex Reveals 14-3-3 Isoform-Specific Binding of Forkhead Box Class O Transcription Factor (FOXO) Phosphoproteins. ACS Omega 2022, 7, 24344–24352. [Google Scholar] [CrossRef]
- Diallo, K.; Oppong, A.K.; Lim, G.E. Can 14-3-3 proteins serve as therapeutic targets for the treatment of metabolic diseases? Pharmacol. Res. 2019, 139, 199–206. [Google Scholar] [CrossRef]
- Deshmukh, P.; Unni, S.; Krishnappa, G.; Padmanabhan, B. The Keap1–Nrf2 pathway: Promising therapeutic target to counteract ROS-mediated damage in cancers and neurodegenerative diseases. Biophys. Rev. 2017, 9, 41–56. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Eguchi, N.; Lau, H.; Ichii, H. The Role of the Nrf2 Signaling in Obesity and Insulin Resistance. Int. J. Mol. Sci. 2020, 21, 6973. [Google Scholar] [CrossRef] [PubMed]
- Asadi, S.; Rahimi, Z.; Sadijam, M.; Shabab, N.; Goodarzi, M.T. Effects of Resveratrol on FOXO1 and FOXO3 a Genes Expression in Adipose Tissue, Serum Insulin, Insulin Resistance and Serum SOD Activity in Type 2 Diabetic Rats. Int. J. Mol. Cell. Med. (IJMCM) 2018, 7, 176–184. [Google Scholar]
- Lim, G.E.; Albrecht, T.; Piske, M.; Sarai, K.; Lee, J.T.C.; Ramshaw, H.S.; Sinha, S.; Guthridge, M.A.; Acker-Palmer, A.; Lopez, A.F.; et al. 14-3-3ζ coordinates adipogenesis of visceral fat. Nat. Commun. 2015, 6, 7671. [Google Scholar] [CrossRef]
- Hayat, J.; Akodad, M.; Moumen, A.; Baghour, M.; Skalli, A.; Ezrari, S.; Belmalha, S. Phytochemical screening, polyphenols, flavonoids and tannin content, antioxidant activities and FTIR characterization of Marrubium vulgare L. from 2 different localities of Northeast of Morocco. Heliyon 2020, 6, e05609. [Google Scholar] [CrossRef] [PubMed]
- de Lima, F.V.I.; Hardiany, N.S.; Dewi, S. Role of Coriander Seed Ethanolic Extract (Coriandrum sativum L.) on the Liver Tissue of High Fat Diet-Induced Rat: Focused on the Oxidative Stress and Cellular Senescence. Master’s Thesis, Faculty of Medicine, Universitas Indonesia, Jakarta, Indonesia, 2022. [Google Scholar]
- Hardiany, N.S.; Fadilah, F.; de Lima, F.V.I.; Dewi, S.; Namirah, I. Flavonoid content and total antioxidant capacity of coriander (Coriandrum sativum L.) seed extract in different level polarity of solvent. AIP Conf. Proc. 2025, 3186, 20063. [Google Scholar]
- Namirah, I.; Hardiany, N.; Arozal, W. The Effects of Ethanolic Extract of C. sativum (Coriandrum sativum L.) Seed on Cellular Senescence in the Cardiovascular Organs of High-Fat Diet-Induced Rats. Ph.D. Dissertation, Faculty of Medicine, University of Indonesia, Jakarta, 2024. [Google Scholar]
- Namirah, I.; Wimbanu, K.S.; Rompies, A.M.E.; Prayogo, Y.S.; Arozal, W.; Fadilah, F.; Hanafi, M.; Hardiany, N.S. The effect of ethanol-based coriander (Coriandrum sativum L.) seed extract on oxidative stress, antioxidant level and cellular senescence in the heart of obese rat. J. Pharm. Pharmacogn. Res. 2024, 12, 1111–1120. [Google Scholar] [CrossRef]
- Pandey, A.K.; Verma, S. An in-silico evaluation of dietary components for structural inhibition of SARS-CoV-2 main protease. J. Biomol. Struct. Dyn. 2022, 40, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wanandi, S.I.; Dewi, S.; Jusman, S.W.A.; Sadikin, M. Expression of manganese superoxide dismutase in rat blood, heart and brain during induced systemic hypoxia. Med. J. Indones. 2011, 20, 27. [Google Scholar] [CrossRef]
- Iskusnykh, I.Y.; Popova, T.N.; Agarkov, A.A.; de Carvalho, M.Â.A.P.; Rjevskiy, S.G. Expression of Glutathione Peroxidase and Glutathione Reductase and Level of Free Radical Processes under Toxic Hepatitis in Rats. J. Toxicol. 2013, 2013, 870628. [Google Scholar] [CrossRef]
- Weydert, C.J.; Cullen, J.J. Measurement of superoxide dismutase, catalase and glutathione peroxidase in cultured cells and tissue. Nat. Protoc. 2010, 5, 51–66. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Douglas, E.V.; Pires, T.L.B.; David, B.A. pkCSM: Predicting Small-Molecule Pharmacokinetic Properties Using Graph-Based Signatures: 1–4. Available online: https://biosig.lab.uq.edu.au/pkcsm/theory (accessed on 4 June 2025).
- Crisman, E.; Duarte, P.; Dauden, E.; Cuadrado, A.; Rodríguez-Franco, M.I.; López, M.G.; León, R. KEAP1-NRF2 protein–protein interaction inhibitors: Design, pharmacological properties and therapeutic potential. Med. Res. Rev. 2023, 43, 237–287. [Google Scholar] [CrossRef]
- Oishi, T.; Matsumaru, D.; Ota, N.; Kitamura, H.; Zhang, T.; Honkura, Y.; Katori, Y.; Motohashi, H. Activation of the NRF2 pathway in Keap1-knockdown mice attenuates progression of age-related hearing loss. NPJ Aging Mech. Dis. 2020, 6, 14. [Google Scholar] [CrossRef]
- McIntyre, R.L.; Liu, Y.J.; Hu, M.; Morris, B.J.; Willcox, B.J.; Donlon, T.A.; Houtkooper, R.H.; Janssens, G.E. Pharmaceutical and nutraceutical activation of FOXO3 for healthy longevity. Ageing Res. Rev. 2022, 78, 101621. [Google Scholar] [CrossRef]
- Pair, F.S.; Yacoubian, T.A. 14-3-3 Proteins: Novel Pharmacological Targets in Neurodegenerative Diseases. Trends Pharmacol. Sci. 2021, 42, 226–238. [Google Scholar] [CrossRef] [PubMed]
- da Sousa, L.R.; Viana, N.R.; Coêlho, A.G.; Barbosa, C.d.O.; Barros, D.S.L.; Martins, M.D.C.d.C.e.; Ramos, R.M.; Arcanjo, D.D.R. Use of Monoterpenes as Potential Therapeutics in Diabetes Mellitus: A Prospective Review. Adv. Pharmacol. Pharm. Sci. 2023, 2023, 1512974. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.S.; Choi, W.J.; Lee, S.; Kim, W.J.; Lee, D.C.; Sohn, U.D.; Shin, H.-S.; Kim, W. Anti-inflammatory, Antioxidant and Antimicrobial Effects of Artemisinin Extracts from Artemisia annua L. Korean J. Physiol. Pharmacol. 2015, 19, 21–27. [Google Scholar]
- Vasileva, L.V.; Savova, M.S.; Amirova, K.M.; Dinkova-Kostova, A.T.; Georgiev, M.I. Obesity and NRF2-mediated cytoprotection: Where is the missing link? Pharmacol. Res. 2020, 156, 104760. [Google Scholar] [CrossRef]
- Gonzalez-Burgos, E.; Gomez-Serranillos, M.P. Terpene Compounds in Nature: A Review of Their Potential Antioxidant Activity. Curr. Med. Chem. 2012, 19, 5319–5341. [Google Scholar]
- Scandar, S.; Zadra, C.; Marcotullio, M.C. Coriander (Coriandrum sativum) Polyphenols and Their Nutraceutical Value against Obesity and Metabolic Syndrome. Molecules 2023, 28, 4187. [Google Scholar] [CrossRef]
- Ji, L.L.; Sheng, Y.C.; Zheng, Z.Y.; Shi, L.; Wang, Z.-T. The involvement of p62–Keap1–Nrf2 antioxidative signaling pathway and JNK in the protection of natural flavonoid quercetin against hepatotoxicity. Free Radic. Biol. Med. 2015, 85, 12–23. [Google Scholar] [CrossRef]
- Grujicic, J.; Allen, A.R. Manganese Superoxide Dismutase: Structure, Function, and Implications in Human Disease. Antioxidants 2025, 14, 848. [Google Scholar] [CrossRef] [PubMed]
- Murugan, R.; Kreiman, G. Multiple transcription auto regulatory loops can act as robust oscillators and decision-making motifs. Comput. Struct. Biotechnol. J. 2022, 20, 5115–5135. [Google Scholar] [CrossRef] [PubMed]
- Jomova, K.; Alomar, S.Y.; Alwasel, S.H.; Nepovimova, E.; Kuca, K.; Valko, M. Several lines of antioxidant defense against oxidative stress: Antioxidant enzymes, nanomaterials with multiple enzyme-mimicking activities, and low-molecular-weight antioxidants. Arch. Toxicol. 2024, 98, 1323–1367. [Google Scholar] [CrossRef]
- Vairetti, M.; Di Pasqua, L.G.; Cagna, M.; Richelmi, P.; Ferrigno, A.; Berardo, C. Changes in Glutathione Content in Liver Diseases: An Update. Antioxidants 2021, 10, 364. [Google Scholar] [CrossRef]
- Chang, C.; Worley, B.L.; Phaëton, R.; Hempel, N. Extracellular Glutathione Peroxidase GPx3 and Its Role in Cancer. Cancers 2020, 12, 2197. [Google Scholar] [CrossRef]
- Esworthy, R.S.; Doroshow, J.H.; Chu, F.F. The beginning of GPX2 and 30 years later. Free Radic. Biol. Med. 2022, 188, 419–433. [Google Scholar] [CrossRef]
- Takio, N.; Yadav, M.; Yadav, H.S. Catalase-mediated remediation of environmental pollutants and potential application–a review. Biocatal. Biotransform. 2021, 39, 389–407. [Google Scholar] [CrossRef]
- Smejkal, G.B.; Kakumanu, S. Enzymes and their turnover numbers. Expert Rev. Proteom. 2019, 16, 543–544. [Google Scholar] [CrossRef] [PubMed]
- Hadwan, M.H.; Hussein, M.J.; Mohammed, R.M.; Hadwan, A.M.; Al-Kawaz, H.S.; Al-Obaidy, S.S.M.; Al Talebi, Z.A. An improved method for measuring catalase activity in biological samples. Biol. Methods Protoc. 2024, 9, bpae015. [Google Scholar] [CrossRef] [PubMed]
- Bizoń, A.; Chojdak-Łukasiewicz, J.; Budrewicz, S.; Pokryszko-Dragan, A.; Piwowar, A. Exploring the Relationship between Antioxidant Enzymes, Oxidative Stress Markers, and Clinical Profile in Relapsing-Remitting Multiple Sclerosis. Antioxidants 2023, 12, 1638. [Google Scholar] [CrossRef] [PubMed]
- Meilhoc, E.; Boscari, A.; Pauly, N.; Lepetit, M.; Frendo, P.; Bruand, C.; Puppo, A.; Brouquisse, R. Oxygen and derived reactive species in legume–rhizobia interactions: Paradoxes and dual roles. J. Exp. Bot. 2025, 76, 3758–3773. [Google Scholar] [CrossRef]
- Xiao, J.; Bei, Y.; Liu, J.; Dimitrova-Shumkovska, J.; Kuang, D.; Zhou, Q.; Li, J.; Yang, Y.; Xiang, Y.; Wang, F.; et al. miR-212 downregulation contributes to the protective effect of exercise against non-alcoholic fatty liver via targeting FGF-21. J. Cell Mol. Med. 2016, 20, 204–216. [Google Scholar] [CrossRef]
- Carbonell, T.; Gomes, A.V. MicroRNAs in the regulation of cellular redox status and its implications in myocardial ischemia-reperfusion injury. Redox Biol. 2020, 36, 101607. [Google Scholar] [CrossRef]
- Guo, J.; Yang, P.; Li, Y.F.; Tang, J.-F.; He, Z.-X.; Yu, S.-G.; Yin, H.-Y. MicroRNA: Crucial modulator in purinergic signalling involved diseases. Purin. Signal. 2023, 19, 329–341. [Google Scholar] [CrossRef]
- Ruiz-Manriquez, L.M.; Carrasco-Morales, O.; Sanchez, Z.E.A.; Osorio-Perez, S.M.; Estrada-Meza, C.; Pathak, S.; Banerjee, A.; Bandyopadhyay, A.; Duttaroy, A.K.; Paul, S. MicroRNA-mediated regulation of key signaling pathways in hepatocellular carcinoma: A mechanistic insight. Front. Genet. 2022, 13, 910733. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Guo, L.; Yang, Y.; Wang, Y.; Xia, S.; Gong, H.; Zhang, B.-K.; Yan, M. Dissecting the Crosstalk Between Nrf2 and NF-κB Response Pathways in Drug-Induced Toxicity. Front. Cell Dev. Biol. 2022, 9, 809952. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Fattah, M.M.; Hassanein, E.H.M.; Sayed, A.M.; Alsufyani, S.E.; El-Sheikh, A.A.K.; Arab, H.H.; Mohamed, W.R. Targeting SIRT1/FoxO3a/Nrf2 and PI3K/AKT Pathways with Rebamipide Attenuates Acetic Acid-Induced Colitis in Rats. Pharmaceuticals 2023, 16, 533. [Google Scholar] [CrossRef]
- Iside, C.; Scafuro, M.; Nebbioso, A.; Altucci, L. SIRT1 Activation by Natural Phytochemicals: An Overview. Front. Pharmacol. 2020, 11, 1225. [Google Scholar] [CrossRef]
- Sousa, C.; Mendes, A.F. Monoterpenes as Sirtuin-1 Activators: Therapeutic Potential in Aging and Related Diseases. Biomolecules 2022, 12, 921. [Google Scholar] [CrossRef]
- Rahman, H.S.; Othman, H.H.; Hammadi, N.I.; Yeap, S.K.; Amin, K.M.; Samad, N.A.; Alitheen, N.B. Novel Drug Delivery Systems for Loading of Natural Plant Extracts and Their Biomedical Applications. Int. J. Nanomed. 2020, 15, 2439–2483. [Google Scholar] [CrossRef] [PubMed]

| Identified Compounds | PubChem ID | ChemSpider ID |
|---|---|---|
| Valeric acid | 7991 | 7701 |
| Caproic acid | 8892 | 8552 |
| Enanthic acid | 8094 | 7803 |
| Caprylic acid | 379 | 370 |
| Pelargonic acid | 8158 | 7866 |
| Capric acid | 2969 | 2863 |
| Isoeugenol | 853433 | 7061 |
| Lauric acid | 3893 | 3756 |
| Myristic acid | 11005 | 10539 |
| Pentadecylic acid | 13849 | 13249 |
| Palmitelaidic acid | 5282745 | 4445872 |
| Palmitic Acid | 985 | 960 |
| Margaric acid | 10465 | 10033 |
| Petroselinic acid | 5281125 | 4444569 |
| Oleic Acid | 445639 | 393217 |
| Vaccenic acid | 5281127 | 4444571 |
| Stearic acid | 5281 | 5091 |
| Arachidic acid | 10467 | 10035 |
| 1-Monooleoylglycerol | 5283468 | 4446588 |
| 3-Hydroxy-2,5-hexadione | 10369937 | - |
| 6-O-Acetyl shanzhiside methyl ester | 48223312 | - |
| Aloeresin C | 70697807 | - |
| Arteannuin | 6543478 | 62026 |
| E-p-Coumaric acid | 637542 | - |
| Eucommiol | 154373 | 136004 |
| Hexitol | 453 | 440 |
| Kaempferol | 5280863 | 4444395 |
| Lactinolide | 14336593 | 58827445 |
| Lobetyolin | 14655097 | 24534044 |
| Madecassoside | 24825675 | 23359979 |
| Quercetin | 5280343 | 4444051 |
| Schizonepetoside E | 46173963 | 10260731 |
| Shionoside B | - | 10252121 |
| Trinexapac-Ethyl | 92421 | 83439 |
| Group | Study Phase | ||
|---|---|---|---|
| Acclimatization Phase | Experimental Phase | ||
| 1 Week | 12 Weeks | 12 Weeks | |
| N (n = 5) | Normal diet | Normal diet | Normal diet |
| NE (n = 5) | Normal diet | Normal diet | Normal diet + coriander seeds ethanolic extract |
| N-FE (n = 5) | Normal diet | Normal diet | High fat diet + coriander seeds ethanolic extract |
| F (n = 5) | Normal diet | High fat diet | High fat diet |
| FE (n = 5) | Normal diet | High fat diet | High fat diet + coriander seeds ethanolic extract |
| Stage | Thermocycling Condition |
|---|---|
| Stage 1 | Reverse transcription: 45 °C 10 min Polymerase activation: 95 °C 2 min |
| Stage 2 (40 cycles) | Denaturation: 95 °C 5 s Annealing: 58 °C 10 s (for MnSOD) 60 °C 10 s (for GPx) 59 °C 10 s (for β-actin) Extension: 72 °C 5 s |
| Stage 3 | 95 °C 1 min |
| Stage 4 | Melting curve analysis: 55 °C 10 s (80 cycles increasing 0.5 °C each cycle) |
| Gene | Primer |
|---|---|
| MnSOD [26] | Forward: AACGTCACCGAGGAGAAGTA Reverse: TGATAGCCTCCAGCAACTCT |
| GPx [27] | Forward: AGTTCGGACATCAGGAGAATGGCA Reverse: TCACCATTCACCTCGCACTTCTCA |
| β-actin [26] | Forward: CACTGGCATTGTGATGGACT Reverse: CTCTCAGCTGTGGTGGTGAA |
| Coriander Seed Extract Compounds | Protein Keap1 | Protein 14-3-3 | ||
|---|---|---|---|---|
| Binding Affinity (kcal/mol) | Inhibition Constant (Ki) | Binding Affinity (kcal/mol) | Inhibition Constant (Ki) | |
| Valeric acid | −4.22 | 811.87 µM | −4.08 | 1.03 mM |
| Caproic acid | −4.40 | 599.74 µM | −4.02 | 1.13 mM |
| Enanthic acid | −4.45 | 544.65 µM | −4.10 | 989.75 µM |
| Caprylic acid | −4.42 | 573.69 µM | −4.52 | 485.93 µM |
| Pelargonic acid | −4.30 | 698.95 µM | −4.49 | 514.94 µM |
| Capric acid | −4.80 | 301.10 µM | −4.17 | 884.98 µM |
| Isoeugenol | −4.73 | 338.67 µM | −4.41 | 589.16 µM |
| Lauric acid | −4.69 | 367.59 µM | −3.55 | 2.50 mM |
| Myristic acid | −4.68 | 372.25 µM | −4.02 | 1.12 mM |
| Pentadecylic acid | −5.26 | 138.77 µM | −3.39 | 3.29 mM |
| Palmitelaidic acid | −4.88 | 264.38 µM | −3.18 | 4.64 mM |
| Palmitic Acid | −4.61 | 416.39 µM | −3.31 | 3.73 mM |
| Margaric acid | −5.16 | 163.83 µM | −2.77 | 9.26 mM |
| Petroselinic acid | −5.87 | 50.20 µM | −3.17 | 4.75 mM |
| Oleic Acid | −4.78 | 314.96 µM | −2.54 | 13.64 mM |
| Vaccenic acid | −4.64 | 396.12 µM | −3.25 | 4.12 mM |
| Stearic acid | −4.53 | 477.72 µM | −2.66 | 11.21 mM |
| Arachidic acid | −4.06 | 1.06 mM | −2.27 | 21.85 mM |
| 1-Monooleoylglycerol | −3.20 | 4.52 mM | −1.42 | 91.24 mM |
| 3-Hydroxy-2,5-hexadione | −4.28 | 734.68 µM | −4.13 | 935.86 µM |
| 6-O-Acetyl shanzhiside methyl ester | −5.17 | 161.44 µM | −3.66 | 2.07 mM |
| Aloeresin C | −7.31 | 4.36 µM | −5.51 | 91.16 µM |
| Arteannuin | −7.44 | 3.50 µM | −6.01 | 39.17 µM |
| E-p-Coumaric acid | −5.00 | 216.39 µM | −5.12 | 175.68 µM |
| Eucommiol | −4.12 | 957.50 µM | −3.13 | 5.08 mM |
| Hexitol | −1.78 | 49.96 mM | −2.20 | 24.49 mM |
| Kaempferol | −6.55 | 15.77 µM | −5.64 | 72.96 µM |
| Lactinolide | −5.45 | 101.36 µM | −5.37 | 115.76 µM |
| Lobetyolin | −5.96 | 43.02 µM | −5.03 | 205.93 µM |
| Madecassoside | −1.62 | 64.73 mM | −2.89 | 7.63 mM |
| Quercetin | −6.49 | 17.65 µM | −5.68 | 68.85 µM |
| Schizonepetoside E | −5.43 | 104.22 µM | −4.72 | 349.55 µM |
| Shionoside B | −8.90 | 298.01 nM | −6.85 | 9.46 µM |
| Trinexapac-Ethyl | −5.87 | 49.44 µM | −5.85 | 51.36 µM |
| Native Ligand of Keap1 ((1S,2R)-2-{[(1S)-1-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)methyl]-3,4-dihydroisoquinolin-2(1H)-yl]carbonyl}cyclohexanecarboxylic acid (C26H26N2O5) | −9.37 | 135.50 nM | ||
| Native Ligand of 14-3-3 [2-[2-oxidanylidene-2-[[3-[2-[2-[2-[3-oxidanylidene-3-(propylamino)propoxy]ethoxy]ethoxy] ethylcarbamoyl]phenyl]amino]ethoxy] phenyl]phosphonic acid (C27 H38 N3 O10 P) | −6.46 | 18.36 uM | ||
| Protein | Compound | Hydrogen Bond | Hydrophobic |
|---|---|---|---|
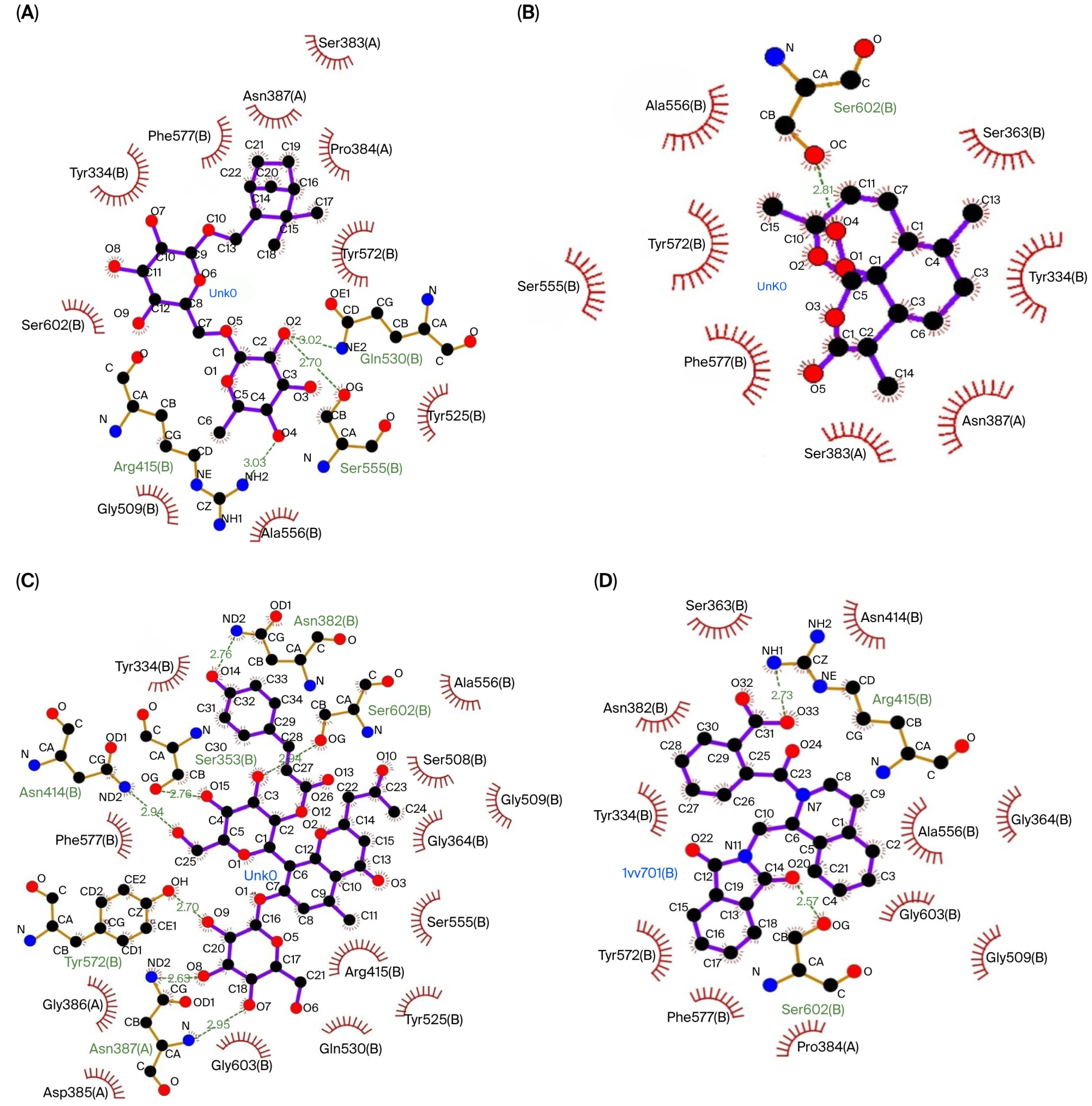
| Keap1 | Shionoside B | Gln530 (3.02 Å), Ser555 (2.70 Å), Arg415 (3.03 Å) | Ser383, Pro384, Tyr572, Tyr525, Ala556, Gly509, Ser602, Tyr334, Phe577, Asn387 |
| Arteannuin | Ser602 (2.81 Å) | Ser383, Tyr572, Ser555, Ala556, Tyr334, Phe577, Asn387, Ser363 | |
| Aloeresin C | Tyr572 (2.70 Å), Ser602 (2.94 Å), Asn387 (2.95 Å, 2.63 Å), Ser363 (2.76 Å), Asn382 (2.76 Å), Asn414 (2.94 Å) | Gln530, Tyr525, Ser555, Ala556, Arg415, Tyr334, Phe577, Ser508, Gly364, Gly509, Gly603, Gly386, Asp385 | |
| Native Ligand | Arg415 (2.73 Å), Ser602 (2.57 Å) | Pro384, Ala556, Tyr334, Phe577, Ser363, Asn382, Gly364, Gly509, Gly603, Asn414, Tyr572 | |
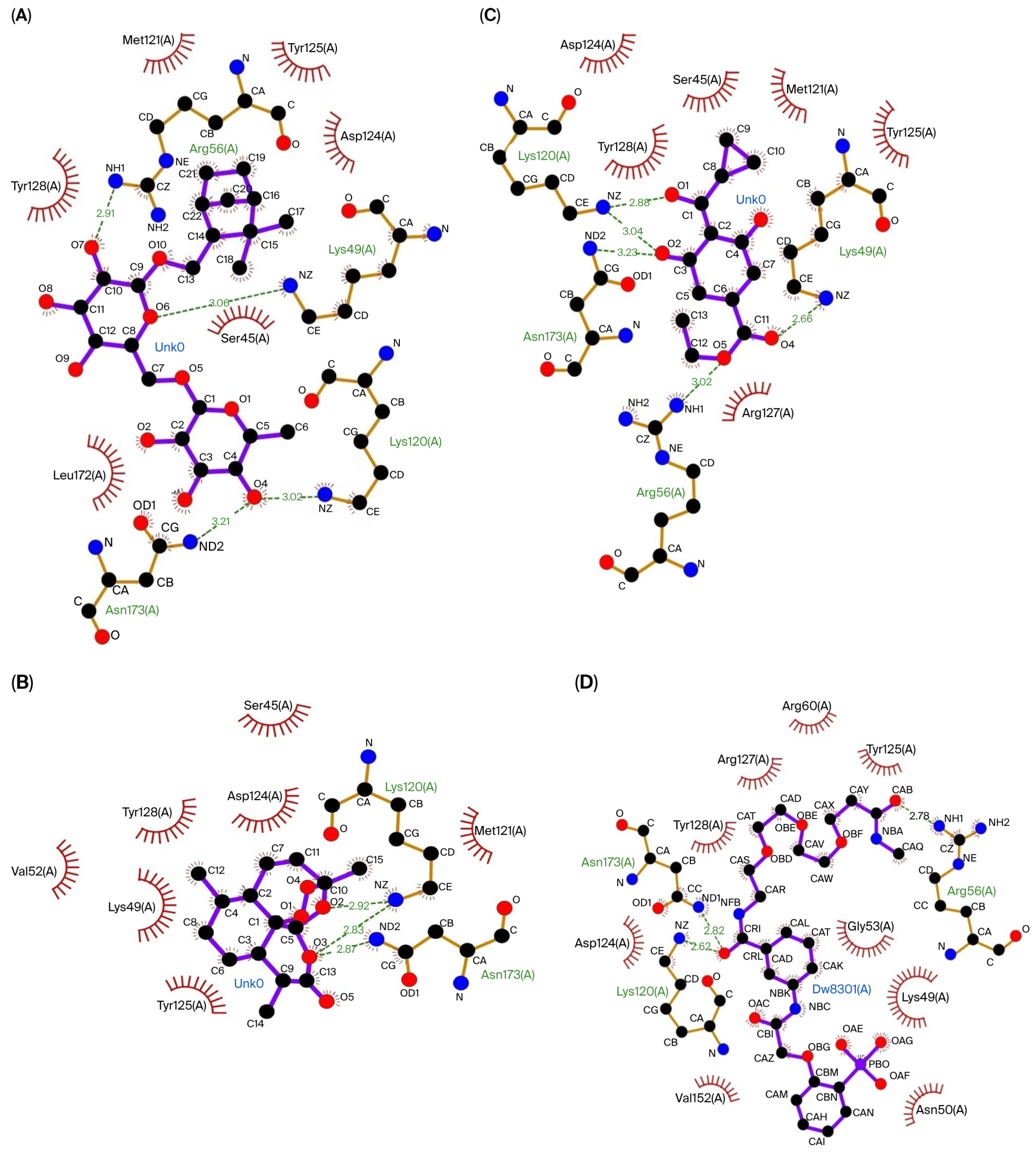
| 14-3-3 | Shionoside B | Arg56 (2.91 Å), Lys49 (3.06 Å), Lys120 (3.02 Å), Asn173 (3.21 Å) | Leu172, Ser45, Tyr128, Asp124, Tyr125, Met121 |
| Arteannuin | Lys120 (2.92 Å; 2.83 Å), Asn173 (2.87 Å) | Met121, Tyr125, Val152, Lys49, Tyr128, Asp124, Ser45 | |
| Trinexapac-Ethyl | Arg56 (3.02 Å), Asn173 (3.23 Å), Lys120 (2.88 Å; 3.04 Å), Lys49 (2.66 Å) | Arg127, Asp124, Ser45, Tyr128, Met121, Tyr125 | |
| Native Ligand | Arg56 (2.78 Å), Lys120 (2.62 Å), Asn173 (2.82 Å) | Gly53, Lys49, Asn50, Val152, Asp124, Tyr128, Arg127, Arg60, Tyr125 |
| Compounds | MW | HBA | HBD | LogP | Drug-Likeness |
|---|---|---|---|---|---|
| Shionoside B | 462.53 g/mol | 10 | 6 | 1.76 | No (1 violation) |
| Arteannuin | 282.33 g/mol | 5 | 0 | 2.72 | Yes (no violation) |
| Aloeresin C | 702.66 g/mol | 16 | 8 | 1.13 | No (3 violation) |
| Trinexapac-Ethyl | 252.26 g/mol | 5 | 1 | 1.72 | Yes (no violation) |
| Parameters | Shionoside B | Arteannuin | Aloeresin C | Trinexapac-Ethyl | |
|---|---|---|---|---|---|
| Absorption | Water solubility (log mol/L) | −1.284 | −3.678 | −2.823 | −1.964 |
| Caco-2 permeability (log Papp in 10−6 cm/s) | −0.425 | 1.295 | −1.01 | 0.477 | |
| Intestinal absorption (human) (% Absorbed) | 34.766 | 97.543 | 7.395 | 83.739 | |
| P-glycoprotein substrate | Yes | No | Yes | No | |
| Distribution | VDss (human) (log L/kg) | 0.16 | 0.457 | 0.347 | −0.096 |
| Fraction unbound (human) (FU) | 0.464 | 0.4 | 0.182 | 0.541 | |
| BBB permeability (log BB) | −0.861 | 0.235 | −2.195 | −0.392 | |
| CNS permeability (log PS) | −4.239 | −2.909 | −4.594 | −2.933 | |
| Metabolism | CYP2D6 substrate | No | No | No | No |
| CYP3A4 substrate | No | Yes | No | No | |
| CYP1A2 inhibitor | No | Yes | No | No | |
| CYP2C19 inhibitor | No | No | No | No | |
| CYP2C9 inhibitor | No | No | No | No | |
| CYP2D6 inhibitor | No | No | No | No | |
| CYP3A4 inhibitor | No | No | No | No | |
| Excretion | Total Clearance (log mL/min/kg) | 1.168 | 0.98 | 0.053 | 0.494 |
| Renal OCT2 substrate | No | No | No | No | |
| Parameters | Shionoside B | Arteannuin | Aloeresin C | Trinexapac-Ethyl |
|---|---|---|---|---|
| Maximum tolerated dose (human) (log mg/kg/day) | −0.023 | 0.065 | 0.23 | 0.704 |
| Oral Rat Acute Toxicity (LD50) (mol/kg) | 2.343 | 2.459 | 2.586 | 2.126 |
| Oral Rat Chronic Toxicity (LOAEL) (log mg/kgBW/day) | 4.678 | 1 | 4.609 | 1.375 |
| Hepatotoxicity | No | No | No | No |
| Neurotoxicity | No | No | No | No |
| Nephrotoxicity | Yes | No | Yes | Yes |
| Cardiotoxicity | Yes | Yes | No | No |
| Carcinogenicity | No | No | No | No |
| Cytotoxicity | No | No | No | No |
| Nrf2 | FOXO3 | |||
|---|---|---|---|---|
| Pearson Correlation | p-Value | Pearson Correlation | p-Value | |
| Nrf2 | 1 | 0.859 ** | 0.000 | |
| FOXO3 | 0.859 ** | 0.000 | 1 | |
| MnSOD relative mRNA expression | 0.119 | 0.285 | −0.038 | 0.428 |
| GPx relative mRNA expression | −0.150 | 0.238 | −0.216 | 0.149 |
| T-SOD specific enzyme activity | 0.558 ** | 0.002 | 0.783 ** | 0.000 |
| MnSOD specific enzyme activity | 0.567 ** | 0.002 | 0.743 ** | 0.00 |
| GPx specific enzyme activity | 0.426 * | 0.017 | 0.672 ** | 0.000 |
| GSH level | 0.458 * | 0.011 | 0.700 ** | 0.000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pertiwi, K.D.; Hardiany, N.S.; Dewi, S.; Tejo, B.A. Regulation of Antioxidant Expression in the Liver Tissue of Obese Rats Treated with Coriander Seed Ethanolic Extract: In Silico and In Vivo Studies. Biologics 2025, 5, 32. https://doi.org/10.3390/biologics5040032
Pertiwi KD, Hardiany NS, Dewi S, Tejo BA. Regulation of Antioxidant Expression in the Liver Tissue of Obese Rats Treated with Coriander Seed Ethanolic Extract: In Silico and In Vivo Studies. Biologics. 2025; 5(4):32. https://doi.org/10.3390/biologics5040032
Chicago/Turabian StylePertiwi, Kartika Diana, Novi Silvia Hardiany, Syarifah Dewi, and Bimo Ario Tejo. 2025. "Regulation of Antioxidant Expression in the Liver Tissue of Obese Rats Treated with Coriander Seed Ethanolic Extract: In Silico and In Vivo Studies" Biologics 5, no. 4: 32. https://doi.org/10.3390/biologics5040032
APA StylePertiwi, K. D., Hardiany, N. S., Dewi, S., & Tejo, B. A. (2025). Regulation of Antioxidant Expression in the Liver Tissue of Obese Rats Treated with Coriander Seed Ethanolic Extract: In Silico and In Vivo Studies. Biologics, 5(4), 32. https://doi.org/10.3390/biologics5040032






