Abstract
Nematophagous fungi (NFs), which are responsible for soil suppression of plant-parasitic nematodes, are multitrophic biocontrol agents. This raises the question of the transition between lifestyles (e.g., endophytism vs. egg parasitism). The NF Pochonia chlamydosporia colonises food crops and promotes their growth and yield. When colonising the plant, P. chlamydosporia induces the plant immunity (PI). However, it also evades the PI. To do this, both endophytic NF and pathogenic fungi (PF) secrete LysM effectors (LysM-effs). LysM effectors have been shown to have diverse functions in different organisms, including the protection of fungal chitin from plant chitinases. P. chlamydosporia is resistant to chitosan, which modulates gene expression in fungi and plants and has antimicrobial properties. P. chlamydosporia chitin deacetylases (CDA) and chitosanases (CSN) also help P. chlamydosporia evade plant immunity, resist exogenous chitosan, and are induced during fungal infection of nematode eggs. NF-chitosan formulations are new biomanagement tools against plant parasitic nematodes, fungal wilt pathogens and insect pests that currently threaten food security crops. Furthermore, omics techniques are useful tools to elucidate the role of CDAs, CSNs, LysM-effs, adhesion proteins and carbohydrate-active enzymes in pathogen–BCA–plant interactions, adhesion and infection to nematode eggs and their modulation by chitosan.
1. Introduction
A plethora of nematophagous, mycoparasitic, plant- and insect-pathogenic, plant-endophytic, and saprophytic fungi belong to the order Hypocreales. Some of these fungi can switch between different lifestyles [1,2]. Nematophagous fungi (NFs), such as Pochonia chlamydosporia and Purpureocillium lilacinum, have been identified in nematode-suppressive soils [3,4]. P. chlamydosporia is a cosmopolitan biocontrol fungus which can parasitize females and eggs of cyst- and root-knot nematodes but also colonises endophytically the roots of many crops [5]. The multitrophic nature of these fungal biocontrol agents (BCAs) is paramount for their efficacy because, unlike conventional agrochemicals, they can be host-specific and persist in the environment. This leads to long-term host management [6] with low environmental impact and no damage to human health, unlike toxic agrochemicals.
Deacetylation of chitin results in the formation of chitosan (Figure 1). This occurs when the degree of deacetylation of chitin reaches approximately 50% due to solubilisation and protonation of the -NH2 functional group at the C-2 position of the D-glucosamine repeat unit [7]. Chitosan is thus a linear polymer of beta-(1-4)-linked N-acetyl-2-amino-2-deoxy-d-glucose and 2-amino-2-deoxy-d-glucose subunits [8]. It is also the only cationic biopolymer, with a wide range of applications, such as flocculant for protein recovery or decontamination [7]. The N-deacetylation of chitin and chitooligosaccharides is catalysed by chitin deacetylases (CDAs). These enzymes have different substrate specificities and give rise to fully or partially deacetylated products with various deacetylation patterns [9]. The main raw materials to obtain chitosan are marine crustacean exoskeletons [10]. Therefore, the multiple applications of this compound add value to the waste of the shellfish industry. As an alternative to toxic agrochemicals, chitosan may reduce current soil, marine, and freshwater pollution rates. Chitosan has antimicrobial properties and modulates the gene expression of plants and fungi [8]. It can be combined with fungal BCAs to protect global food security crops [11].
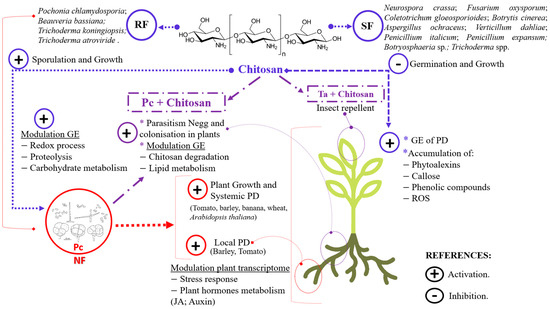
Figure 1.
Effects of chitosan on biocontrol fungi and plants. Responses of nematophagous fungi (NFs, red) to chitosan (completely deacetylated as portrayed) and plants to NF combined with Chitosan (purple) are displayed. Abbreviations: RF—chitosan-resistant fungi; SF—chitosan-sensitive fungi; NFs—nematophagous fungi; Pc—Pochonia chlamydosporia [12]; Negg—nematode eggs; GE—gene expression; PD—plant defences; JA—jasmonic acid; ROS—reactive oxygen species; Ta—Trichoderma atroviride.
Polysaccharide deacetylases are widely distributed in fungi and other organisms [13,14,15]. They have been reported to have diverse functions in developmental processes other than virulence [9,16,17,18,19,20,21,22]. For instance, the fungal pathogen Magnaporthe oryzae expresses CDAs during appressorium development. Additionally, chitosan is a cell wall component of germ tubes and appressoria of M. oryzae [17]. Alternatively, chitosan could promote the avoidance of plant immunity (PI) and fitness/virulence of fungal pathogens, including BCAs such as P. chlamydosporia [8]. Moreover, fungal effectors (F-effs) prevent pathogen recognition by plants, suppressing host immune responses (IR), and manipulating host cell physiology to facilitate colonisation [23]. Many effectors and their modes of action have been well identified and characterized as LysM domains effectors (LysM-effs) in fungi [23]. Analysing fungi with different lifestyles, our research group has shown that LysM-effs, CDAs, chitosanases (CSNs), and other hydrolases contribute to fungal lifestyle [24,25,26,27,28,29].
This article describes recent advances in NF gene expression in relation to their multi-trophic lifestyles and their relevance as BCAs. The importance of their combination with natural elicitors, such as chitosan, and their modulatory and regulatory potential on fungi and plants is highlighted. The mechanisms of action and effects of NFs, PFs, and chitosan on plants are discussed in a cross-sectional and integrative manner, considering the relevance of F-effs, CDAs, CSNs, and plant cell wall-degrading enzymes in the evolution of NFs to evade PI.
2. Nematophagous Fungi and Chitosan: Growth and Defence Modulators in Plants
P. chlamydosporia is an endophyte of both mono- and dicotyledonous crops of global economic importance, such as banana [30], tomato [5], wheat, potato [31], and barley [32]. P. chlamydosporia strain 123 (Pc123) promotes growth in Musa acuminata cv. ‘Dwarf Cavendish’ [30], tomato [5], and barley [32]. Several P. chlamydosporia strains reduce the flowering and fruiting times of Arabidopsis thaliana [33] and Solanum lycopersicum [34]. In addition to this, NF also induces systemic [35,36] as well as local [5,32] defences in plants. The colonisation of barley roots by Pc123 induces genes involved in stress response (PR1), plant IR, auxin biosynthesis, auxin-mediated transcriptional regulation, and jasmonic acid (JA) metabolism [35]. Zavala-Gonzalez et al. [33] have also proposed that JA pathways modulate P. chlamydosporia colonisation in A. thaliana. This would be related to JA plant priming induction and regulation of plant–microorganism symbiosis [37]. Considering the effects of P. chlamydosporia on plants, other endophytic NFs could also help plants to cope to abiotic and biotic stresses (Figure 1).
Chitosan is a natural compound with multiple actions with the ability to modulate gene expression in fungi and plants (Figure 1) [8]. The composition of the plasma membrane determines the sensitivity of fungi to chitosan. Fungi with low-fluidity membranes, such as P. chlamydosporia, are resistant to chitosan [38]. Fungal plasma membrane permeabilisation leads to the accumulation of reactive oxygen species (ROS) and triggers cell death [8] in the chitosan sensitive fungus Neurospora crassa [39]. Trichoderma spp. have a versatile response to chitosan. Chitosan promotes growth and sporulation of T. koningiopsis, while T. citronoviride, T. pseudokoningii, T. neocrassum, and T. harzianum are sensitive to this compound [40]. Chitosan inhibits germination, alters germ tube elongation, and reduces mycelium growth of many plants fungal pathogens, such as Fusarium oxysporum f. sp. lycopersici, Colletotrichum gloeosporioides, Botrytis cinerea, Aspergillus ochraceus, Verticillium dahliae, Botryosphaeria sp., Penicillium italicum, and Penicillium expansum [41,42,43,44,45,46]. In contrast, several biocontrol fungi (Pc123, P. rubescens, and Beauveria bassiana) use chitosan as a nutrient [42]. In P. chlamydosporia, chitosan modulates redox processes, carbohydrate metabolism, and proteolysis [28]. It also enhances sporulation of some entomopathogenic fungi (EPFs) (B. bassiana) and NF (P. chlamydosporia) [42]. Chitosan activates plant defence genes through the octadecanoid pathway [47]. Among other events, this biopolymer induces phytoalexin synthesis and accumulation [48,49], lignification and deposition of callose, phenolic compounds, and ROS [50,51]. All this evidence suggests that the combined use of chitosan with BCAs (Figure 1) can improve the integrated management of pests and diseases caused by nematodes, wilt phytopathogens, and insects.
NFs are more efficient biocontrol agents when combined with chitosan (Figure 1) [11,27,52,53]. Chitosan stimulates P. chlamydosporia appressorium differentiation, proteolytic activity, and nematode egg-parasitism [11]. This biopolymer also promotes the colonisation of tomato roots by P. chlamydosporia [52]. Chitosan activates P. chlamydosporia ROS detoxification metabolism and modulates expression of genes involved in chitosan degradation, lipid metabolism, nematode egg parasitism, and endophytism [28]. Foliar applications T. atroviride spores and chitosan show a pronounced insect-repellent effect [53]. These studies manifest the potential of chitosan combined with NFs against plant pests and diseases.
3. Plant Defence Avoidance
3.1. Plant Immunity (PI)
Plants have developed immune receptors to detect pathogens and thus prevent infections [54]. There are two main types: pattern recognition receptors (PRRs) and nucleotide-rich leucine repeat receptors [55,56]. Cell membrane PRRs include extracellular ligand-binding, transmembrane, and intracellular kinase domains [57]. They are the primary defence line against pathogens. PRRs can specifically recognize microbe-associated molecular patterns (MAMPs), such as fungal chitin and β-glucans activating pattern-triggered immunity (PTI) [58,59,60,61].
Once MAMPs are recognized, plants activate secondary metabolism and secrete degradative enzymes such as glucanases and chitinases, which release β-glucan and chitin oligomers from fungal cell walls (CWs) [56]. In turn, fungi have evolved to evade plant defences (PDs). During colonisation of plant tissue, some pathogenic fungi (PF) can modify their CW-transforming chitin into chitosan [19,62]. This mechanism protects PF hyphae from plant chitinases because the presence of chitin fragments induces a rapid response of plant cells. However, after this initial stimulus, these cells become completely refractory, resulting in a slow recovery of the ability to respond to chitin oligomers [63]. Meanwhile, some chitin oligomers with high levels of acetylation induce alkalinisation and ROS production in the plant, the main PTI responses [64,65,66]. PI also involves the overexpression of pathogenesis-related (PR) proteins in response to microbial pathogens, such as the PR-4 family, which includes class I and II chitinases [67,68]. In addition, chitosan induces the expression of PR proteins (NPR1) in roots [69] and leaves (PR1 and PR5 [70]). Chitosan is a less efficient MAMP than chitin. For this reason, plant chitinases have a lower affinity for chitosan than for chitin [19,65].
Virulence is directly linked to the deacetylation of chitin oligomers, whose N-acetyl group contributes to the perception of host lysine motif (LysM)-containing receptors for ligand-induced immunity [71,72]. When a fungus invades plants, plant LysM proteins on the cell membrane detect and bind MAMPs from the surface of fungus and activate PTI. The analysis of expression of MaLysMs in Musa acuminata after root inoculation with the banana wilt fungus Fusarium oxysporum f. sp. cubense Tropical Race 4 (FocTR4), showed that MaLysM1 was down- and MaLysM11-1 up-regulated [73].
Alternatively, LysM-domain proteins expressed as extracellular proteins in fungi are involved in pathogenicity and the invasion of plant cells, as well as inactivation of the plant IR; these proteins are fungal LysM-effs (Table 1) [8].

Table 1.
LysM effectors, Chitin Deacetylases (CDAs) and Chitosanases (CSNs) of Nematophagous fungi (NFs), plant pathogenic fungi (PFs), Entomopathogenic fungi (EPFs), and Mycoparasites (Ms). Omics strategies used for their characterisation are also given.
3.2. Secreted Proteins and Effectors of Nematophagous Fungi
Cytoplasmic effectors act within host cells, whereas apoplastic ones do so in the extracellular matrix [59,96]. A wide range of fungal pathogens secrete LysM-effs (Table 1). Cladosporium fulvum Ecp6, a protein with three LysM domains, mopes chitin oligosaccharides released from the cell walls of invading hyphae to evade chitin-mediated PTI [74,75]. Zymoseptoria tritici Mg1LysM and Mg3LysM could also protect hyphae from plant hydrolytic enzymes [76]. However, only Mg3LysM can block chitin-triggered stimulation of PDs. ChELP1 and ChELP2 from Colletotrichum higginsianum, M. oryzae Slp1, Rhizoctonia solani RsLys, V. dahliae Vd2LysM interfere with the activation of induced immunity through chitin and contribute to virulence [77,79,80,81,82]. LysM-effs contribute to fungal lifestyle [29]. Therefore, NF with a multitrophic lifestyle encode a larger number of putative LysM-effs than endoparasites [29]. These NFs include the endophytes P. chlamydosporia (parasite of nematode eggs and females), Arthrobotrys oligospora (nematode trapping fungus), and Pleurotus ostreatus (toxin-producing NF) [34,87,97]. EPFs, such as B. bassiana and Metarhizium robertsii [89,98], but also mycoparasites (Trichoderma spp.), possess a high number of putative LysM-effs encoded in their genomes [29]. T. atroviride Tal6 interacts with N–acetylglucosamine to protect its hyphae from plant chitinases, thereby preventing detection of the fungus as an evasive response to PDs [91]. Similarly, Rhizophagus irregularis RiSLM, binds to chitin oligosaccharides and effectively interferes with the IR triggered by chitin, protecting its cell wall and evading PI [90,99]. Pc123 putative LysM-effs are constitutively expressed. However, Pc123 Lys1 is the most highly expressed when banana roots are present [29]. It could therefore be a key effector for shielding chitin from Pc123 cell wall.
Many F-effs generate transcriptome specific patterns in planta [96]. Previous studies on Foc TR4 transformants overexpressing key transcription factors (SGE1 and FTF1), show abundance of Lys-M-effectors, cerato-platanin effectors and SIX-effectors (SIX6, SIX9, and SIX13) [100]. Colletrotichum gloeosporioides Cg2LysM knock-out mutants showed affected fungal growth and development and reduced virulence to rubber trees [83]. Likewise, Tal6 protein inhibits Trichoderma spp. germination [92]. These studies indicate that LysM-effs from phytopathogenic and endophytic fungi play an important role in PD evasion and are growth and development regulators.
3.3. Enzymes Involved in the Degradation of Chitin and Chitosan
CDAs are involved in diverse biological processes. Activity in vivo of many fungal CDAs involved in chitin deacetylation has been identified (Table 1). However, mode of action and substrate specificity are available for only a few CDAs [9]. These enzymes are involved in CW development and morphology [16], germling adhesion [17], spore formation [18], PI evasion [19], and fungal autolysis [20]. Furthermore, fungi with chitosan as well as chitin in their CWs secrete periplasmic CDAs, which catalyse chitosan biosynthesis from chitin [9]. At random, the sequential or processive mechanisms of fungal CDAs show diverse chitooligosaccharide specificities, resulting in chitosan oligosaccharides with various acetylation patterns [21].
To successfully colonise host plants, endophytic fungi must evade PI. PesCDA from the endophyte Pestalotiopsis sp. deacetylates chitosan oligomers abolishing their elicitor activity in rice cells [22]. Pc123 was shown to express CDAs and CSNs during nematode egg infection. Chitosan immunolocalization in Pc123 appressoria strongly suggests avoidance of release of chitooligosasaccarides during nematode egg infection which would elicit defences from nearby root cells [26]. Chitooligosaccharides could originate from fungal CW, nematode eggshell, or (more likely) both. The activity of CDAs has also been demonstrated in Metarhizium anisopliae [93]. Pc123 CDA1 and CDA2 genes are significantly induced with nematode eggs [26]. CDA2 has been characterised as a protein containing a carbohydrate esterase catalytic domain (CE4) flanked by two carbohydrate-binding modules (CBM18) and chitin-binding domains [27]. Chitin promotes the expression of chitinases, while chitosan is an elicitor of both chitinases and CSNs [101]. Therefore, it has been hypothesised that upon nematode egg infection, PcCDAs generate chitosan and induce expression of CSNs [26].
Chitosan biodegradation is carried out by CSNs [102]. The activity of these enzymes has been linked to defence against pathogens in plants [103] and in soil fungi [104]. However, CSN expression is also associated with damage caused by the pathogen Fusarium solani [95]. Pc123 CSNs expression is maximized with fungus, chitosan, and nematode eggs together [25,27]. Pc123 genome encodes 11 putative CSNs [25] from GH75 family. This is a high number compared to that of similar fungi that only encode 3 (Metarhizium acridum, M. anisopliae, Trichoderma reesei), 5 (T. virens), and 6 CSNs (T. atroviride). Furthermore, evolutionary studies of these putative enzymes have confirmed that gene expression is due to recent duplication events in the closely related paralogous genes csn3, csn4, and csn5 of P. chlamydosporia [28]. There is also evidence for alternative splicing in csn3 [105]. This suggests that CSN isoforms may have different localisations or functions [94]. Furthermore, csn3 is induced six-fold when Pc123 infects nematode eggs [26] and doubled when infection occurs in the presence of chitosan [28]. The dentification and characterisation of proteins with important activities would lead to a better understanding of the infection process nematode eggs, fungal resistance to chitosan, evasion of PI. Since both chitin and chitosan are found in CW, it may even provide future evidence for the role of these enzymes in fungal–fungal interactions.
4. Host Infection and Colonisation by Nematophagous Fungi
Pc123 genome includes many genes encoding glycosyl hydrolases other than CSNs [25]. The transition between nematophagous, saprophytic, and endophytic lifestyles of this fungus is associated with the expression of genes encoding carbohydrate-active enzymes that stimulate the degradation of polysaccharides present in the plant cell wall, such as cellulose, xylan, and pectin [26]. VCP1 serine protease and SCP1 serine carboxypeptidase are secreted barley roots endophytically colonised by P. chlamydosporia [24,106]. Both enzymes also have been immunolocalised and characterised in M. javanica eggs (Mj-eggs) infected by this fungus [11]. More than 40% of P. chlamydosporia gene expression during endophytic colonisation of barley root corresponds to glycoside hydrolase (GH) and carbohydrate esterase (CE) families [25]. This is also confirmed for the expression of GHs when the fungus parasitises Mj-eggs, which higher than that of related fungal species such as M. acridum and M. anisopliae [26]. The combined action of endoglucanases and cellobiohydrolases is involved in cellulose degradation [107]. The study of genes encoding cellohydrolase families of GHs suggests that Pc, like the saprophytes N. crassa and T. reesi, encodes enzymes of the GH6 and GH7 families, which is different from EPF such as B. bassiana, M. anisopliae, M. acridum, and Cordyceps militaris, which have completely lost these enzymes and others such as the lytic polysaccharide monooxygenases of the AA9 family, which are essential for the degradation of cellulose into soluble mono- and disaccharides. The same occur for the GH67 family involved in xylan degradation. In terms of pectinolytic activity, the enzymes encoded by Pc123 are quite reduced, with only three endopolygalacturonases belonging to the GH8 family, and they do not possess any pectin esterase or pectin lyase, compared to other fungi such as N. crassa and A. oligospora, which have an extensive pectinolytic apparatus [26].
Comparison of genes involved in the endophytic phase in Pc123 [25] with the nutritional transition in Pc170 [84] also revealed that some secreted proteins may be involved in multiple lifestyle transitions, including peptidases, CAZymes (e.g., glycoside hydrolases: GH16, GH17, GH25, GH72; glycosyltransferase: GT31; Acetyl-xylanesterase: CE5; and glyoxal oxidase: AA5), among others. In the Pc170 genome, genes potentially involved in nematode egg infection and fungal adaptation have been identified, including around of 71 genes encoding secreted proteins, including cellulase (GH5), GH30 proteins, and copper/zinc superoxide dismutase [84]. Among these gene pairs, two genes encoding secreted GH30 O-glycosyl hydrolases are found; however, they are absent in many Hypocreales fungi, but their homologues are found in endophytic and mycoparasitic fungi (Trichoderma spp.) and in nematophagous fungi (P. lilacinum and H. minnesotensis). Expression of GH30 genes was observed in barley root colonisation in Pc123 [32] and is associated with plant wall degradation [108,109] and may therefore be related to the endophytic lifestyle of the fungus. In addition, 32 genes were found to be under positive selection when comparing the genomes of Pc170 and Pc123, most of which are of unknown function but are thought to play an important role in the parasitism and adaptation of the fungus, based on the functions of 10 genes, including the appressorium specific CAS1-protein-encoding gene, related to host infection.
As mentioned above, Pc123 gene expression levels are induced in the presence of chitosan and Mj-eggs [28]. Under these conditions, genes coding for carbohydrate metabolism (glucokinase), chitosan, and sugar degradation (GH2, GH3, and GH75), membrane transport (MFS transporters), and adhesion proteins (FLO1) are overexpressed. Thus, Pc123 is thought to bind to the eggshell of M. javanica eggs by binding carbohydrates, lipids, and peptides [28]. The flocculation protein FLO1 is a mannose-binding glycoprotein present in yeasts and associated with hyphal adhesion [110]. Therefore, FLO1 may be relevant for adhesion to the nematode eggshell.
The importance of carbohydrates in the metabolism of P. chlamydosporia has already been demonstrated. Previous studies of P. chlamydosporia IMI 380407 have observed the expression of GAL4 [111], a specific fungal activator of the GAL system. Thus, galactose is present in the fungal wall as a glycoprotein [112] and acts as a promoter of filamentous growth in Candida albicans [113] and cellulase gene expression in the anamorphic fungus T. reesei [114]. In turn, nematode-trapping fungi recognise the nematode cuticle, which contains galactose residues, including N-acetyl-D-galactosamine and D-galactose [115,116,117,118,119]. Thus, since galactose could be released from the eggshell, the study of GAL4 in Pc will show promoters that regulate this sugar metabolism [111].
5. Multi-Omics Tools for Understanding Integrated Pest Management
5.1. Biocontrol Mechanisms of Fungal BCAs
Fungal BCAs may act by direct antagonism, antibiosis, competition, induced resistance, or a combination of these [6]. In hyperparasitism [120] or direct antagonism, a fungus is parasitised by other fungi. Trichoderma spp. are mycoparasites of many economically important plant pathogens [121,122]. For instance, Trichoderma strains are antagonists of the yerba mate root rot pathogen F. oxysporum [123] and parasitise R. solani hyphae to control citrus seedling wetness [121,124]. Fungi secrete secondary metabolites with antibiotic activity, including volatiles [125]. Pc, B. bassiana, and M. robertsii volatile organic compounds (VOCs) act as repellents for economically important insect pests [126]. Yeasts and filamentous fungi can inhibit pathogens by competing for nutrients. Trichoderma spp. have improved water and nutrient uptake when associated with the plant root system, which confers protection to the host against pathogens [127,128,129]. Competing for iron, Trichoderma spp. can effectively control growth of F. oxysporum and Pythium spp. in soil [130]. Induced plant resistance is also involved in the control of plant pathogens by beneficial microorganisms [6]. Non-pathogenic F. oxysporum sensu lato, an endophytic fungus, promote JA, salicylic acid, and ethylene in tomato, which control pathogenicity of F. oxysporum f. sp. lycopersici [131].
Secretion of extracellular hydrolytic enzymes, as discussed above for Pc, is also involved in the mechanism of action of fungal BCAs [132].
5.2. Omics to Elucidate BCA–Plant–Pathogen Interaction
We have reviewed the importance of multi-omics (genomics, transcriptomics, proteomics, and metabolomics) in the production of NF LysM-effs, extracellular enzymes, and secondary metabolites, including VOCs. They can be induced by elicitors such as chitosan, phytopathogens (nematode eggs and endophytic PFs) or plants (Table 1). Therefore, multi-omics is a key tool for the understanding of the molecular mechanisms of NFs in terms of the biocontrol of phytopathogens and colonisation of crops [106]. It is now even possible to quantify the expression of genes encoding enzymes and transcription factors and estimate NF biomass using qPCR.
The use of genetic engineering techniques has led to improvements in fungal strains resulting in more effective modes of action and the generation of fungi-resistant crops [132]. In addition, obtaining fungal protoplasts has become a widely used tool to elucidate the importance and predict the behaviour of specific genes in PFs [133] and NFs [134]. Thus, these CW-free cells have allowed us to evaluate the effect of chitosan or their genetic transformation to obtain knockout mutants of genes encoding enzymes [134] or transcriptional regulators [133].
Among the genetic transformation methods in filamentous fungi, the most common include protoplast-mediated transformation (PMT), Agrobacterium-mediated transformation (AMT), electroporation transformation, biolistic transformation, and shock wave-mediated transformation (SWMT) [135,136]. Although genome editing technology has been increasingly developed in recent years, new technologies used in fungi include clustered regularly interspaced short palindromic repeats (CRISPR) technology [137]. Among the third generation DNA technology tools using endonucleases for fungal genome editing is CRISPR-associated protein 9 (Cas9) [135]. This technology has been implemented in several species of filamentous fungi as pathogens and fungal species of industrial interest [138,139]. In N. crassa, Cas9, and sgRNA (single RNA) constructs have been introduced into fungal conidia using donor plasmids by electroporation [140]. In T. reesei, CRISPR-Cas9 was used in two steps: a fungal strain expressing Cas9 was created and then transformed with sgRNA transcripts generated in vitro [141]. More recently, a method based on a Cas9–sgRNA ribonucleoprotein (RNP) complex has been used to edit the genomes of species from different kingdoms. The RNP strategy consists of a purified Cas9 protein and a sgRNA transcript synthesised in vitro, and this complex is then transfected into host cells [142]. According to Wang and Coleman [143], Cas9–sgRNA complexes have several advantages over the use of plasmids. First, sgRNA transcription and Cas9 protein expression do not depend on host machinery. In addition, the assembled RNPs have immediate excision activity and the excision efficiency of sgRNAs can be tested in vitro, allowing the selection of the most appropriate sgRNAs for the target gene. The Cas9–sgRNA-mediated CRISPR–Cas9 method has been used in several fungal species, such as the saprophyte A. fumigatus [144] or the phytopathogen Fusarium proliferatum [145]. In view of the above, advances in multi-omics and new recombinant DNA technologies are essential to elucidate the gene expression and enzyme functions of the mechanisms of action of each fungal species and the mechanisms of interaction between BCA and pathogen, BCA and plant, and the triple interaction BCA–plant–pathogen. This would ultimately reveal the specific characteristics of multitrophic lifestyles in NFs.
5.3. Efficient and Stable BCA Formulates against Pests
There are currently several commercial products that use biocontrol fungi to manage plant pests and diseases. Fungal BCAs have several advantages, including being widely distributed on our planet, many are easy to grow and maintain under laboratory conditions, have high host specificity, are resistant and can spread [6]. As discussed in this work they also evade host IRs. In addition, compared to chemicals fungicides, fungal BCAs do not cause negative impacts on the environment and soil biodiversity. The beneficial effects of using BCAs such as P. chlamydosporia would be greatly enhanced in combination with compounds such as chitosan, which acts as an elicitor of PI, limiting the growth, germination, and hyphal morphology of economically important phytopathogens [8], and as a nutrient for BCAs themselves [49].
P. chlamydosporia-combined chitosan has potential for the integrated management of root-knot nematodes in the field [52]. The search for new formulations, such as the implementation of microencapsulates for the integrated management of plant diseases and pests and containing biological and chemical agents, is a challenge for the research of our laboratory, which is looking for a formulation containing P. chlamydosporia (or metabolites secreted by this fungus) and chitosan with the ability to suppress the presence of nematodes and pathogenic wilt fungi in agricultural ecosystems and insect pests. In this sense, our group has recently patented registered a formulation of P. chlamydosporia and chitosan coacervates. Also, as mentioned above, our group has found that the VOCs secreted by the NF P. chlamydosporia and the EPFs B. bassiana and M. robertsii are repellents for the black banana weevil [126].
6. Closing Remarks and Future Perspectives
Multi-omics is a useful tool for understanding the morphology, physiology, and biocontrol potential of P. chlamydosporia and other NFs. We can now envisage the relationship between gene expression in nematode egg infection (parasitism), the evasion of PDs (endophytism), or growth in soil (saprophytism). These processes are modified in the presence of chitosan.
The induction of LysM-effs, CDAs, CSNs, and enzymes involved in plant colonisation is likely to be correlated with NF lifestyle switches. They are therefore involved in protection against wilt fungi (such as FocTR4) and plant parasitic nematodes (such as M. javanica). Some of them (eg. NF LysM-effs) might even be used in the future as additives combined with NFs and chitosan for smart plant protection.
Finally, investigating the mechanisms involved in pathogen–plant, BCA–pathogen, BCA–plant, and BCA–plant–pathogen interactions may help designing new BCA–chitosan formulations or metabolite combinations to safely manage pest and diseases which affect food. This is paramount in our current scenario of global change. Our scarce knowledge on the mechanisms of host defence evasion of NFs is the current bottleneck. Environmental multi-omics is probably a key solution.
Author Contributions
Conceptualization, C.M.B., F.L.-M. and L.V.L.-L.; writing—original draft preparation, C.M.B.; writing—review and editing, L.V.L.-L., F.L.-M. and C.M.B. All authors have read and agreed to the published version of the manuscript.
Funding
This project has been funded by the PID-2020-119734RB-I00 project of the Spanish Ministry of Science and Innovation.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bushley, K.E.; Raja, R.; Jaiswal, P.; Cumbie, J.S.; Nonogaki, M.; Boyd, A.E.; Owensby, C.A.; Knaus, B.J.; Elser, J.; Miller, D.; et al. The Genome of Tolypocladium inflatum: Evolution, Organization, and Expression of the Cyclosporin Biosynthetic Gene Cluster. PLoS Genet. 2013, 9, e1003496. [Google Scholar] [CrossRef] [PubMed]
- Sung, G.-H.; Poinar, G.O.; Spatafora, J.W. The oldest fossil evidence of animal parasitism by fungi supports a Cretaceous diversification of fungal–arthropod symbioses. Mol. Phylogenet. Evol. 2008, 49, 495–502. [Google Scholar] [CrossRef]
- Bent, E.; Loffredo, A.; McKenry, M.V.; Becker, J.O.; Borneman, J. Detection and Investigation of Soil Biological Activity against Meloidogyne incognita. J. Nematol. 2008, 40, 109–118. [Google Scholar] [PubMed]
- Lamovsek, J.; Urek, G.; Trdan, S. Biological control of root-knot nematodes (Meloidogyne spp.): Microbes against the pests. Acta Agron. Slov. 2013, 101, 263–275. [Google Scholar] [CrossRef]
- Bordallo, J.J.; Lopez-Llorca, L.V.; Jansson, H.B.; Salinas, J.; Persmark, L.; Asensio, L. Colonization of plant roots by egg-parasitic and nematode-trapping fungi. New Phytol. 2002, 154, 491–499. [Google Scholar] [CrossRef]
- Singh, S.; Sharma, A. Fungi as Biological Control Agents. In Biofertilizers for Sustainable Agriculture and Environment; Giri, B., Prasad, R., Wu, Q.S., Varma, A., Eds.; Springer: Cham, The Netherlands, 2019; Volume 55. [Google Scholar]
- Rinaudo, M. Chitin and chitosan: Properties and applications. Prog. Polym. Sci. 2006, 31, 603–632. [Google Scholar] [CrossRef]
- Lopez-Moya, F.; Suarez-Fernandez, M.; Lopez-Llorca, L.V. Molecular Mechanisms of Chitosan Interactions with Fungi and Plants. Int. J. Mol. Sci. 2019, 20, 332. [Google Scholar] [CrossRef]
- Grifoll-Romero, L.; Pascual, S.; Aragunde, H.; Biarnés, X.; Planas, A. Chitin Deacetylases: Structures, Specificities, and Biotech Applications. Polymer 2018, 10, 352. [Google Scholar] [CrossRef]
- Younes, I.; Rinaudo, M. Chitin and Chitosan Preparation from Marine Sources. Structure, Properties and Applications. Mar. Drugs 2015, 13, 1133–1174. [Google Scholar] [CrossRef]
- Escudero, N.; Ferreira, S.R.; Lopez-Moya, F.; Naranjo-Ortiz, M.A.; Marin-Ortiz, A.I.; Thornton, C.R.; Lopez-Llorca, L.V. Chitosan enhances parasitism of Meloidogyne javanica eggs by the nematophagous fungus Pochonia chlamydosporia. Fungal Biol. 2016, 120, 572–585. [Google Scholar] [CrossRef]
- Zare, R.; Gams, W.; Evans, H.C. A revision of Verticillium section Prostrata. V. The genus Pochonia, with notes on Rotiferophthora. Nova Hedwig. 2001, 73, 51–86. [Google Scholar] [CrossRef]
- Berlemont, R. Distribution and diversity of enzymes for polysaccharide degradation in fungi. Sci. Rep. 2017, 7, 222. [Google Scholar] [CrossRef] [PubMed]
- Andreou, A.; Giastas, P.; Christoforides, E.; Eliopoulos, E.E. Structural and Evolutionary Insights within the Polysaccharide Deacetylase Gene Family of Bacillus anthracis and Bacillus cereus. Genes 2018, 9, 386. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Gao, L.; Xue, C.; Mao, X. Marine-polysaccharide degrading enzymes: Status and prospects. Compr. Rev. Food Sci. Food Saf. 2020, 19, 2767–2796. [Google Scholar] [CrossRef] [PubMed]
- Baker, L.G.; Specht, C.A.; Donlin, M.J.; Lodge, J.K. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryot. Cell 2007, 6, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Geoghegan, I.A.; Gurr, S.J. Chitosan Mediates Germling Adhesion in Magnaporthe oryzae and Is Required for Surface Sensing and Germling Morphogenesis. PLoS Pathog. 2016, 12, e1005703. [Google Scholar] [CrossRef]
- Christodoulidou, A.; Bouriotis, V.; Thireos, G. Two Sporulation-specific Chitin Deacetylase-encoding Genes Are Required for the Ascospore Wall Rigidity of Saccharomyces cerevisiae. J. Biol. Chem. 1996, 271, 31420–31425. [Google Scholar] [CrossRef]
- Sánchez-Vallet, A.; Mesters, J.R.; Thomma, B.P. The battle for chitin recognition in plant-microbe interactions. FEMS Microbiol. Rev. 2015, 39, 171–183. [Google Scholar] [CrossRef]
- White, S.; McIntyre, M.; Berry, D.R.; McNeil, B. The autolysis of industrial filamentous fungi. Crit. Rev. Biotechnol. 2002, 22, 1–14. [Google Scholar] [CrossRef]
- Zhao, Y.; Park, R.-D.; Muzzarelli, R.A. Chitin deacetylases: Properties and applications. Mar. Drugs 2010, 8, 24–46. [Google Scholar] [CrossRef]
- Cord-Landwehr, S.; Melcher, R.L.; Kolkenbrock, S.; Moerschbacher, B.M. A chitin deacetylase from the endophytic fungus Pestalotiopsis sp. efficiently inactivates the elicitor activity of chitin oligomers in rice cells. Sci. Rep. 2016, 6, 38018. [Google Scholar] [CrossRef]
- Liu, T.; Chen, Y.; Tian, S.; Li, B. Crucial Roles of Effectors in Interactions between Horticultural Crops and Pathogens. Horticulturae 2023, 9, 250. [Google Scholar] [CrossRef]
- Lopez-Llorca, L.V.; Gómez-Vidal, S.; Monfort, E.; Larriba, E.; Casado-Vela, J.; Elortza, F.; Jansson, H.B.; Salinas, J.; Martín-Nieto, J. Expression of serine proteases in egg-parasitic nematophagous fungi during barley root colonization. Fungal Genet. Biol. 2010, 47, 342–351. [Google Scholar] [CrossRef]
- Larriba, E.; Jaime, M.D.; Carbonell-Caballero, J.; Conesa, A.; Dopazo, J.; Nislow, C.; Martín-Nieto, J.; Lopez-Llorca, L.V. Sequencing and functional analysis of the genome of a nematode egg-parasitic fungus, Pochonia chlamydosporia. Fungal Genet. Biol. 2014, 65, 69–80. [Google Scholar] [CrossRef]
- Aranda-Martinez, A.; Lenfant, N.; Escudero, N.; Zavala-Gonzalez, E.A.; Henrissat, B.; Lopez-Llorca, L.V. CAZyme content of Pochonia chlamydosporia reflects that chitin and chitosan modification are involved in nematode parasitism. Environ. Microbiol. 2016, 18, 4200–4215. [Google Scholar] [CrossRef]
- Aranda-Martinez, A.; Grifoll-Romero, L.; Aragunde, H.; Sancho-Vaello, E.; Biarnés, X.; Lopez- Llorca, L.V.; Planas, A. Expression and specificity of a chitin deacetylase from the nematophagous fungus Pochonia chlamydosporia potentially involved in pathogenicity. Sci. Rep. 2018, 8, 2170. [Google Scholar] [CrossRef]
- Suarez-Fernandez, M.; Sambles, C.; Lopez-Moya, F.; Nueda, M.J.; Studholme, D.J.; Lopez-Llorca, L.V. Chitosan modulates Pochonia chlamydosporia gene expression during nematode egg parasitism. Environ. Microbiol. 2021, 23, 4980–4997. [Google Scholar] [CrossRef]
- Suarez-Fernandez, M.; Aragon-Perez, A.; Lopez-Llorca, L.V.; Lopez-Moya, F. Putative LysM Effectors Contribute to Fungal Lifestyle. Int. J. Mol. Sci. 2021, 22, 3147. [Google Scholar] [CrossRef]
- Mingot-Ureta, C.; Lopez-Moya, F.; Lopez-Llorca, L.V. Isolates of the Nematophagous Fungus Pochonia chlamydosporia Are Endophytic in Banana Roots and Promote Plant Growth. Agronomy 2020, 10, 1299. [Google Scholar] [CrossRef]
- Manzanilla-López, R.H.; Esteves, I.; Powers, S.J.; Kerry, B.R. Effects of crop plants on abundance of Pochonia chlamydosporia and other fungal parasites of root-knot and potato cyst nematodes. Ann. Appl. Biol. 2011, 159, 118–129. [Google Scholar] [CrossRef]
- Maciá-Vicente, J.G.; Rosso, L.C.; Ciancio, A.; Jansson, H.-B.; Lopez-Llorca, L.V. Colonisation of barley roots by endophytic Fusarium equiseti and Pochonia chlamydosporia: Effects on plant growth and disease. Ann. Appl. Biol 2009, 155, 391–401. [Google Scholar] [CrossRef]
- Zavala-Gonzalez, E.A.; Rodríguez-Cazorla, E.; Escudero, N.; Aranda-Martinez, A.; Martínez-Laborda, A.; Ramírez-Lepe, M.; Vera, A.; Lopez-Llorca, L.V. Arabidopsis thaliana root colonization by the nematophagous fungus Pochonia chlamydosporia is modulated by jasmonate signaling and leads to accelerated flowering and improved yield. New Phytol. 2017, 213, 351–364. [Google Scholar] [CrossRef]
- Zavala-González, E.A.; Escudero, N.; Lopez-Moya, F.; Aranda-Martinez, A.; Exposito, A.; Ricaño-Rodríguez, J.; Naranjo-Ortiz, M.A.; Ramírez-Lepe, M.; Lopez-Llorca, L.V. Some isolates of the nematophagous fungus Pochonia chlamydosporia promote root growth and reduce flowering time of tomato. Ann. Appl. Biol. 2015, 166, 472–483. [Google Scholar] [CrossRef]
- Larriba, E.; Jaime, M.D.; Nislow, C.; Martín-Nieto, J.; Lopez-Llorca, L.V. Endophytic colonization of barley (Hordeum vulgare) roots by the nematophagous fungus Pochonia chlamydosporia reveals plant growth promotion and a general defense and stress transcriptomic response. J. Plant Res. 2015, 128, 665–678. [Google Scholar] [CrossRef]
- Ghahremani, Z.; Escudero, N.; Saus, E.; Gabaldón, T.; Sorribas, F.J. Pochonia chlamydosporia induces plant-dependent systemic resistance to Meloidogyne incognita. Front. Plant Sci. 2019, 10, 945. [Google Scholar] [CrossRef]
- Jung, S.C.; Martinez-Medina, A.; Lopez-Raez, J.A.; Pozo, M.J. Mycorrhiza-induced resistance and priming of plant defenses. J. Chem. Ecol. 2019, 38, 651–664. [Google Scholar] [CrossRef]
- Palma-Guerrero, J.; Lopez-Jimenez, J.A.; Pérez-Berná, A.J.; Huang, I.-C.; Jansson, H.-B.; Salinas, J.; Villalaín, J.; Read, N.D.; Lopez-Llorca, L.V. Membrane fluidity determines sensitivity of filamentous fungi to chitosan. Mol. Microbiol. 2010, 75, 1021–1032. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Moya, F.; Kowbel, D.; Nueda, M.J.; Palma-Guerrero, J.; Glass, N.L.; Lopez- Llorca, L.V. Neurospora crassa transcriptomics reveals oxidative stress and plasma membrane homeostasis biology genes as key targets in response to chitosan. Mol. BioSyst. 2016, 12, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Zavala-González, E.A.; Lopez-Moya, F.; Aranda-Martinez, A.; Cruz-Valerio, M.; Lopez-Llorca, L.V.; Ramírez-Lepe, M. Tolerance to chitosan by Trichoderma species is associated with low membrane fluidity. J. Basic Microbiol. 2016, 56, 792–800. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Tian, S.; Meng, X.; Xu, Y. Effects of chitosan on control of postharvest diseases and physiological responses of tomato fruit. Postharvest Biol. Technol. 2007, 44, 300–306. [Google Scholar] [CrossRef]
- Palma-Guerrero, J.; Jansson, H.-B.; Salinas, J.; Lopez-Llorca, L.V. Effect of chitosan on hyphal growth and spore germination of plant pathogenic and biocontrol fungi. J. Appl. Microbiol. 2008, 104, 541–553. [Google Scholar] [CrossRef]
- Lee, C.G.; Koo, J.C.; Park, J.K. Antifungal Effect of Chitosan as Ca2+ Channel Blocker. Plant Pathol. J. 2016, 32, 242–250. [Google Scholar] [CrossRef]
- Wang, Y.; Li, B.; Zhang, X.; Peng, N.; Mei, Y.; Liang, Y. Low molecular weight chitosan is an effective antifungal agent against Botryosphaeria sp. and preservative agent for pear (Pyrus) fruits. Int. J. Biol. Macromol. 2017, 95, 1135–1143. [Google Scholar] [CrossRef] [PubMed]
- Xoca-Orozco, L.-Á.; Cuellar-Torres, E.A.; González-Morales, S.; Gutiérrez-Martínez, P.; López-García, U.; Herrera-Estrella, L.; Vega-Arreguín, J.; Chacón-López, A. Transcriptomic Analysis of Avocado Hass (Persea americana Mill) in the Interaction System Fruit-Chitosan-Colletotrichum. Front. Plant Sci. 2017, 8, 956. [Google Scholar] [CrossRef]
- Meng, D.; Garba, B.; Ren, Y.; Yao, M.; Xia, X.; Li, M.; Wang, Y. Antifungal activity of chitosan against Aspergillus ochraceus and its possible mechanisms of action. Int. J. Biol. Macromol. 2020, 158, 1063–1070. [Google Scholar] [CrossRef]
- Riseh, R.S.; Hassanisaadi, M.; Vatankhah, M.; Babaki, S.A.; Barka, E.A. Chitosan as a potential natural compound to manage plant diseases. Int. J. Biol. Macromol. 2022, 220, 998–1009. [Google Scholar] [CrossRef]
- Orzali, L.; Corsi, B.; Forni, C.; Riccioni, L. Chitosan in agriculture: A new challenge for managing plant disease. In Biological Activities and Application of Marine Polysaccharides; Shalaby, E., Ed.; InTech: London, UK, 2017; pp. 17–36. [Google Scholar]
- Hadwiger, L.A. Multiple effects of chitosan on plant systems: Solid science or hype. Plant Sci. 2013, 208, 42–49. [Google Scholar] [CrossRef]
- Luna, E.; Pastor, V.; Robert, J.; Flors, V.; Mauch-Mani, B.; Ton, J. Callose deposition: A multifaceted plant defense response. Mol. Plant-Microbe Interact. 2011, 24, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Narasimhamurthy, K.; Udayashankar, A.C.; De Britto, S.; Lavanya, S.N.; Abdelrahman, M.; Soumya, K.; Shetty, H.S.; Srinivas, C.; Jogaiah, S. Chitosan and chitosan-derived nanoparticles modulate enhanced immune response in tomato against bacterial wilt disease. Int. J. Biol. Macromol. 2022, 220, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Escudero, N.; Lopez-Moya, F.; Ghahremani, Z.; Zavala-Gonzalez, E.A.; Alaguero-Cordovilla, A.; Ros-Ibañez, C.; Lacasa, A.; Sorribas, F.J.; Lopez-Llorca, L.V. Chitosan Increases Tomato Root Colonization by Pochonia chlamydosporia and Their Combination Reduces Root-Knot Nematode Damage. Front. Plant Sci. 2017, 8, 1415. [Google Scholar] [CrossRef]
- Kappel, L.; Kosa, N.; Gruber, S. The Multilateral Efficacy of Chitosan and Trichoderma on Sugar Beet. J. Fungi 2022, 8, 137. [Google Scholar] [CrossRef] [PubMed]
- Nürnberger, T.; Kemmerling, B. Receptor protein kinases—Pattern recognition receptors in plant immunity. Trends Plant Sci. 2006, 11, 519–522. [Google Scholar] [CrossRef]
- Lu, Y.; Tsuda, K. Intimate Association of PRR- and NLR-Mediated Signalling in Plant Immunity. Mol. Plant-Microbe Interact. 2021, 34, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.-M.; Zhang, Y. Plant Immunity: Danger Perception and Signalling. Cell 2020, 181, 978–989. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, M.; Yu, L.; Zhou, Z.; Liang, X.; Liu, Z.; Cai, G.; Gao, L.; Zhang, X.; Wang, Y.; et al. The FLS2-Associated Kinase BIK1 Directly Phosphorylates the NADPH Oxidase RbohD to Control Plant Immunity. Cell Host Microbe 2014, 15, 329–338. [Google Scholar] [CrossRef]
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-Microbe Interactions: Shaping the Evolution of the Plant Immune Response. Cell 2006, 124, 803–814. [Google Scholar] [CrossRef]
- Jones, J.; Dangl, J. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Newman, M.-A.; Sundelin, T.; Nielsen, J.T.; Erbs, G. MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front. Plant Sci. 2013, 4, 139. [Google Scholar] [CrossRef]
- Yu, X.; Feng, B.; He, P.; Shan, L. From Chaos to Harmony: Responses and Signalling upon Microbial Pattern Recognition. Annu. Rev. Phytopathol. 2017, 55, 109–137. [Google Scholar] [CrossRef]
- Geoghegan, I.; Steinberg, G.; Gurr, S. The Role of the Fungal Cell Wall in the Infection of Plants. Trends Microbiol. 2017, 25, 957–967. [Google Scholar] [CrossRef]
- Felix, G.; Regenass, M.; Boller, T. Specific perception of subnanomolar concentrations of chitin fragments by tomato cells: Induction of extracellular alkalinization, changes in protein phosphorylation, and establishment of a refractory state. Plant J. 1993, 4, 307–316. [Google Scholar] [CrossRef]
- Baureithel, K.; Felix, G.; Boller, T. Specific, high affinity binding of chitin fragments to tomato cells and membranes. Competitive inhibition of binding by derivates of chitooligosaccharides and Nod factor of Rhizobium. J. Biol. Chem. 1994, 269, 17931–17938. [Google Scholar]
- Vander, P.; Vårum, K.M.; Domard, A.; Eddine El Gueddari, N.; Moerschbacher, B.M. Comparison of the ability of partially N-acetylated chitosans and chitooligosaccharides to elicit resistance reactions in wheat leaves. Plant Physiol. 1998, 118, 1353–1359. [Google Scholar] [CrossRef]
- Li, P.; Linhardt, R.J.; Cao, Z. Structural characterization of oligochitosan elicitor from Fusarium sambucinum and its elicitation of defensive responses in Zanthoxylum bungeanum. Int. J. Mol. Sci. 2016, 7, 2076. [Google Scholar] [CrossRef]
- van Loon, L.C.; Rep, M.; Pieterse, C.M. Significance of Inducible Defense-related Proteins in Infected Plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef]
- Bravo, J.M.; Campo, S.; Murillo, I.; Coca, M.; San Segundo, B. Fungus- and wound-induced accumulation of mRNA containing a class II chitinase of the pathogenesis-related protein 4 (PR-4) family of maize. Plant Mol. Biol. 2003, 52, 745–759. [Google Scholar] [CrossRef]
- Lopez-Moya, F.; Escudero, N.; Zavala-González, E.A.; Esteve-Bruna, D.; Blázquez, M.A.; Alabadí, D.; Lopez-Llorca, L.V. Induction of auxin biosynthesis and WOX5 repression mediate changes in root development in Arabidopsis exposed to chitosan. Sci. Rep. 2017, 7, 16813. [Google Scholar] [CrossRef] [PubMed]
- Beatrice, C.; Linthorst, J.H.; Cinzia, F.; Luca, R. Enhancement of PR1 and PR5 gene expressions by chitosan treatment in kiwifruit plants inoculated with Pseudomonas syringae pv. actinidiae. Eur. J. Plant Pathol. 2017, 148, 163–179. [Google Scholar] [CrossRef]
- Gao, F.; Zhang, B.-S.; Zhao, J.-H.; Huang, J.-F.; Jia, P.-S.; Wang, S.; Zhang, J.; Zhou, J.-M.; Guo, H.-S. Deacetylation of chitin oligomers increases virulence in soil-borne fungal pathogens. Nat. Plants 2019, 5, 1167–1176. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.-P.; Li, J.-J.; Dhar, N.; Li, J.-P.; Chen, J.-Y.; Jian, W.; Dai, X.-F.; Yang, X.-F. Lysin Motif (LysM) Proteins: Interlinking Manipulation of Plant Immunity and Fungi. Int. J. Mol. Sci. 2021, 22, 3114. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Zhang, C.; Wang, M.; Zhang, C.; Xu, X.; Huang, Y.; Chen, Y.; Lin, Y.; Lai, Z. Genome-wide identification, evolution analysis of LysM gene family members and their expression analysis in response to biotic and abiotic stresses in banana (Musa L.). Gene 2022, 845, 146849. [Google Scholar] [CrossRef]
- Bolton, M.D.; van Esse, H.P.; Vossen, J.H.; de Jonge, R.; Stergiopoulos, I.; Stulemeijer, I.J.; van Den Berg, G.C.M.; Borrás-Hidalgo, O.; Dekker, H.L.; de Koster, C.G.; et al. The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol. Microbiol. 2008, 69, 119–136. [Google Scholar] [CrossRef]
- de Jonge, R.; van Esse, H.P.; Kombrink, A.; Shinya, T.; Desaki, Y.; Bours, R.; van Der Krol, S.; Shibuya, N.; Joosten, M.H.A.J.; Thomma, B.P. Conserved Fungal LysM Effector Ecp6 Prevents Chitin-Triggered Immunity in Plants. Science 2010, 329, 953–955. [Google Scholar] [CrossRef]
- Marshall, R.; Kombrink, A.; Motteram, J.; Loza-Reyes, E.; Lucas, J.; Hammond-Kosack, K.E.; Thomma, B.P.; Rudd, J.J. Analysis of Two in Planta Expressed LysM Effector Homologs from the Fungus Mycosphaerella graminicola Reveals Novel Functional Properties and Varying Contributions to Virulence on Wheat. Plant Physiol. 2011, 156, 756–769. [Google Scholar] [CrossRef]
- Tian, H.; MacKenzie, C.I.; Rodriguez-Moreno, L.; van den Berg, G.C.; Chen, H.; Rudd, J.J.; Mesters, J.R.; Thomma, B.P. Three LysM effectors of Zymoseptoria tritici collectively disarm chitin-triggered plant immunity. Mol. Plant Pathol. 2021, 22, 683–693. [Google Scholar] [CrossRef]
- Lee, W.-S.; Rudd, J.J.; Hammond-Kosack, K.E.; Kanyuka, K. Mycosphaerella graminicola LysM Effector-Mediated Stealth Pathogenesis Subverts Recognition Through Both CERK1 and CEBiP Homologues in Wheat. Mol. Plant-Microbe Interact. 2014, 27, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Mentlak, T.A.; Kombrink, A.; Shinya, T.; Ryder, L.S.; Otomo, I.; Saitoh, H.; Terauchi, R.; Nishizawa, Y.; Shibuya, N.; Thomma, B.P.; et al. Effector-Mediated Suppression of Chitin-Triggered Immunity by Magnaporthe oryzae Is Necessary for Rice Blast Disease. Plant Cell 2012, 24, 322–335. [Google Scholar] [CrossRef] [PubMed]
- Takahara, H.; Hacquard, S.; KombrinK, A.; Hughes, H.B.; Halder, V.; Robin, G.P.; Hiruma, K.; Neumann, U.; Shinya, T.; Kombrink, E.; et al. Colletotrichum higginsianum extracellular LysM proteins play dual roles in appressorial function and suppression of chitin-triggered plant immunity. New Phytol. 2016, 211, 1323–1337. [Google Scholar] [CrossRef] [PubMed]
- Kombrink, A.; Rovenich, H.; Shi-Kunne, X.; Rojas-Padilla, E.; van den Berg, G.C.; Domazakis, E.; de Jonge, R.; Valkenburg, D.-J.; Sánchez-Vallet, A.; Seidl, M.F.; et al. Verticillium dahliae LysM effectors differentially contribute to virulence on plant hosts. Mol. Plant Pathol. 2017, 18, 596–608. [Google Scholar] [CrossRef] [PubMed]
- Dölfors, F.; Holmquist, L.; Dixelius, C.; Tzelepis, G. A LysM effector protein from the basidiomycete Rhizoctonia solani contributes to virulence through suppression of chitin-triggered immunity. Mol. Genet. Genom. 2019, 294, 1211–1218. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Liao, Z.; Feng, L.; An, B.; He, C.; Wang, Q.; Luo, H. Colletotrichum gloeosporioides Cg2LysM contributed to virulence toward rubber tree through affecting invasive structure and inhibiting chitin-triggered plant immunity. Front. Microbiol. 2023, 14, 1129101. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Qin, F.; Shen, B.; Shi, Q.; Liu, C.; Zhang, X.; Jiao, Y.; Lu, J.; Gao, Y.; Suarez-Fernandez, M.; et al. Genome and secretome analysis of Pochonia chlamydosporia provide new insight into egg-parasitic mechanisms. Sci. Rep. 2018, 8, 1123. [Google Scholar] [CrossRef]
- Lee, Y.Y.; Vidal-Diez de Ulzurrum, G.; Schwarz, E.M. Genome sequence of the oyster mushroom Pleurotua ostreatus strain PC9. G3 2021, 11, jkaa008. [Google Scholar] [CrossRef]
- Boontawon, T.; Nakazawa, T.; Inoue, C.; Osakabe, K.; Kawauchi, M.; Sakamoto, M.; Hond, Y. Efficient genome editing with CRISPR/Cas9 in Pleurotus ostreatus. AMB Expr. 2021, 11, 30. [Google Scholar] [CrossRef]
- Zhang, Y.; Qiao, M.; Xu, J.; Cao, Y.; Zhang, K.-Q.; Yu, Z.-F. Genetic diversity and recombination in natural populations of the nematode-trapping fungus Arthrobotrys oligospora from China. Ecol. Evol. 2013, 3, 312–325. [Google Scholar] [CrossRef] [PubMed]
- Cen, K.; Li, B.; Lu, Y.; Zhang, S.; Wang, C. Divergent LysM effectors contribute to the virulence of Beauveria bassiana by evasion of insect immune defenses. PLoS Pathog. 2017, 13, e1006604. [Google Scholar] [CrossRef]
- Sasan, R.K.; Bidochka, M.J. The insect-pathogenic fungus Metarhizium robertsii (Clavicipitaceae) is also an endophyte that stimulates plant root development. Am. J. Bot. 2012, 99, 101–107. [Google Scholar] [CrossRef]
- Zeng, T.; Rodriguez-Moreno, L.; Mansurkhodzaev, A.; Wang, P.; van den Berg, W.; Gasciolli, V.; Cottaz, S.; Fort, S.; Thomma, B.P.; Bono, J.-J.; et al. A lysin motif effector subverts chitin-triggered immunity to facilitate arbuscular mycorrhizal symbiosis. New Phytol. 2020, 225, 448–460. [Google Scholar] [CrossRef]
- Romero-Contreras, Y.J.; Ramírez-Valdespino, C.A.; Guzmán-Guzmán, P.; Macías-Segoviano, J.I.; Villagómez-Castro, J.C.; Olmedo-Monfil, V. Tal6 From Trichoderma atroviride Is a LysM Effector Involved in Mycoparasitism and Plant Association. Front. Microbiol. 2019, 10, 2231. [Google Scholar] [CrossRef]
- Seidl-Seiboth, V.; Zach, S.; Frischmann, A.; Spadiut, O.; Dietzsch, C.; Herwig, C.; Ruth, C.; Rodler, A.; Jungbauer, A.; Kubicek, C.P. Spore germination of Trichoderma atroviride is inhibited by its LysM protein TAL6. FEBS J. 2013, 280, 1226–1236. [Google Scholar] [CrossRef]
- Nahar, P.; Ghormade, V.; Deshpande, M.V. The extracellular constitutive production of chitin deacetylase in Metarhizium anisopliae: Possible edge to entomopathogenic fungi in the biological control of insect pests. J. Invertebr. Pathol. 2004, 85, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Hiramatsu, S.; Songsri, P.; Fujie, M. Alternative Expression of a Chitosanase Gene Produces Two Different Proteins in Cells Infected with Chlorella Virus CVK2. Virology 1997, 230, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, B.; Li, C.; Bao, X. Knock down of chitosanase expression in phytopathogenic fungus Fusarium solani and its effect on pathogenicity. Curr. Genet. 2010, 56, 275–281. [Google Scholar] [CrossRef] [PubMed]
- de Jonge, R.; Bolton, M.D.; Thomma, B.P. How filamentous pathogens co-opt plants: The ins and outs of fungal effectors. Curr. Opin. Plant Biol. 2011, 14, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Bharagava, R.N.; Chowdhary, P. Emerging and Eco-Friendly Approaches for Waste Management; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- McKinnon, A.C.; Saari, S.; Moran-Diez, M.E.; Meyling, N.V.; Raad, M.; Glare, T.R. Beauveria bassiana as an endophyte: A critical review on associated methodology and biocontrol potential. BioControl 2017, 62, 1–17. [Google Scholar] [CrossRef]
- Gust, A.A.; Willmann, R.; Desaki, Y.; Grabherr, H.M.; Nürnberger, T. Plant LysM proteins: Modules mediating symbiosis and immunity. Trends Plant Sci. 2012, 17, 495–502. [Google Scholar] [CrossRef]
- Zhao, S.; An, B.; Guo, Y.; Hou, X.; Luo, H.; He, C.; Wang, Q. Label free proteomics and systematic analysis of secretome reveals effector candidates regulated by SGE1 and FTF1 in the plant pathogen Fusarium oxysporum f. sp. cubense tropical race 4. BMC Genom. 2020, 21, 275. [Google Scholar] [CrossRef]
- Palma-Guerrero, J.; Gómez-Vidal, S.; Tikhonov, V.E.; Salinas, J.; Jansson, H.-B.; Lopez-Llorca, L.V. Comparative analysis of extracellular proteins from Pochonia chlamydosporia grown with chitosan or chitin as main carbon and nitrogen sources. Enzym. Microb. Technol. 2010, 46, 568–574. [Google Scholar] [CrossRef]
- Thadathil, N.; Velappan, S.P. Recent developments in chitosanase research and its biotechnological applications: A review. Food Chem. 2014, 150, 392–399. [Google Scholar] [CrossRef]
- Grenier, J.; Asselin, A. Some Pathogenesis-Related Proteins Are Chitosanases with Lytic Activity against Fungal Spores. Mol. Plant-Microbe Interact. 1990, 3, 401–407. [Google Scholar] [CrossRef]
- Hirano, Y.; Yamamoto, R.; Dannoura, M.; Aono, K.; Igarashi, T.; Ishii, M.; Yamase, K.; Makita, N.; Kanazawa, Y. Detection frequency of Pinus thunbergii roots by ground-penetrating radar is related to root biomass. Plant Soil 2012, 360, 363–373. [Google Scholar] [CrossRef]
- Sambles, C.; Suarez-Fernandez, M.; Lopez-Moya, F.; Lopez-Llorca, L.V.; Studholme, D.J. Chitosan induces differential transcript usage of chitosanase 3 encoding gene (csn3) in the biocontrol fungus Pochonia chlamydosporia 123. BMC Genom. 2022, 23, 101. [Google Scholar] [CrossRef]
- Larriba, E.; Martín-Nieto, J.; Lopez-Llorca, L.V. Gene cloning, molecular modeling, and phylogenetics of serine protease P32 and serine carboxypeptidase SCP1 from nematophagous fungi Pochonia rubescens and Pochonia chlamydosporia. Can. J. Microbiol. 2012, 58, 815–827. [Google Scholar] [CrossRef]
- Medie, F.M.; Davies, G.J.; Drancourt, M.; Henrissat, B. Genome analyses highlight the different biological roles of cellulases. Nat. Rev. Microbiol. 2012, 10, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Glass, N.L.; Schmoll, M.; Cate, J.H.; Coradetti, S. Plant cell wall deconstruction by ascomycete fungi. Annu. Rev. Microbiol. 2013, 67, 477–498. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. NAR 2014, 42, 490–495. [Google Scholar] [CrossRef]
- Moreno-García, J.; Martín-García, F.J.; Ogawa, M.; García-Martínez, T.; Moreno, J.; Mauricio, J.C.; Bisson, L.F. FLO1, FLO5 and FLO11 flocculation gene expression impacts Saccharomyces cerevisiae attachment to Penicillium chrysogenum in a co-immobilization technique. Front. Microbiol. 2018, 9, 2586. [Google Scholar] [CrossRef] [PubMed]
- Rosso, L.C.; Finetti-Sialer, M.M.; Hirsch, P.R.; Ciancio, A.; Kerry, B.R.; Clark, I.M. Transcriptome analysis shows differential gene expression in the saprotrophic to parasitic transition of Pochonia chlamydosporia. Appl. Microbiol. Biot. 2011, 90, 1981–1994. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Herrera, J. Fungal Cell Wall: Structure, Synthesis, and Assembly; CRC Press: Boca Raton, FL, USA, 1992. [Google Scholar]
- Brown, V.; Sabina, J.; Johnston, M. Specialized sugar sensing in diverse fungi. Curr. Biol. 2009, 19, 436–441. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Karaffa, L.; Fekete, E.; Gamauf, C.; Szentirmai, A.; Kubicek, C.P.; Seiboth, B. D-galactose induces cellulase gene expression in Hypocrea jecorina at low growth rates. Microbiology 2006, 152, 1507–1514. [Google Scholar] [CrossRef] [PubMed]
- Nordbring-Hertz, B.; Friman, E.; Mattiasson, B. A recognition mechanism in the adhesion of nematodes to nematode-trapping fungi. In Lectins-Biology, Biochemistry and Clinical Biochemistry; Bog-Hansen, T.C., Ed.; Walter de Gruyter: Berlin, Germany, 1982; Volume 2, pp. 83–90. [Google Scholar]
- Premachandran, D.; Pramer, D. Role of N-acetylgalactosamine specific protein in trapping of nematodes by Arthrobotrys oligospora. Appl. Environ. Microbiol. 1984, 47, 1358–1359. [Google Scholar] [CrossRef]
- Sharon, E.; Spiegel, Y. Glycoprotein characterization of the gelatinous matrix in the root-knot nematode Meloidogyne javanica. J. Nematol. 1993, 25, 585–589. [Google Scholar]
- Clarke, A.J. The composition of the cyst wall of the beet cyst-nematode Heterodera schachtii. Biochem. J. 1970, 118, 315–318. [Google Scholar] [CrossRef][Green Version]
- Forrest, J.M.S.; Robertson, W.M. Characterization and localization of saccharides on the head region of four populations of the potato cyst nematode Globodera rostochiensis and G. pallida. J. Nematol. 1986, 18, 23. [Google Scholar] [PubMed]
- Baker, K.; Cook, R.J. Biological Control of Plant Pathogens; W.H. Freeman and Company: San Francisco, CA, USA, 1974. [Google Scholar]
- Harman, G.E.; Howell, C.R.; Viterbo, A.; Chet, I.; Lorito, M. Trichoderma species opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2004, 2, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Motlagh, M.R.; Samimi, Z. Evaluation of Trichoderma spp., as biological agents in some of plant pathogens. Ann. Biol. Res. 2013, 4, 173–179. [Google Scholar]
- Quevedo, A.C.; Muniz, M.F.; Savian, L.G.; Sarzi, J.S.; Saldanha, M.A. Ação antagonista in vitro de Trichoderma spp. sobre Fusarium oxysporum. Ciência Florest. 2022, 32, 2288–2303. [Google Scholar] [CrossRef]
- Asad, S.A.; Ali, N.; Hameed, A.; Khan, S.A.; Ahmad, R.; Bilal, M.; Shahzad, M.; Tabassum, A. Biocontrol efficacy of different isolates of Trichoderma against soil borne pathogen Rhizoctonia solani. Pol. J. Microbiol. 2014, 63, 95–103. [Google Scholar] [CrossRef]
- Inamdar, A.A.; Morath, S.; Bennett, J.W. Fungal Volatile Organic Compounds: More Than Just a Funky Smell? Annu. Rev. Microbiol. 2020, 74, 101–116. [Google Scholar] [CrossRef]
- Lozano-Soria, A.; Picciotti, U.; Lopez-Moya, F.; Lopez-Cepero, J.; Porcelli, F.; Lopez-Llorca, L.V. Volatile Organic Compounds from Entomopathogenic and Nematophagous Fungi, Repel Banana Black Weevil (Cosmopolites sordidus). Insects 2020, 11, 509. [Google Scholar] [CrossRef]
- Benítez, T.; Rincón, A.M.; Limón, M.C.; Codón, A.C. Biocontrol mechanisms of Trichoderma strains. Int. Microbiol. 2004, 7, 249–260. [Google Scholar]
- Harman, G.E. Overview of Mechanisms and Uses of Trichoderma spp. Phytopathology 2006, 96, 190–194. [Google Scholar] [CrossRef]
- Contreras-Cornejo, H.A.; López-Bucio, J.S.; Méndez-Bravo, A.; Macías-Rodríguez, L.; Ramos-Vega, M.; Guevara-García, Á.A.; López-Bucio, J. Mitogen-Activated Protein Kinase 6 and Ethylene and Auxin Signaling Pathways Are Involved in Arabidopsis Root-System Architecture Alterations by Trichoderma atroviride. Mol. Plant-Microbe Interact. 2015, 28, 701–710. [Google Scholar] [CrossRef]
- Tjamos, E.C. Selective Elimination of Soilborne Plant Pathogens and Enhancement of Antagonists by Steaming, Sublethal Fumigation and Soil Solarization. In Biological Control of Plant Diseases; Tjamos, E.C., Papavizas, G.C., Cook, R.J., Eds.; NATO ASI Series; Springer: Boston, MA, USA, 1992; Volume 230. [Google Scholar]
- Constantin, M.E.; de Lamo, F.J.; Vlieger, B.V.; Rep, M.; Takken, F.L. Endophyte-Mediated Resistance in Tomato to Fusarium oxysporum Is Independent of ET, JA, and SA. Front. Plant Sci. 2019, 10, 979. [Google Scholar] [CrossRef]
- Afshan, N.-U.-S. Recent Advancement in Fungal Biocontrol Agents. In Plant Mycobiome; Rashad, Y.M., Baka, Z.A., Moussa, T.A., Eds.; Springer: Cham, Switzerland, 2023; pp. 203–223. [Google Scholar]
- Uchida, M.; Konishi, T.; Fujigasaki, A.; Kita, K.; Arie, T.; Teraoka, T.; Kanda, Y.; Mori, M.; Arazoe, T.; Kamakura, T. Dysfunctional Pro1 leads to female sterility in rice blast fungi. iScience 2023, 26, 107020. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Xiao, J.; Dai, L.; Huang, Y.; Mao, Z.; Lin, R.; Yao, Y.; Xie, B. Development of a high-efficiency gene knockout system for Pochonia chlamydosporia. Microbiol. Res. 2014, 170, 18–26. [Google Scholar] [CrossRef]
- Li, D.; Tang, Y.; Lin, J.; Cai, W. Methods for genetic transformation of filamentous fungi. Microb. Cell Fact. 2017, 16, 168. [Google Scholar] [CrossRef] [PubMed]
- Lichius, A.; Ruiz, D.M.; Zeilinger, S. Genetic Transformation of Filamentous Fungi: Achievements and Challenges. In Grand Challenges in Fungal Biotechnology. Grand Challenges in Biology and Biotechnology; Nevalainen, H., Ed.; Springer: Cham, Switzerland, 2020. [Google Scholar]
- Zhang, M.; Jiang, S.; Zheng, J.; Zheng, Z.; Li, X.; Pan, L.; Luo, S. Construction of an integration vector carrying hygromycin B resistance gene and its genetic transformation in Rhizopus oryzae. Chin. J. Biotechnol. 2015, 31, 1203–1218. [Google Scholar]
- Nødvig, C.S.; Nielsen, J.B.; Kogle, M.E.; Mortensen, U.H. A CRISPR-Cas9 System for Genetic Engineering of Filamentous Fungi. PLoS ONE 2015, 10, e0133085. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.-Q.; Liu, G.-N.; Ji, R.-Y.; Shi, K.; Song, P.; Ren, L.-J.; Huang, H.; Ji, X.-J. CRISPR/Cas9-based genome editing of the filamentous fungi: The state of the art. Appl. Microbiol. Biotechnol. 2017, 101, 7435–7443. [Google Scholar] [CrossRef]
- Matsu-ura, T.; Baek, M.; Kwon, J.; Hong, C. Efficient gene editing in Neurospora crassa with CRISPR technology. Fungal Biol. Biotechnol. 2015, 2, 4. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Chen, L.; Jiang, Y.; Zhou, Z.; Zou, G. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov. 2015, 1, 15007. [Google Scholar] [CrossRef] [PubMed]
- Astudillo-Echeverría, A.; Pazmiño-Centeno, D.; Naranjo-Briceño, L. Uso de CRISPR/Cas9 como herramienta de edición de genomas en hongos filamentosos: Una revisión del estado actual y últimas tendencias. Genética Médica Y Genómica 2021, 5. [Google Scholar]
- Wang, Q.; Coleman, J.J. Progress and Challenges: Development and Implementation of CRISPR/Cas9 Technology in Filamentous Fungi. Comput. Struct. Biotechnol. J. 2019, 17, 761–769. [Google Scholar] [CrossRef]
- Al Abdallah, Q.; Ge, W.; Fortwendel, J.R. A simple and universal system for gene manipulation in Aspergillus fumigatus: In vitro-assembled Cas9-guide RNA ribonucleoproteins coupled with microhomology repair templates. Msphere 2017, 2, e00446-17. [Google Scholar] [CrossRef]
- Ferrara, M.; Haidukowski, M.; Logrieco, A.F.; Leslie, J.F.; Mulè, G. A CRISPR-Cas9 System for Genome Editing of Fusarium proliferatum. Sci. Rep. 2019, 9, 19836. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).