Chytrids in Soil Environments: Unique Adaptations and Distributions
Abstract
1. Introduction
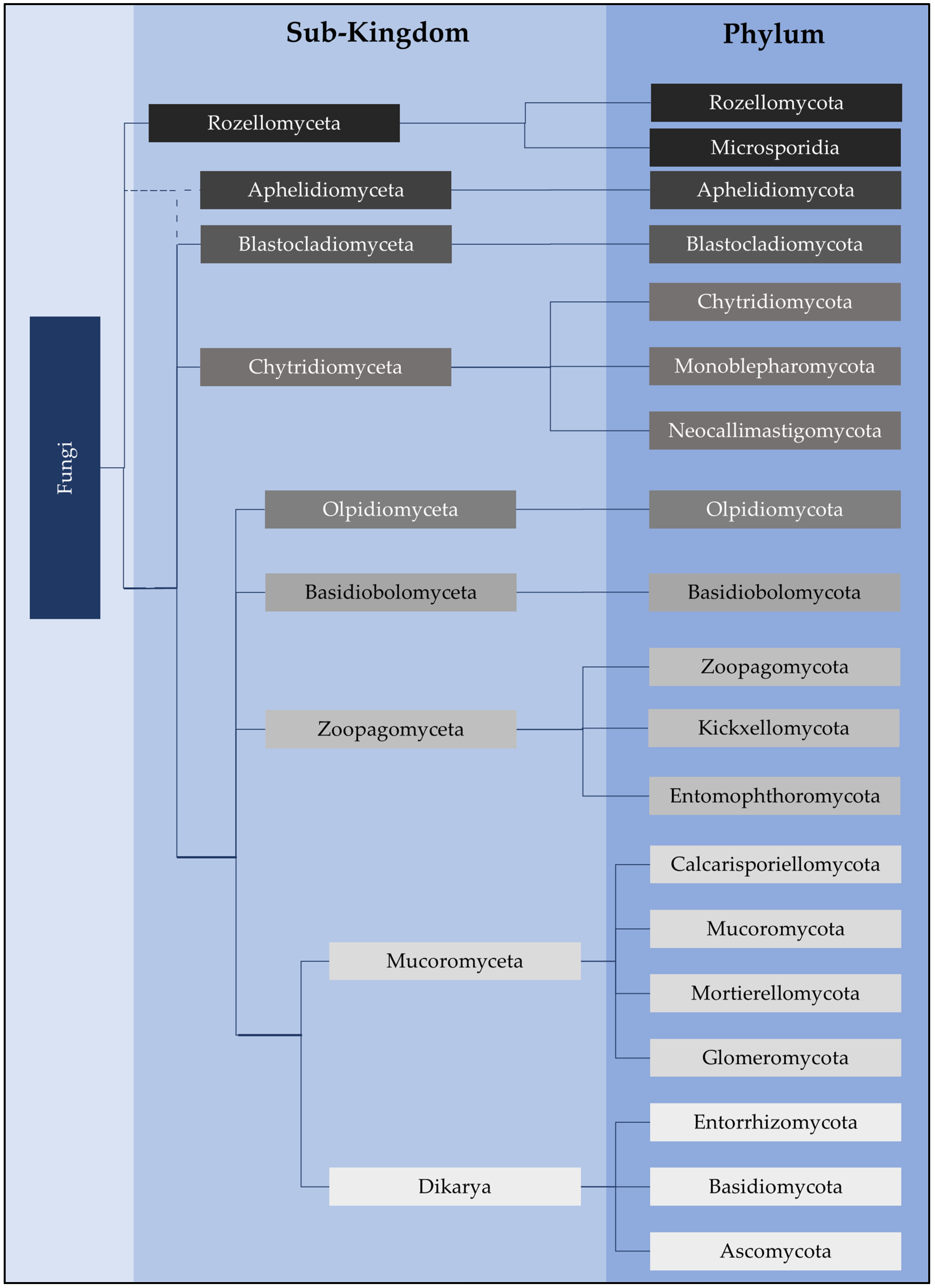
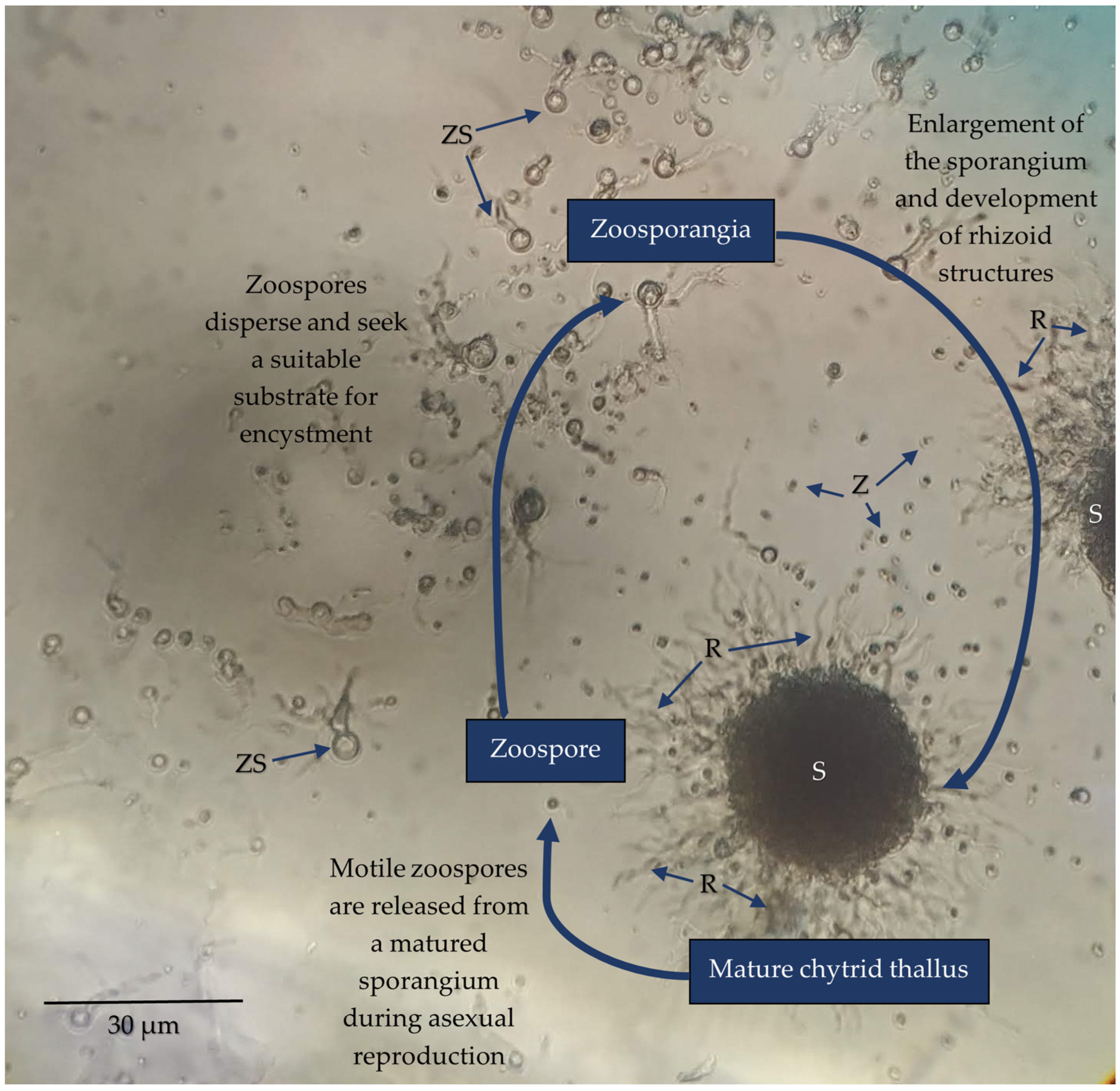
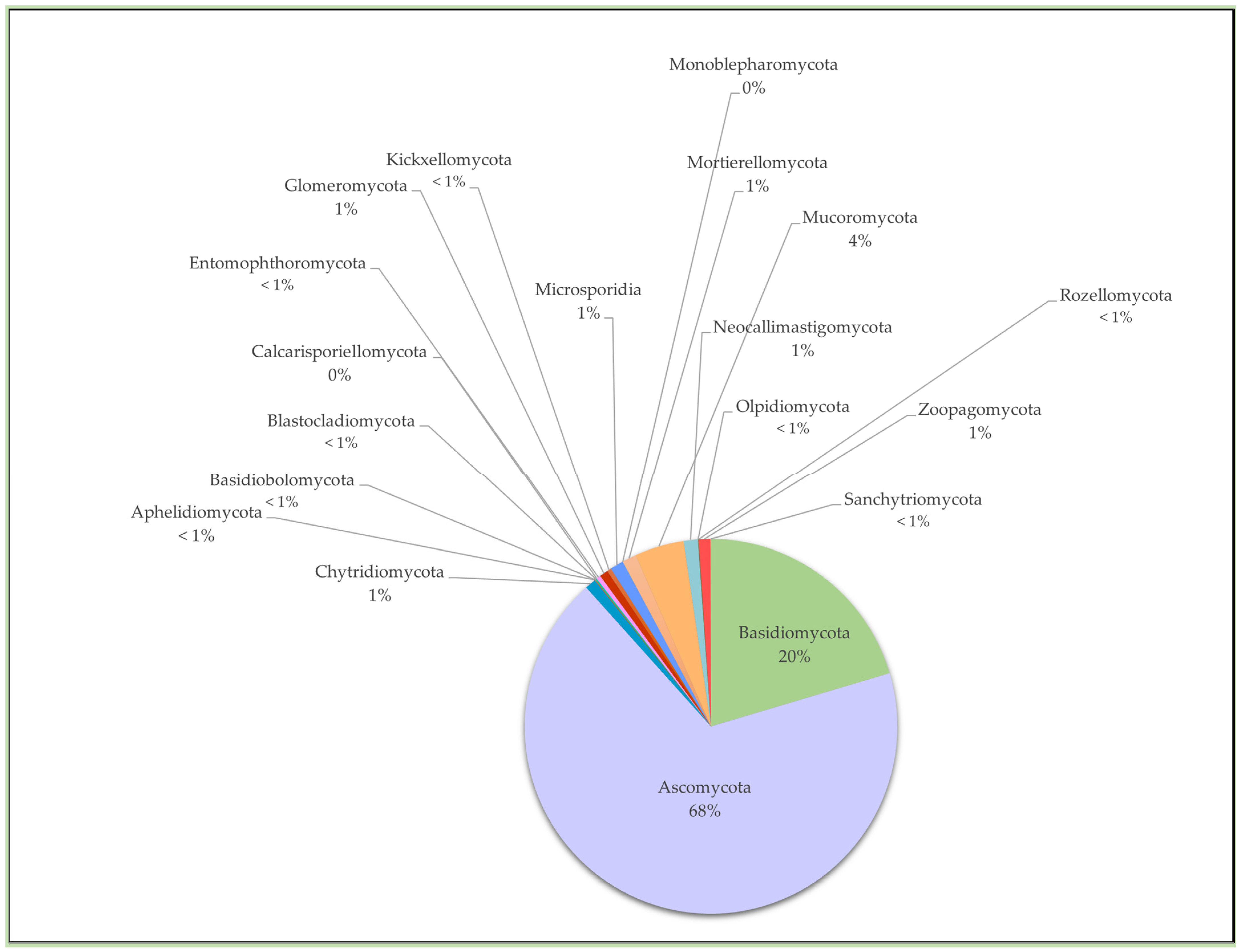
2. Chytrid Presence and Diversity in Soils
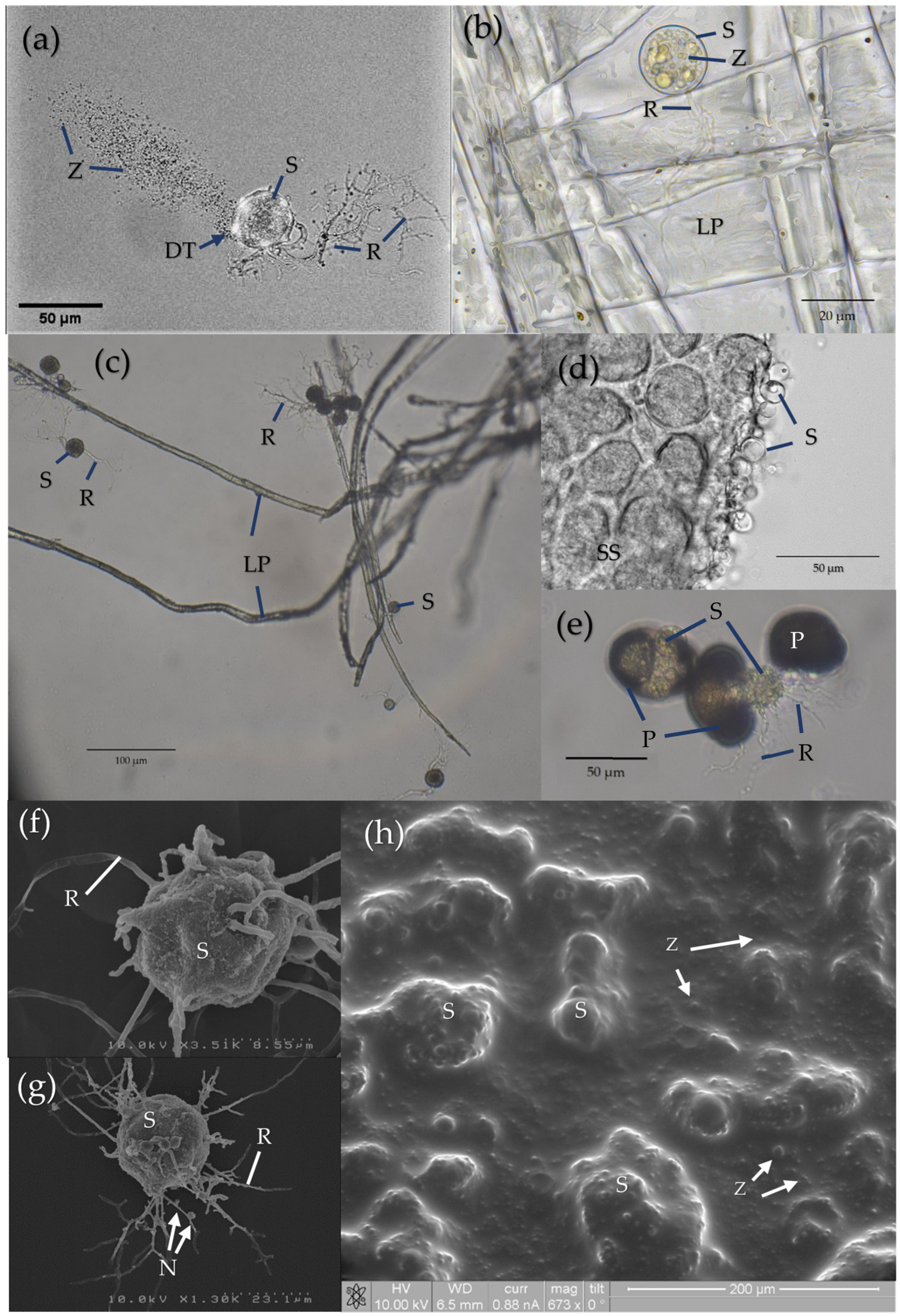
3. The Soil Environment at the Microscale: Chytrid Distribution and Adaptations
4. Chytrid Mechanisms and Adaptations to Extreme Conditions and Environmental Gradients in Soil
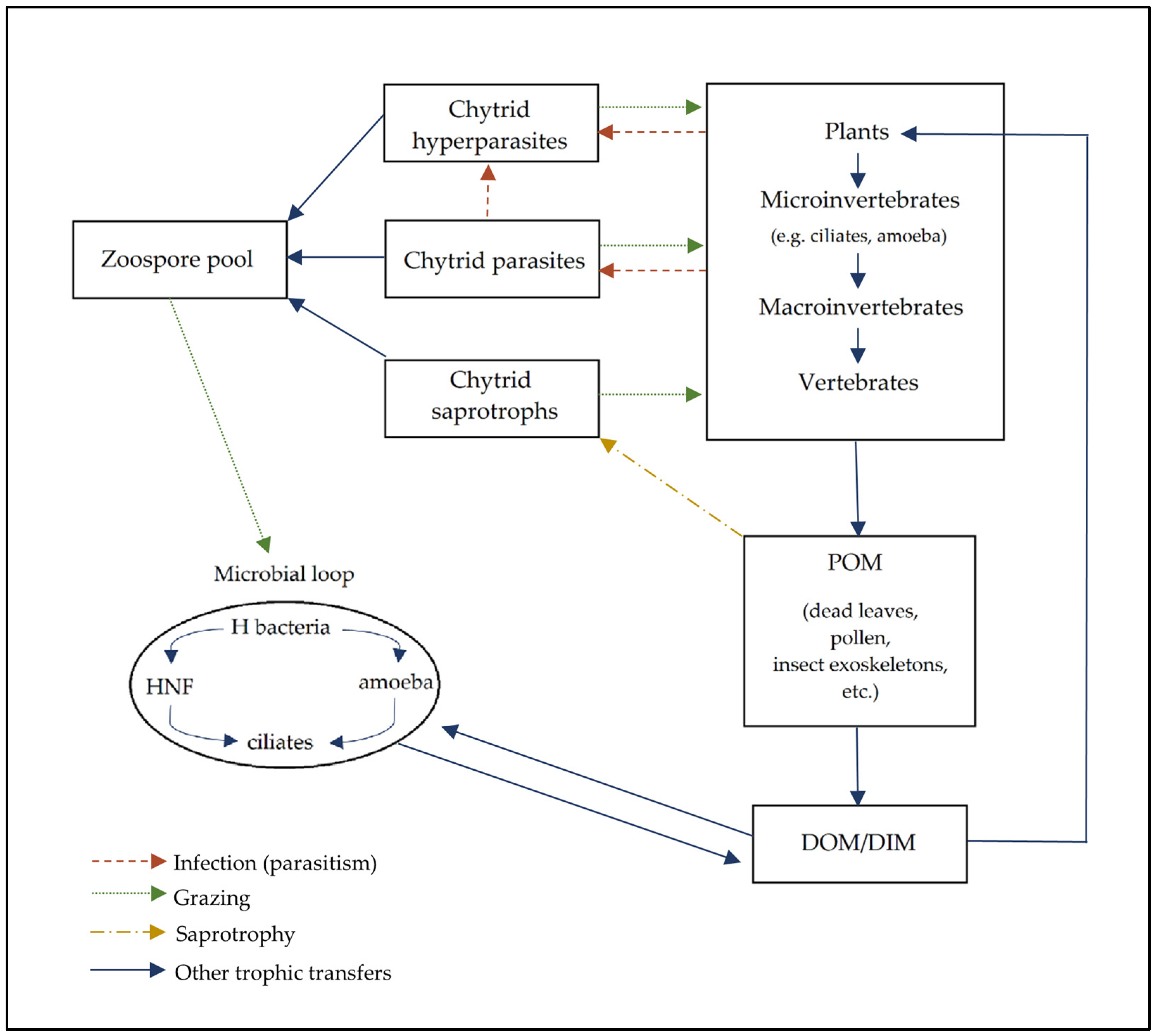
5. Chytrid Roles in Soil Communities
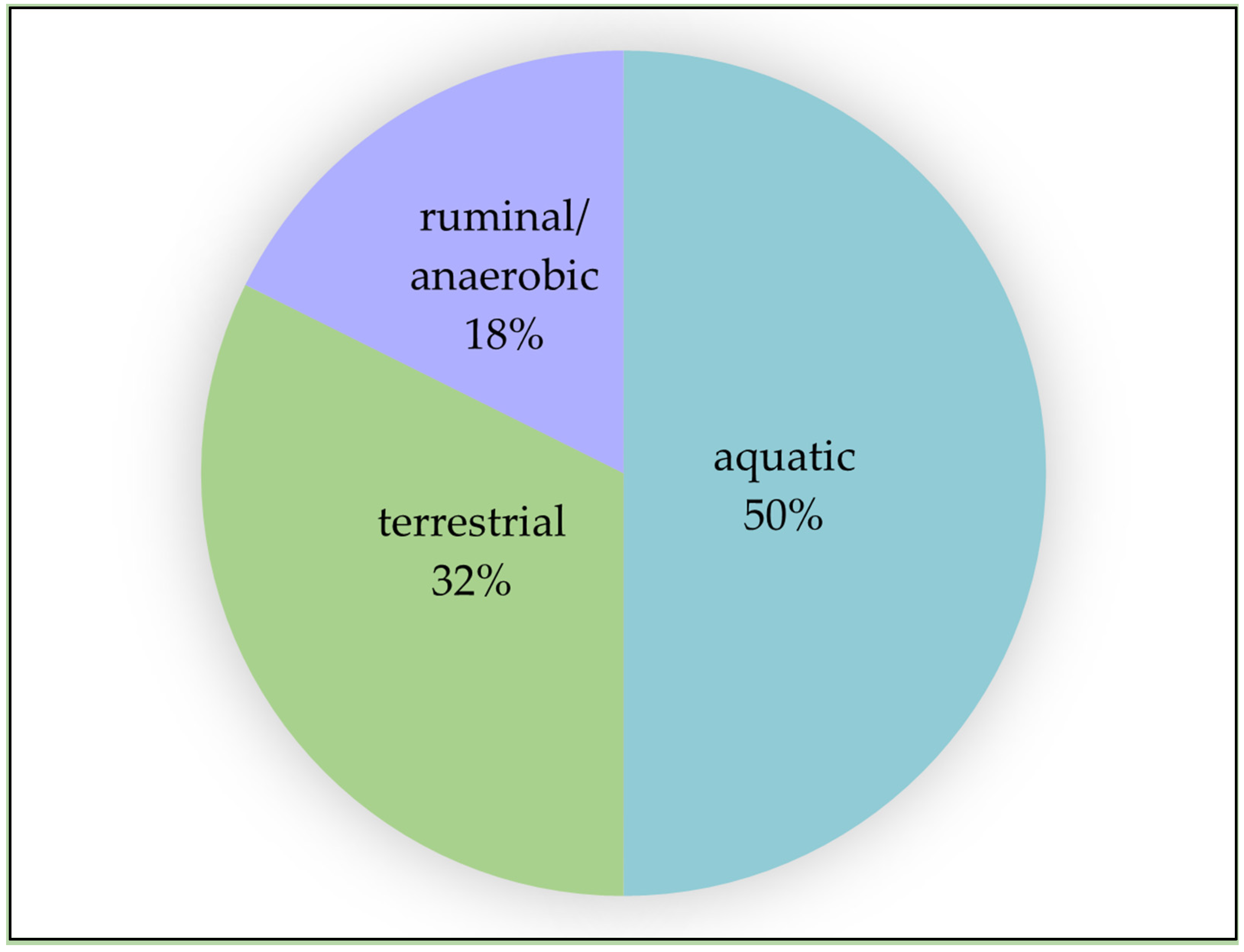
6. Further Research
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sparrow, F.K. Aquatic Phycomycetes, 2nd ed.; University of Michigan Press: Ann Arbor, MI, USA, 1960. [Google Scholar]
- Willoughby, L.G. The Ecology of some lower fungi at Esthwaite water. Trans. Br. Mycol. Soc. 1961, 44, 305–332, IN1–IN2. [Google Scholar] [CrossRef]
- Kagami, M.; de Bruin, A.; Ibelings, B.W.; Van Donk, E. Parasitic chytrids: Their effects on phytoplankton communities and food-web dynamics. Hydrobiologia 2007, 578, 113–129. [Google Scholar] [CrossRef]
- Naranjo-Ortiz, M.A.; Gabaldón, T. Fungal evolution: Diversity, taxonomy and phylogeny of the Fungi. Biol. Rev. Camb. Philos. Soc. 2019, 94, 2101–2137. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Puusepp, R.; Nilsson, R.H.; James, T. Novel soil-inhabiting clades fill gaps in the fungal tree of life. Microbiome 2017, 5, 42. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Steenwyk, J.L.; Chang, Y.; Wang, Y.; James, T.Y.; Stajich, J.E.; Spatafora, J.W.; Groenewald, M.; Dunn, C.W.; Hittinger, C.T.; et al. A genome-scale phylogeny of the kingdom Fungi. Curr. Biol. 2021, 31, 1653–1665.e5. [Google Scholar] [CrossRef] [PubMed]
- James, T.Y.; Stajich, J.E.; Hittinger, C.T.; Rokas, A. Toward a Fully Resolved Fungal Tree of Life. Annu. Rev. Microbiol. 2020, 74, 291–313. [Google Scholar] [CrossRef] [PubMed]
- Baldauf, S.L. The Deep Roots of Eukaryotes. Sci. Am. Assoc. Adv. Sci. 2003, 300, 1703–1706. [Google Scholar] [CrossRef]
- Baldauf, S.L. An overview of the phylogeny and diversity of eukaryotes. J. Syst. Evol. 2008, 46, 263–273. [Google Scholar]
- Ruggiero, M.A.; Gordon, D.P.; Orrell, T.M.; Bailly, N.; Bourgoin, T.; Brusca, R.C.; Cavalier-Smith, T.; Guiry, M.D.; Kirk, P.M. A higher level classification of all living organisms. PLoS ONE 2015, 10, e0119248. [Google Scholar] [CrossRef]
- Tedersoo, L.; Sánchez-Ramírez, S.; Kõljalg, U.; Bahram, M.; Döring, M.; Schigel, D.; May, T.; Ryberg, M.; Abarenkov, K. High-level classification of the Fungi and a tool for evolutionary ecological analyses. Fungal Div. 2018, 90, 135–159. [Google Scholar] [CrossRef]
- Powell, M.J.; Letcher, P.M. Chytridiomycota, Monoblepharidomycota, and Neocallimastigomycota. In The Mycota, 2nd ed.; Esser, K., McLaughlin, D.J., Spatafora, J.W., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; Volume 7, pp. 141–175. [Google Scholar] [CrossRef]
- Strassert, J.F.H.; Wurzbacher, C.; Hervé, V.; Antany, T.; Brune, A.; Radek, R. Long rDNA amplicon sequencing of insect-infecting nephridiophagids reveals their affiliation to the Chytridiomycota and a potential to switch between hosts. Sci. Rep. 2021, 11, 396. [Google Scholar] [CrossRef] [PubMed]
- Hibbett, D.S.; Binder, M.; Bischoff, J.F.; Blackwell, M.; Cannon, P.F.; Eriksson, O.E.; Huhndorf, S.; James, T.; Kirk, P.M.; Lücking, R.; et al. A higher-level phylogenetic classification of the Fungi. Mycol. Res. 2007, 111, 509–547. [Google Scholar] [CrossRef] [PubMed]
- James, T.Y.; Letcher, P.M.; Longcore, J.E.; Mozley-Standridge, S.E.; Porter, D.; Powell, M.J.; Griffith, G.W.; Vilgalys, R. A molecular phylogeny of the flagellated fungi (Chytridiomycota) and description of a new phylum (Blastocladiomycota). Mycologia 2006, 98, 860–871. [Google Scholar] [CrossRef] [PubMed]
- Powell, M.J. Chytridiomycota. In Handbook of the Protists, 2nd ed.; Archibald, J., Simpson, A., Slamovits, C., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 1523–1558. [Google Scholar] [CrossRef]
- Porter, T.M.; Martin, W.; James, T.Y.; Longcore, J.E.; Gleason, F.H.; Adler, P.H.; Letcher, P.M.; Vilgalys, R. Molecular phylogeny of the Blastocladiomycota (Fungi) based on nuclear ribosomal DNA. Fungal Biol. 2011, 115, 381–392. [Google Scholar] [CrossRef] [PubMed]
- James, T.Y.; Kauff, F.; Schoch, C.L.; Matheny, P.B.; Hofstetter, V.; Cox, C.J.; Celio, G.; Gueidan, C.; Fraker, E.; Miadlikowska, J. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 2006, 443, 818–822. [Google Scholar] [CrossRef]
- Gleason, F.H.; Crawford, J.W.; Neuhauser, S.; Henderson, L.E.; Lilje, O. Resource seeking strategies of zoosporic true fungi in heterogeneous soil habitats at the microscale level. Soil Biol. Biochem. 2012, 45, 79–88. [Google Scholar] [CrossRef]
- Held, A.A. Attraction and attachment of zoospores of the parasitic chytrid Rozella allomycis in response to host-dependent factors. Arch. Microbiol. 1974, 95, 97–114. [Google Scholar] [CrossRef]
- Mitchell, R.T.; Deacon, J.W. Selective accumulation of zoospores of chytridiomycetes and oomycetes on cellulose and chitin. Trans. Br. Mycol. Soc. 1986, 86, 219–223. [Google Scholar] [CrossRef]
- Gleason, F.H.; Lilje, O. Structure and function of fungal zoospores: Ecological implications. Fungal Ecol. 2009, 2, 53–59. [Google Scholar] [CrossRef]
- Weete, J.D.; Laseter, J.L. Distribution of sterols in the fungi I. Fungal spores. Lipids 1974, 9, 575–581. [Google Scholar] [CrossRef]
- Beakes, G.W.; Canter, H.M.; Jaworski, G.H.M. Comparative ultrastructural ontogeny of zoosporangia of Zygorhizidium affluens and Z. planktonicum, chytrid parasites of the diatom Asterionella formosa. Mycol. Res. 1992, 96, 1047–1059. [Google Scholar] [CrossRef]
- Barr, D.J.S. Phlyctochytrium reinboldtae (Chytridiales): Morphology and physiology. Can. J. Bot. 1970, 48, 479–484. [Google Scholar] [CrossRef]
- Scheele, B.C.; Skerratt, L.F.; Grogan, L.F.; Hunter, D.A.; Clemann, N.; McFadden, M.; Newell, D.; Hoskin, C.J.; Gillespie, G.R.; Heard, G.W. After the epidemic: Ongoing declines, stabilizations and recoveries in amphibians afflicted by chytridiomycosis. Biol. Conserv. 2017, 206, 37–46. [Google Scholar] [CrossRef]
- Scheele, B.C.; Pasmans, F.; Skerratt, L.F.; Berger, L.; Martel, A.; Beukema, W.; Acevedo, A.A.; Burrowes, P.A.; Carvalho, T.; Catenazzi, A.; et al. Amphibian fungal panzootic causes catastrophic and ongoing loss of biodiversity. Science 2019, 363, 1459–1463. [Google Scholar] [CrossRef] [PubMed]
- Tessa, G.; Sotgiu, G.; Bovero, S.; Angelini, C.; Favelli, M.; Gazzaniga, E.; Giacoma, C.; Garner, T.W.J. Cryptic but direct costs of an epidemic caused by Batrachochytrium dendrobatidis in the endangered Sardinian newt Euproctus platycephalus (Amphibia, Caudata). Amphibia-Reptilia 2023, 44, 83–94. [Google Scholar] [CrossRef]
- O’Hanlon, S.J.; Rieux, A.; Farrer, R.A.; Rosa, G.M.; Waldman, B.; Bataille, A.; Kosch, T.A.; Murray, K.; Brankovics, B.; Fumagalli, M.; et al. A 20th Century Out-of-Asia Origin of a panzootic threat to global amphibian biodiversity. Science 2018, 360, 621–627. [Google Scholar] [CrossRef] [PubMed]
- Weldon, C.; Channing, A.; Misinzo, G.; Cunningham, A.A. Disease driven extinction in the wild of the Kihansi spray toad, Nectophrynoides Asperginis. Afr. J. Herpetol. 2020, 69, 151–164. [Google Scholar] [CrossRef]
- Klawonn, I.; Van den Wyngaert, S.; Iversen, M.H.; Walles, T.J.W.; Flintrop, C.M.; Cisternas-Novoa, C.; Nejstgaard, J.C.; Kagami, M.; Grossart, H.-P. Fungal parasitism on diatoms alters formation and bio–physical properties of sinking aggregates. Commun. Biol. 2023, 6, 206. [Google Scholar] [CrossRef]
- Kagami, M.; Gurung, T.; Yoshida, T.; Urabe, J. To sink or to be lysed? Contrasting fate of two large phytoplankton species in Lake Biwa. Limnol. Oceanogr. 2006, 51, 2775–2786. [Google Scholar] [CrossRef]
- Gerphagnon, M.; Colombet, J.; Latour, D.; Sime-Ngando, T. Spatial and temporal changes of parasitic chytrids of cyanobacteria. Sci. Rep. 2017, 7, 6056. [Google Scholar] [CrossRef]
- Ibelings, B.W.; Gsell, A.S.; Mooij, W.M.; Van Donk, E.; Van Den Wyngaert, S.; De Senerpont Domis, L.N. Chytrid infections and diatom spring blooms: Paradoxical effects of climate warming on fungal epidemics in lakes. Freshw. Biol. 2011, 56, 754–766. [Google Scholar] [CrossRef]
- Van den Wyngaert, S.; Ganzert, L.; Seto, K.; Rojas-Jimenez, K.; Agha, R.; Berger, S.A.; Woodhouse, J.; Padisak, J.; Wurzbacher, C.; Kagami, M.; et al. Seasonality of parasitic and saprotrophic zoosporic fungi: Linking sequence data to ecological traits. ISME J. 2022, 16, 2242–2254. [Google Scholar] [CrossRef] [PubMed]
- Gsell, A.S.; Wolinska, J.; Preuß, K.; Teurlincx, S.; Özkundakci, D.; Hilt, S.; van Donk, E.; Ibelings, B.W.; Adrian, R. Long-term trends and seasonal variation in host density, temperature, and nutrients differentially affect chytrid fungi parasitising lake phytoplankton. Freshw. Biol. 2022, 67, 1532–1542. [Google Scholar] [CrossRef]
- Lepelletier, F.; Karpov, S.A.; Alacid, E.; Le Panse, S.; Bigeard, E.; Garcés, E.; Jeanthon, C.; Guillou, L. Dinomyces arenysensis gen. et sp. nov. (Rhizophydiales, Dinomycetaceae fam. nov.), a Chytrid Infecting Marine Dinoflagellates. Protist 2014, 165, 230–244. [Google Scholar] [CrossRef] [PubMed]
- Hanrahan-Tan, D.G.; Henderson, L.; Kertesz, M.A.; Lilje, O. The Effects of Nitrogen and Phosphorus on Colony Growth and Zoospore Characteristics of Soil Chytridiomycota. J. Fungi 2022, 8, 341. [Google Scholar] [CrossRef] [PubMed]
- Henderson, L.; Marano, A.V.; Truszewski, E.; Gleason, F.H.; Lilje, O. Copper (II), lead (ll) and zinc (ll) reduce the rate of attachment in three zoosporic true fungi from soils of NSW, Australia. Nova Hedwig. 2019, 108, 435–447. [Google Scholar] [CrossRef]
- Henderson, L.E.; Lilje, E.; Robinson, K.; Gleason, F.H.; Lilje, O. Effects of Toxic Metals on Chytrids, Fungal-Like Organisms, and Higher Fungi. In The Fungal Community: Its organisation and Role in the Ecosystem, 4th ed.; Dighton, J., White, J.F., Eds.; CRC Press: Boca Raton, FL, USA, 2017; Volume 32, pp. 487–512. [Google Scholar]
- Freeman, K.R.; Martin, A.P.; Karki, D.; Lynch, R.C.; Mitter, M.S.; Meyer, A.F.; Longcore, J.E.; Simmons, D.R.; Schmidt, S.K. Evidence That Chytrids Dominate Fungal Communities in High-Elevation Soils. Proc. Natl. Acad. Sci. USA 2009, 106, 18315–18320. [Google Scholar] [CrossRef]
- Liu, H.; Cheng, J.; Jin, H.; Xu, Z.; Yang, X.; Min, D.; Xu, X.; Shao, X.; Lu, D.; Qin, B. Characterization of Rhizosphere and Endophytic Microbial Communities Associated with Stipa purpurea and Their Correlation with Soil Environmental Factors. Plants 2022, 11, 363. [Google Scholar] [CrossRef]
- Khan, S.; Chen, N.; Zhang, C.; Wang, L.; Han, C.; Lu, K.; Li, Y.; Rafiq, M.; Iqbal, A.; Zhao, C. Soil fungal taxonomic diversity along an elevation gradient on the semi-arid Xinglong Mountain, Northwest China. Arch. Microbiol. 2020, 202, 2291–2302. [Google Scholar] [CrossRef]
- Shi, Y.; Xiang, X.; Shen, C.; Chu, H.; Neufeld, J.D.; Walker, V.K.; Grogan, P. Vegetation-associated impacts on arctic tundra bacterial and microeukaryotic communities. Appl. Environ. Microbiol. 2015, 81, 492–501. [Google Scholar] [CrossRef]
- Botnen, S.S.; Thoen, E.; Eidesen, P.B.; Krabberød, A.K.; Kauserud, H. Community composition of arctic root-associated fungi mirrors host plant phylogeny. FEMS Microbiol. Ecol. 2020, 96, 1. [Google Scholar] [CrossRef] [PubMed]
- An, Z.; Guo, F.; Chen, Y.; Bai, G.; Chen, Z. Rhizosphere bacterial and fungal communities during the growth of Angelica sinensis seedlings cultivated in an Alpine uncultivated meadow soil. PeerJ 2020, 2020, e8541. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, J.; Zhou, Y.; Huang, Y.; Tang, X. Comparative Analysis on Rhizosphere Soil and Endophytic Microbial Communities of Two Cultivars of Cyperus esculentus L. Var. Sativus. J. Soil Sci. Plant Nutr. 2022, 22, 2156–2168. [Google Scholar] [CrossRef]
- Sun, R.; Yi, Z.; Fu, Y.; Liu, H. Dynamic changes in rhizosphere fungi in different developmental stages of wheat in a confined and isolated environment. Appl. Microbiol. Biotechnol. 2022, 106, 441–453. [Google Scholar] [CrossRef] [PubMed]
- Gleason, F.H.; Midgley, D.J.; Letcher, P.M.; McGee, P.A. Can soil Chytridiomycota survive and grow in different osmotic potentials? Mycol. Res. 2006, 110, 869–875. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, D.J.; Martiny, J.B.H. Patterns of fungal diversity and composition along a salinity gradient. ISME J. 2011, 5, 379–388. [Google Scholar] [CrossRef]
- Wang, J.; Huang, R.; Zhu, L.; Guan, H.; Lin, L.; Fang, H.; Yang, M.; Ji, S.; Zou, X.; Li, X. The Effects of Biochar on Microbial Community Composition in and Beneath Biological Soil Crusts in a Pinus massoniana Lamb. Plantation. Forests 2022, 13, 1141. [Google Scholar] [CrossRef]
- Abed, R.M.M.; Tamm, A.; Hassenrück, C.; Al-Rawahi, A.N.; Rodríguez-Caballero, E.; Fiedler, S.; Maier, S.; Weber, B. Habitat-dependent composition of bacterial and fungal communities in biological soil crusts from Oman. Sci. Rep. 2019, 9, 6468. [Google Scholar] [CrossRef]
- García-Carmona, M.; Lepinay, C.; García-Orenes, F.; Baldrian, P.; Arcenegui, V.; Cajthaml, T.; Mataix-Solera, J. Moss biocrust accelerates the recovery and resilience of soil microbial communities in fire-affected semi-arid Mediterranean soils. Sci. Total Environ. 2022, 846, 157467. [Google Scholar] [CrossRef]
- Gleason, F.H.; Letcher, P.M.; Commandeur, Z.; Jeong, C.E.; McGee, P.A. The growth response of some Chytridiomycota to temperatures commonly observed in the soil. Mycol. Res. 2005, 109, 717–722. [Google Scholar] [CrossRef]
- Letcher, P.M.; McGee, P.A.; Powell, M.J. Distribution and diversity of zoosporic fungi from soils of four vegetation types in New South Wales, Australia. Can. J. Bot. 2004, 82, 1490–1500. [Google Scholar] [CrossRef]
- Marčiulynas, A.; Marčiulynienė, D.; Mishcherikova, V.; Franić, I.; Lynikienė, J.; Gedminas, A.; Menkis, A. High Variability of Fungal Communities Associated with the Functional Tissues and Rhizosphere Soil of Picea abies in the Southern Baltics. Forests 2022, 13, 1103. [Google Scholar] [CrossRef]
- Bai, Z.; Wu, X.; Lin, J.-J.; Xie, H.-T.; Yuan, H.-S.; Liang, C. Litter-, soil- and C:N-stoichiometry-associated shifts in fungal communities along a subtropical forest succession. Catena Giess. 2019, 178, 350–358. [Google Scholar] [CrossRef]
- Yu, P.; Ning, C.; Chen, J.; Zhu, F.; Sun, Y.; Shen, A.; Zeng, W.; Jiang, L. The Effects of Suillus luteus Inoculation on the Diversity of Fungal Communities and Their Structures in the Soil under Pinus massoniana Located in a Mining Area. Forests 2022, 13, 2162. [Google Scholar] [CrossRef]
- Longcore, J.E. Zoosporic fungi from Australian and New Zealand tree-canopy detritus. Aust. J. Bot. 2005, 53, 259–272. [Google Scholar] [CrossRef]
- Letcher, P.M.; Longcore, J.E.; Powell, M.J. Dendrochytridium crassum gen. et sp. nov., a taxon in Chytridiales with unique zoospore ultrastructure. Mycologia 2014, 106, 145–153. [Google Scholar] [CrossRef]
- Orlovich, D.A.; Draffin, S.J.; Daly, R.A.; Stephenson, S.L. Piracy in the high trees: Ectomycorrhizal fungi from an aerial ‘canopy soil’ microhabitat. Mycologia 2013, 105, 52–60. [Google Scholar] [CrossRef]
- van Elsas, J.D.; Boersma, F.G.H. A review of molecular methods to study the microbiota of soil and the mycosphere. Eur. J. Soil Biol. 2011, 47, 77–87. [Google Scholar] [CrossRef]
- Joos, L.; Beirinckx, S.; Haegeman, A.; Debode, J.; Vandecasteele, B.; Baeyen, S.; Goormachtig, S.; Clement, L.; De Tender, C. Daring to be differential: Metabarcoding analysis of soil and plant-related microbial communities using amplicon sequence variants and operational taxonomical units. BMC Genom. 2020, 21, 733. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Polme, S.; Koljalg, U.; Yorou, N.S.; Wijesundera, R.; Villarreal Ruiz, L.; Vasco, A.; Pham Quang, T.; Suija, A.; et al. Global diversity and geography of soil fungi. Sci. Am. Assoc. Adv. Sci. 2014, 346, 1078. [Google Scholar] [CrossRef]
- Leff, J.W.; Jones, S.E.; Prober, S.M.; Barberán, A.; Borer, E.T.; Firn, J.L.; Harpole, W.S.; Hobbie, S.E.; Hofmockel, K.S.; Knops, J.M.H.; et al. Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proc. Natl. Acad. Sci. USA 2015, 112, 10967–10972. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.; Mu, M.; Xie, H.; Wu, Y.; Zhou, Y.; Li, W. Rhizospheric fungal diversities and soil biochemical factors of Fritillaria taipaiensis over five cultivation years. Horticulturae 2021, 7, 560. [Google Scholar] [CrossRef]
- Hannula, S.E.; Di Lonardo, D.P.; Christensen, B.T.; Crotty, F.V.; Elsen, A.; Erp, P.J.; Hansen, E.M.; Rubæk, G.H.; Tits, M.; Toth, Z.; et al. Inconsistent effects of agricultural practices on soil fungal communities across 12 European long-term experiments. Eur. J. Soil Sci. 2021, 72, 1902–1923. [Google Scholar] [CrossRef]
- Moussa, T.A.A.; Al-Zahrani, H.S.; Almaghrabi, O.A.; Abdelmoneim, T.S.; Fuller, M.P. Comparative metagenomics approaches to characterize the soil fungal communities of western coastal region, Saudi Arabia. PLoS ONE 2017, 12, e0185096. [Google Scholar] [CrossRef]
- Procter, A.C.; Ellis, J.C.; Fay, P.A.; Polley, H.W.; Jackson, R.B. Fungal community responses to past and future atmospheric CO2 differ by soil type. Appl. Environ. Microbiol. 2014, 80, 7364–7377. [Google Scholar] [CrossRef]
- Beng, K.C.; Wolinska, J.; Funke, E.; Van den Wyngaert, S.; Gsell, A.S.; Monaghan, M.T. Temporal dynamics of freshwater planktonic parasites inferred using a DNA metabarcoding time-series. Parasitology 2021, 148, 1602–1611. [Google Scholar] [CrossRef] [PubMed]
- Comeau, A.M.; Vincent, W.F.; Bernier, L.; Lovejoy, C. Novel chytrid lineages dominate fungal sequences in diverse marine and freshwater habitats. Sci. Rep. 2016, 6, 30120. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, B.D.; Nilsson, R.H.; Tedersoo, L.; Abarenkov, K.; Carlsen, T.; Kjøller, R.; Kõljalg, U.; Pennanen, T.; Rosendahl, S.; Stenlid, J.; et al. Fungal community analysis by high-throughput sequencing of amplified markers—A user’s guide. New Phytol. 2013, 199, 288–299. [Google Scholar] [CrossRef]
- Bellemain, E.; Carlsen, T.; Brochmann, C.; Coissac, E.; Taberlet, P.; Kauserud, H. ITS as an environmental DNA barcode for fungi: An in silico approach reveals potential PCR biases. BMC Microbiol. 2010, 10, 189. [Google Scholar] [CrossRef]
- Mondini, A.; Donhauser, J.; Itcus, C.; Marin, C.; Perșoiu, A.; Lavin, P.; Frey, B.; Purcarea, C. High-throughput sequencing of fungal communities across the perennial ice block of Scărișoara Ice Cave. Ann. Glaciol. 2018, 59, 134–146. [Google Scholar] [CrossRef]
- Mondini, A.; Anwar, M.Z.; Anwar, M.Z.; Ellegaard-Jensen, L.; Lavin, P.; Lavin, P.; Jacobsen, C.S.; Purcarea, C. Heat Shock Response of the Active Microbiome From Perennial Cave Ice. Front. Microbiol. 2022, 12, 809076. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Anslan, S.; Bahram, M.; Kõljalg, U.; Abarenkov, K. Identifying the ‘unidentified’ fungi: A global-scale long-read third-generation sequencing approach. Fungal Div. 2020, 103, 273–293. [Google Scholar] [CrossRef]
- Blaalid, R.; Khomich, M. Current knowledge of Chytridiomycota diversity in Northern Europe and future research needs. Fungal Biol. Rev. 2021, 36, 42–51. [Google Scholar] [CrossRef]
- PlutoF. GLOBAL Soil Organisms Occurrence Dataset. Available online: https://www.gbif.org/dataset/9f0e1ca6-fb08-4c72-9a4a-1e3b7a528c10/metrics (accessed on 27 January 2023).
- Voigt, K.; James, T.Y.; Kirk, P.M.; Santiago, A.L.C.M.d.A.; Waldman, B.; Griffith, G.W.; Fu, M.; Radek, R.; Strassert, J.F.H.; Wurzbacher, C.; et al. Early-diverging fungal phyla: Taxonomy, species concept, ecology, distribution, anthropogenic impact, and novel phylogenetic proposals. Fungal Divers. 2021, 109, 59–98. [Google Scholar] [CrossRef]
- Katra, I.; Arotsker, L.; Krasnov, H.; Zaritsky, A.; Kushmaro, A.; Ben-Dov, E. Richness and diversity in dust stormborne biomes at the southeast mediterranean. Sci. Rep. 2014, 4, 5265. [Google Scholar] [CrossRef]
- Gleason, F.H.; Letcher, P.M.; McGee, P.A. Some Chytridiomycota in soil recover from drying and high temperatures. Mycol. Res. 2004, 108, 583–589. [Google Scholar] [CrossRef]
- Ortega-Arbulú, A.S.; Pichler, M.; Vuillemin, A.; Orsi, W.D. Effects of organic matter and low oxygen on the mycobenthos in a coastal lagoon. Environ. Microbiol. 2019, 21, 374–388. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Grossart, H.-P.; Wurzbacher, C.; James, T.Y.; Kagami, M. Discovery of dark matter fungi in aquatic ecosystems demands a reappraisal of the phylogeny and ecology of zoosporic fungi. Fungal Ecol. 2016, 19, 28–38. [Google Scholar] [CrossRef]
- Penton, C.R.; Louis, D.S.; Cole, J.R.; Yiqi, L.U.O.; Liyou, W.U.; Schuur, E.A.G.; Jizhong, Z.; Tiedje, J.M. Fungal Diversity in Permafrost and Tallgrass Prairie Soils under Experimental Warming Conditions. Appl. Environ. Microbiol. 2013, 79, 7063–7072. [Google Scholar] [CrossRef]
- Perini, L.; Gostinčar, C.; Gunde-Cimerman, N. Fungal and bacterial diversity of Svalbard subglacial ice. Sci. Rep. 2019, 9, 20230. [Google Scholar] [CrossRef]
- Kilias, E.S.; Junges, L.; Šupraha, L.; Leonard, G.; Metfies, K.; Richards, T.A. Chytrid fungi distribution and co-occurrence with diatoms correlate with sea ice melt in the Arctic Ocean. Commun. Biol. 2020, 3, 183. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Or, D. Microscale pH variations during drying of soils and desert biocrusts affect HONO and NH3 emissions. Nat. Commun. 2019, 10, 3944. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.U.; Hooda, P.S.; Blackwell, M.S.A.; Busquets, R. Microbial Biomass Responses to Soil Drying-Rewetting and Phosphorus Leaching. Front. Environ. Sci. 2019, 7, 1–9. [Google Scholar] [CrossRef]
- Morris, E.K.; Morris, D.J.P.; Vogt, S.; Gleber, S.C.; Bigalke, M.; Wilcke, W.; Rillig, M.C. Visualizing the dynamics of soil aggregation as affected by arbuscular mycorrhizal fungi. ISME J. 2019, 13, 1639–1646. [Google Scholar] [CrossRef]
- Smucker, A.J.M.; Park, E.J.; Dorner, J.; Horn, R. Soil Micropore Development and Contributions to Soluble Carbon Transport within Macroaggregates. Vadose Zone J. 2007, 6, 282–290. [Google Scholar] [CrossRef]
- Ritz, K.; Young, I.M. Interactions between soil structure and fungi. Mycologist 2004, 18, 52–59. [Google Scholar] [CrossRef]
- Rabbi, S.M.F.; Daniel, H.; Lockwood, P.V.; Macdonald, C.; Pereg, L.; Tighe, M.; Wilson, B.R.; Young, I.M. Physical soil architectural traits are functionally linked to carbon decomposition and bacterial diversity. Sci. Rep. 2016, 6, 33012. [Google Scholar] [CrossRef] [PubMed]
- Otten, W.; Hall, D.; Harris, K.; Ritz, K.; Young, I.M.; Gilligan, C.A. Soil Physics, Fungal Epidemiology and the Spread of Rhizoctonia Solani. New Phytol. 2001, 151, 459–468. [Google Scholar] [CrossRef]
- Hassink, J.; Bouwman, L.A.; Zwart, K.B.; Brussaard, L. Relationships between habitable pore space, soil biota and mineralization rates in grassland soils. Soil Biol. Biochem. 1993, 25, 47–55. [Google Scholar] [CrossRef]
- Meng, M.; Chen, H.Y.H.; Lin, J.; Liu, X.; Guo, X.; Yuan, Y.; Zhang, J. Long term forest conversion affected soil nanoscale pores in subtropical China. Catena Giess. 2020, 185, 104289. [Google Scholar] [CrossRef]
- Geisen, S.; Mitchell, E.A.D.; Adl, S.; Bonkowski, M.; Dunthorn, M.; Ekelund, F.; Fernández, L.D.; Jousset, A.; Krashevska, V.; Singer, D.; et al. Soil protists: A fertile frontier in soil biology research. FEMS Microbiol. Rev. 2018, 42, 293–323. [Google Scholar] [CrossRef] [PubMed]
- Meisterfeld, R. Testate amoebae with filopodia. In The llustrated Guide to the Protozoa; Lee, J.J.L.G., Bradbury, P., Eds.; Society of Protozoologists: Lawrence, KS, USA, 2002; Volume 2, pp. 827–860. [Google Scholar]
- Paterson, R.A. Observations on two species of Rhizophydium from Northern Michigan. Trans. Br. Mycol. Soc. 1963, 46, 530–536, IN9. [Google Scholar] [CrossRef]
- Longcore, J.E. Rhizophydium brooksianum sp. nov., a multipored chytrid from soil. Mycologia 2004, 96, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Fukumasu, J.; Jarvis, N.; Koestel, J.; Kätterer, T.; Larsbo, M. Relations between soil organic carbon content and the pore size distribution for an arable topsoil with large variations in soil properties. Eur. J. Soil Sci. 2022, 73, e13212. [Google Scholar] [CrossRef]
- Keuschnig, C.; Martins, J.M.F.; Navel, A.; Simonet, P.; Larose, C. Micro-fractionation shows microbial community changes in soil particles below 20 μm. Front. Ecol. Evol. 2022, 10, 1–15. [Google Scholar] [CrossRef]
- Mangalassery, S.; Sjögersten, S.; Sparkes, D.L.; Sturrock, C.J.; Mooney, S.J. The effect of soil aggregate size on pore structure and its consequence on emission of greenhouse gases. Soil Tillage Res. 2013, 132, 39–46. [Google Scholar] [CrossRef]
- Miller, R.M.; Jastrow, J.D. Mycorrhizal Fungi Influence Soil Structure. In Arbuscular Mycorrhizas: Physiology and Function; Kapulnik, Y., Douds, D.D., Eds.; Springer: Dordrecht, The Netherlands, 2000; pp. 3–18. [Google Scholar] [CrossRef]
- Rillig, M.C.; Mummey, D.L. Mycorrhizas and Soil Structure. New Phytol. 2006, 171, 41–53. [Google Scholar] [CrossRef]
- Rillig, M.C.; Mardatin, N.F.; Leifheit, E.F.; Antunes, P.M. Mycelium of arbuscular mycorrhizal fungi increases soil water repellency and is sufficient to maintain water-stable soil aggregates. Soil Biol. Biochem. 2010, 42, 1189–1191. [Google Scholar] [CrossRef]
- Thomas, R.S.; Franson, R.L.; Bethlenfalvay, G.J. Separation of vesicular-arbuscular mycorrhizal fungus and root effects on soil aggregation. Soil Sci. Soc. Am. J. 1993, 57, 77–81. [Google Scholar] [CrossRef]
- Wilson, G.W.T.; Rice, C.W.; Rillig, M.C.; Springer, A.; Hartnett, D.C. Soil aggregation and carbon sequestration are tightly correlated with the abundance of arbuscular mycorrhizal fungi: Results from long-term field experiments. Ecol. Lett. 2009, 12, 452–461. [Google Scholar] [CrossRef] [PubMed]
- Samaei, F.; Asghari, S.; Aliasgharzad, N. The effects of two arbuscular mycorrhizal fungi on some physical properties of a sandy loam soil and nutrients uptake by spring barley. J. Soil Environ. 2015, 1, 1–9. [Google Scholar]
- Laundon, D.; Chrismas, N.; Wheeler, G.; Cunliffe, M. Chytrid rhizoid morphogenesis resembles hyphal development in multicellular fungi and is adaptive to resource availability. Proc. R. Society. B Biol. Sci. 2020, 287, 20200433. [Google Scholar] [CrossRef]
- Prostak, S.M.; Robinson, K.A.; Titus, M.A.; Fritz-Laylin, L.K. The actin networks of chytrid fungi reveal evolutionary loss of cytoskeletal complexity in the fungal kingdom. Curr. Biol. 2021, 31, 1192–1205.e6. [Google Scholar] [CrossRef]
- Shen, Q.; Kirschbaum, M.U.F.; Hedley, M.J.; Arbestain, M.C. Testing an alternative method for estimating the length of fungal hyphae using photomicrography and image processing. PLoS ONE 2016, 11, e0157017. [Google Scholar] [CrossRef] [PubMed]
- Lienhard, P.; Terrat, S.; Prévost-Bouré, N.C.; Nowak, V.; Régnier, T.; Sayphoummie, S.; Panyasiri, K.; Tivet, F.; Mathieu, O.; Levêque, J.; et al. Pyrosequencing evidences the impact of cropping on soil bacterial and fungal diversity in Laos tropical grassland. Agron. Sustain. Dev. 2014, 34, 525–533. [Google Scholar] [CrossRef]
- van Der Walt, A.J.; Johnson, R.M.; Cowan, D.A.; Seely, M.; Ramond, J.-B.; Kelly, R.M. Unique microbial phylotypes in Namib Desert dune and gravel plain fairy circle soils. Appl. Environ. Microbiol. 2016, 82, 4592–4601. [Google Scholar] [CrossRef]
- Doniger, T.; Kerfahi, D.; Wachtel, C.; Marais, E.; Maggs-Kölling, G.; Sherman, C.; Adams, J.M.; Steinberger, Y. Plant Gender Affects Soil Fungal Microbiota Associated with Welwitschia mirabilis, an Unusual Desert Gymnosperm. Microb. Ecol. 2022. [Google Scholar] [CrossRef]
- Gleason, F.H.; Letcher, P.M.; McGee, P.A. Freeze tolerance of soil chytrids from temperate climates in Australia. Mycol Res 2008, 112, 976–982. [Google Scholar] [CrossRef]
- Booth, T.; Barrett, P. Occurrence and distribution of zoosporic fungi from Devon Island, Canadian Eastern Arctic. Can. J. Bot. 1971, 49, 359–369. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, N.-F.; Zhang, Y.-Q.; Liu, H.-Y.; Yu, L.-Y. Diversity and distribution of aquatic fungal communities in the Ny-Ålesund Region, Svalbard (High Arctic). Microb. Ecol. 2016, 71, 543–554. [Google Scholar] [CrossRef] [PubMed]
- Bridge, P.D.; Newsham, K.K. Soil fungal community composition at Mars Oasis, a southern maritime Antarctic site, assessed by PCR amplification and cloning. Fungal Ecol. 2009, 2, 66–74. [Google Scholar] [CrossRef]
- Li, W.; Jiang, L.; Zhang, Y.; Teng, D.; Wang, H.; Wang, J.; Lv, G. Structure and driving factors of the soil microbial community associated with Alhagi sparsifolia in an arid desert. PLoS ONE 2021, 16, e0254065. [Google Scholar] [CrossRef]
- Longcore, J.E.; Barr, D.J.S.; Désaulniers, N. Powellomyces, a new genus in the Spizellomycetales. Can. J. Bot. 1995, 73, 1385–1390. [Google Scholar] [CrossRef]
- Henderson, L.; Pilgaard, B.; Gleason, F.H.; Lilje, O. Copper (II) lead (II), and zinc (II) reduce growth and zoospore release in four zoosporic true fungi from soils of NSW, Australia. Fungal Biol. 2015, 119, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Jia, T.; Wang, R.; Fan, X.; Chai, B. A comparative study of fungal community structure, diversity and richness between the soil and the phyllosphere of native grass species in a copper tailings dam in Shanxi Province, China. Appl. Sci. 2018, 8, 1297. [Google Scholar] [CrossRef]
- Passarini, M.R.Z.; Ottoni, J.R.; Costa, P.E.d.S.; Hissa, D.C.; Falcão, R.M.; Melo, V.M.M.; Balbino, V.Q.; Mendonça, L.A.R.; Lima, M.G.d.S.; Coutinho, H.D.M.; et al. Fungal community diversity of heavy metal contaminated soils revealed by metagenomics. Arch. Microbiol. 2022, 204, 255. [Google Scholar] [CrossRef]
- Canter, H.M.; Lund, J.W.G. Studies on plankton parasites III. Examples of the interaction between parasitism and other factors determining the growth of diatoms. Ann. Bot. 1951, 15, 359–371. [Google Scholar]
- Ibelings, B.W.; De Bruin, A.; Kagami, M.; Rijkeboer, M.; Brehm, M.; Donk, E.V. Host parasite interactions between freshwater phytoplankton and chytrid fungi (Chytridiomycota). J. Phycol. 2004, 40, 437–453. [Google Scholar] [CrossRef]
- Gleason, F.; Macarthur, D. The chytrid epidemic revisited. Inoculum 2008, 59, 1–3. [Google Scholar]
- Ward, M.W. Observations on Rhizophlyctis rosea. J. Elisha Mitchell Sci. Soc. 1939, 55, 353–360. [Google Scholar]
- McGee, P.A.; Daynes, C.N.; Gleason, F.H.; Marano, A.V.; Barrera, M.D.; Steciow, M.M. Rhizophlyctis rosea (Rhizophlyctidales, Chytridiomycota) in soil: Frequency, abundance and density of colonization of lens paper baits. Nova Hedwig. 2011, 93, 73–84. [Google Scholar] [CrossRef]
- Phuphumirat, W.; Gleason, F.H.; Phongpaichit, S.; Mildenhall, D.C. The infection of pollen by zoosporic fungi in tropical soils and its impact on pollen preservation: A preliminary study. Nova Hedwig. 2011, 92, 233–244. [Google Scholar] [CrossRef]
- Gleason, F.H.; Daynes, C.N.; McGee, P.A. Some zoosporic fungi can grow and survive within a wide pH range. Fungal Ecol. 2010, 3, 31–37. [Google Scholar] [CrossRef]
- Fernandes, M.L.P.; Bastida, F.; Jehmlich, N.; Martinović, T.; Větrovský, T.; Baldrian, P.; Delgado-Baquerizo, M.; Starke, R. Functional soil mycobiome across ecosystems. J. Proteom. 2022, 252, 104428. [Google Scholar] [CrossRef] [PubMed]
- Picard, K.T.; Letcher, P.M.; Powell, M.J. Evidence for a facultative mutualist nutritional relationship between the green coccoid alga Bracteacoccus sp. (Chlorophyceae) and the zoosporic fungus Rhizidium phycophilum (Chytridiomycota). Fungal Biol. 2013, 117, 319–328. [Google Scholar] [CrossRef]
- Gleason, F.H.; Kagami, M.; Lefevre, E.; Sime-Ngando, T. The ecology of chytrids in aquatic ecosystems: Roles in food web dynamics. Fungal Biol. Rev. 2008, 22, 17–25. [Google Scholar] [CrossRef]
- Kagami, M.; Miki, T.; Takimoto, G. Mycoloop: Chytrids in aquatic food webs. Front. Microbiol. 2014, 5, 166. [Google Scholar] [CrossRef]
- Gleason, F.H.; Lilje, O.; Marano, A.V.; Sime-Ngando, T.; Sullivan, B.K.; Kirchmair, M.; Neuhauser, S. Ecological functions of zoosporic hyperparasites. Front. Microbiol. 2014, 5, 244. [Google Scholar] [CrossRef]
- Deacon, J.W.; Saxena, G. Orientated zoospore attachment and cyst germination in Catenaria anguillulae, a facultative endoparasite of nematodes. Mycol. Res. 1997, 101, 513–522. [Google Scholar] [CrossRef]
- Gleason, F.H.; Marano, A.V.; Johnson, P.; Martin, W.W. Blastocladian parasites of invertebrates. Fungal Biol. Rev. 2010, 24, 56–67. [Google Scholar] [CrossRef]
- Longcore, J.E.; Simmons, D.R.; Letcher, P.M. Synchytrium microbalum sp. nov. is a saprobic species in a lineage of parasites. Fungal Biol. 2016, 120, 1156–1164. [Google Scholar] [CrossRef]
- Dijk, L.J.A.; Ehrlén, J.; Tack, A.J.M. The relationship between pathogen life-history traits and metapopulation dynamics. New Phytol. 2022, 233, 2585–2598. [Google Scholar] [CrossRef] [PubMed]
- Lange, L.; Lenne, J.M.; Olson, L.W. Ultrastructural studies of zoosporangium and resting sporangium of Synchytrium Desmodii. J. Phytopathol. 1989, 125, 361–371. [Google Scholar] [CrossRef]
- Przetakiewicz, J. The viability of winter sporangia of Synchytrium endobioticum (Schilb.) Perc. from Poland. Am. J. Potato Res. 2015, 92, 704–708. [Google Scholar] [CrossRef]
- Laidlaw, W.M.R. A method for the detection of the resting sporangia of potato wart disease (Synchytrium endobioticum) in the soil of old outbreak sites. Potato Res. 1985, 28, 223–232. [Google Scholar] [CrossRef]
- Barr, D.J. Rhizophydium graminis (Chytridiales): Morphology, host range, and temperature effect. Can. Plant Dis. Surv. 1973, 53, 191–193. [Google Scholar]
- Ledingham, G. Rhizophydium graminis n. sp., a parasite of wheat roots. Can. J. Res. 2011, 14, 117–121. [Google Scholar] [CrossRef]
- Karling, J.S. Parasitism Among the Chytrids. II Chytriomyces verrucosus sp. nov. and Phlyctochytrium Synchytrii. Bull. Torrey Bot. Club 1960, 87, 326–336. [Google Scholar] [CrossRef]
- Kaczmarek, A.; Boguś, M.I. Fungi of entomopathogenic potential in Chytridiomycota and Blastocladiomycota, and in fungal allies of the Oomycota and Microsporidia. IMA Fungus 2021, 12, 29. [Google Scholar] [CrossRef]
- Hajek, A.E.; Longcore, J.E.; Rabern Simmons, D.; Peters, K.; Humber, R.A. Chytrid mycoparasitism of entomophthoralean azygospores. J. Invertebr. Pathol. 2013, 114, 333–336. [Google Scholar] [CrossRef]
- Wakefield, W.S.; Powell, M.J.; Letcher, P.M.; Barr, D.J.S.; Churchill, P.F.; Longcore, J.E.; Chen, S.-F. A molecular phylogenetic evaluation of the Spizellomycetales. Mycologia 2010, 102, 596–604. [Google Scholar] [CrossRef] [PubMed]
- Daft, G.C.; Tsao, P.H. Parasitism of Phytophthora cinnamomi and P. parasitica spores by Catenaria anguillulae in a soil environment. Trans. Br. Mycol. Soc. 1984, 82, 485–490. [Google Scholar] [CrossRef]
- Sykes, E.E.; Porter, D. Infection and Development of the Obligate Parasite Catenaria allomycis on Allomyces Arbuscula. Mycologia 1980, 72, 288–300. [Google Scholar] [CrossRef]
- Sneh, B. Parasitism of Oospores of Phytophthora megasperma var. sojae, P. cactorum, Pythium sp., and Aphanomyces euteiches in Soil by Oomycetes, Chytridiomycetes, Hyphomycetes, Actinomycetes, and Bacteria. Phytopathology 1977, 77, 622. [Google Scholar] [CrossRef]
- Jiao, S.; Wang, J.; Wei, G.; Chen, W.; Lu, Y. Dominant role of abundant rather than rare bacterial taxa in maintaining agro-soil microbiomes under environmental disturbances. Chemosphere 2019, 235, 248–259. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, J.; Ye, Z.; Wang, J.; Qu, L.; Zhang, T. Spatial factors and plant attributes influence soil fungal community distribution patterns in the lower reaches of the Heihe River Basin, Northwest China. Environ. Microbiol. 2021, 23, 2499–2508. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.N.; Huang, Z.J.; Li, M.Q.; Min, W. Response of soil fungal community structure and diversity to saline water irrigation in alluvial grey desert soils. Appl. Ecol. Environ. Res. 2020, 18, 4969–4985. [Google Scholar] [CrossRef]
- Dacal, M.; García-Palacios, P.; Asensio, S.; Wang, J.; Singh, B.K.; Maestre, F.T. Climate change legacies contrastingly affect the resistance and resilience of soil microbial communities and multifunctionality to extreme drought. Funct. Ecol. 2022, 36, 908–920. [Google Scholar] [CrossRef]
- Zhao, B.; Xing, P.; Wu, Q.L. Interactions between bacteria and fungi in macrophyte leaf litter decomposition. Environ. Microbiol. 2021, 23, 1130–1144. [Google Scholar] [CrossRef]
- Rousk, J.; Bååth, E. Growth of saprotrophic fungi and bacteria in soil. FEMS Microbiol. Ecol. 2011, 78, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Roberts, C.; Allen, R.; Bird, K.E.; Cunliffe, M. Chytrid fungi shape bacterial communities on model particulate organic matter. Biol. Lett. 2020, 16, 20200368. [Google Scholar] [CrossRef] [PubMed]
- Busi, S.B.; Bourquin, M.; Fodelianakis, S.; Michoud, G.; Kohler, T.J.; Peter, H.; Pramateftaki, P.; Styllas, M.; Tolosano, M.; De Staercke, V.; et al. Genomic and metabolic adaptations of biofilms to ecological windows of opportunity in glacier-fed streams. Nat. Commun. 2022, 13, 2168. [Google Scholar] [CrossRef]
- St Wilken, E.; Monk, J.M.; Leggieri, P.A.; Lawson, C.E.; Lankiewicz, T.S.; Seppälä, S.; Daum, C.G.; Jenkins, J.; Lipzen, A.M.; Mondo, S.J.; et al. Experimentally Validated Reconstruction and Analysis of a Genome-Scale Metabolic Model of an Anaerobic Neocallimastigomycota Fungus. mSystems 2021, 6, e00002-21. [Google Scholar] [CrossRef] [PubMed]
- Van de Vossenberg, B.T.L.H.; Warris, S.; Nguyen, H.D.T.; van Gent-Pelzer, M.P.E.; Joly, D.L.; van de Geest, H.C.; Bonants, P.J.M.; Smith, D.S.; Lévesque, A.C.; van der Lee, T.A.J. Comparative genomics of chytrid fungi reveal insights into the obligate biotrophic and pathogenic lifestyle of Synchytrium Endobioticum. Sci. Rep. 2019, 9, 8672. [Google Scholar] [CrossRef]
- Olive, L.S. Caulochytrium protostelioides sp. nov., a new chytrid with aerial sporangia. Am. J. Bot. 1980, 67, 568–574. [Google Scholar] [CrossRef]
- Ahrendt, S.R.; Quandt, C.A.; Ciobanu, D.; Clum, A.; Salamov, A.; Andreopoulos, B.; Cheng, J.-F.; Woyke, T.; Pelin, A.; Henrissat, B.; et al. Leveraging single-cell genomics to expand the fungal tree of life. Nat. Microbiol. 2018, 3, 1417–1428. [Google Scholar] [CrossRef]
- Kadłubowska, J.Z. Rare species of fungi parasiting on algae I. Parasites of Spirogyra and Mougeotia. Acta Mycol. 1998, 33, 247–254. [Google Scholar] [CrossRef]
- Haitjema, C.H.; Gilmore, S.P.; Henske, J.K.; Solomon, K.V.; de Groot, R.; Kuo, A.; Mondo, S.J.; Salamov, A.A.; LaButti, K.; Zhao, Z.; et al. A parts list for fungal cellulosomes revealed by comparative genomics. Nat. Microbiol. 2017, 2, 17087. [Google Scholar] [CrossRef]
- Forget, L.; Ustinova, J.; Wang, Z.; Huss, V.A.R.; Lang, B.F. Hyaloraphidium curvatum: A linear mitochondrial genome, tRNA editing, and an evolutionary link to lower fungi. Mol. Biol. Evol. 2002, 19, 310–319. [Google Scholar] [CrossRef]
- Simmons, D.R. Phylogeny of Powellomycetaceae fam. nov. and description of Geranomyces variabilis gen. et comb. nov. Mycologia 2011, 103, 1411–1420. [Google Scholar] [CrossRef]
- Vélez, C.G.; Letcher, P.M.; Schultz, S.; Mataloni, G.; Lefèvre, E.; Powell, M.J. Three new genera in Chytridiales from aquatic habitats in Argentina. Mycologia 2013, 105, 1251–1265. [Google Scholar] [CrossRef]
- Wang, S.-K.; Zuo, X.-A.; Zhao, X.-Y.; Li, Y.-Q.; Zhou, X.; Lv, P.; Luo, Y.-Q.; Yun, J.-Y. Responses of soil fungal community to the sandy grassland restoration in Horqin Sandy Land, northern China. Environ. Monit. Assess. 2016, 188, 21. [Google Scholar] [CrossRef]
- Bernstein, L.B. A Biosystematic Study of Rhizophlyctis rosea with Emphasis on Zoospore Variability. J. Elisha Mitchell Sci. Soc. 1968, 84, 84–93. [Google Scholar]
- Federici, B.A. Species-specific gating of gametangial dehiscence as a temporal reproductive isolating mechanism in Coelomomyces. Proc. Natl. Acad. Sci. USA 1983, 80, 604–607. [Google Scholar] [CrossRef]
- Saranak, J.; Foster, K.W. Rhodopsin guides fungal phototaxis. Nature 1997, 387, 465–466. [Google Scholar] [CrossRef]
- Ahrendt, S.R.; Medina, E.M.; Chang, C.-E.A.; Stajich, J.E. Exploring the binding properties and structural stability of an opsin in the chytrid using comparative and molecular modeling. PeerJ 2017, 5, e3206. [Google Scholar] [CrossRef]
- Broser, M. Far-Red Absorbing Rhodopsins, Insights From Heterodimeric Rhodopsin-Cyclases. Front. Mol. Biosci. 2021, 8, 806922. [Google Scholar] [CrossRef]
- Broser, M.; Spreen, A.; Konold, P.E.; Peter, E.; Adam, S.; Borin, V.; Schapiro, I.; Seifert, R.; Kennis, J.T.M.; Bernal Sierra, Y.A.; et al. NeoR, a near-infrared absorbing rhodopsin. Nat. Commun. 2020, 11, 5682. [Google Scholar] [CrossRef]
- Lilje, O.; Lilje, E.; Marano, A.V.; Gleason, F.H. Three dimensional quantification of biological samples using micro-computer aided tomography (microCT). J. Microbiol. Methods 2013, 92, 33–41. [Google Scholar] [CrossRef]
- Soufan, R.; Delaunay, Y.; Gonod, L.V.; Shor, L.M.; Garnier, P.; Otten, W.; Baveye, P.C. Pore-scale monitoring of the effect of microarchitecture on fungal growth in a two-dimensional soil-like micromodel. Front. Environ. Sci. 2018, 6, 1–11. [Google Scholar] [CrossRef]
- Schloter, M.; Nannipieri, P.; Sorensen, S.J.; van Elsas, J.D. Microbial indicators for soil quality. Biol. Fertil. Soils 2018, 54, 1–10. [Google Scholar] [CrossRef]
- Heeger, F.; Bourne, E.C.; Baschien, C.; Yurkov, A.; Bunk, B.; Spröer, C.; Overmann, J.; Mazzoni, C.J.; Monaghan, M.T. Long-read DNA metabarcoding of ribosomal RNA in the analysis of fungi from aquatic environments. Mol. Ecol. Resour. 2018, 18, 1500–1514. [Google Scholar] [CrossRef] [PubMed]
- Lange, L.; Pilgaard, B.; Herbst, F.-A.; Busk, P.K.; Gleason, F.; Pedersen, A.G. Origin of fungal biomass degrading enzymes: Evolution, diversity and function of enzymes of early lineage fungi. Fungal Biol. Rev. 2019, 33, 82–97. [Google Scholar] [CrossRef]
- Chang, Y.; Wang, S.; Sekimoto, S.; Aerts, A.L.; Choi, C.; Clum, A.; LaButti, K.M.; Lindquist, E.A.; Yee Ngan, C.; Ohm, R.A.; et al. Phylogenomic Analyses Indicate that Early Fungi Evolved Digesting Cell Walls of Algal Ancestors of Land Plants. Genome Biol. Evol. 2015, 7, 1590–1601. [Google Scholar] [CrossRef]
- Huang, Y.; Zheng, X.; Pilgaard, B.; Holck, J.; Muschiol, J.; Li, S.; Lange, L. Identification and characterization of GH11 xylanase and GH43 xylosidase from the chytridiomycetous fungus, Rhizophlyctis Rosea. Appl. Microbiol. Biotechnol. 2019, 103, 777–791. [Google Scholar] [CrossRef]
- Lange, L.; Barrett, K.; Pilgaard, B.; Gleason, F.; Tsang, A. Enzymes of early-diverging, zoosporic fungi. Appl. Microbiol. Biotechnol. 2019, 103, 6885–6902. [Google Scholar] [CrossRef]
- Song, N.; Xu, H.; Yan, Z.; Yang, T.; Wang, C.; Jiang, H.-L. Improved lignin degradation through distinct microbial community in subsurface sediments of one eutrophic lake. Renew. Energy 2019, 138, 861–869. [Google Scholar] [CrossRef]
- Chen, Z.; Fei, Y.-h.; Liu, W.-S.; Ding, K.; Lu, J.; Cai, X.; Cui, T.; Tang, Y.-T.; Wang, S.; Chao, Y.; et al. Untangling microbial diversity and assembly patterns in rare earth element mine drainage in South China. Water Res. 2022, 225, 119172. [Google Scholar] [CrossRef]
- Ravindran, R.; Jaiswal, A.K. A comprehensive review on pre-treatment strategy for lignocellulosic food industry waste: Challenges and opportunities. Bioresour. Technol. 2016, 199, 92–102. [Google Scholar] [CrossRef]
- Wongwilaiwalin, S.; Rattanachomsri, U.; Laothanachareon, T.; Eurwilaichitr, L.; Igarashi, Y.; Champreda, V. Analysis of a thermophilic lignocellulose degrading microbial consortium and multi-species lignocellulolytic enzyme system. Enzym. Microb. Technol. 2010, 47, 283–290. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hanrahan-Tan, D.G.; Lilje, O.; Henderson, L. Chytrids in Soil Environments: Unique Adaptations and Distributions. Encyclopedia 2023, 3, 642-664. https://doi.org/10.3390/encyclopedia3020046
Hanrahan-Tan DG, Lilje O, Henderson L. Chytrids in Soil Environments: Unique Adaptations and Distributions. Encyclopedia. 2023; 3(2):642-664. https://doi.org/10.3390/encyclopedia3020046
Chicago/Turabian StyleHanrahan-Tan, Deirdre G., Osu Lilje, and Linda Henderson. 2023. "Chytrids in Soil Environments: Unique Adaptations and Distributions" Encyclopedia 3, no. 2: 642-664. https://doi.org/10.3390/encyclopedia3020046
APA StyleHanrahan-Tan, D. G., Lilje, O., & Henderson, L. (2023). Chytrids in Soil Environments: Unique Adaptations and Distributions. Encyclopedia, 3(2), 642-664. https://doi.org/10.3390/encyclopedia3020046








