SARS-CoV-2 Intermittent Virulence as a Result of Natural Selection
Abstract
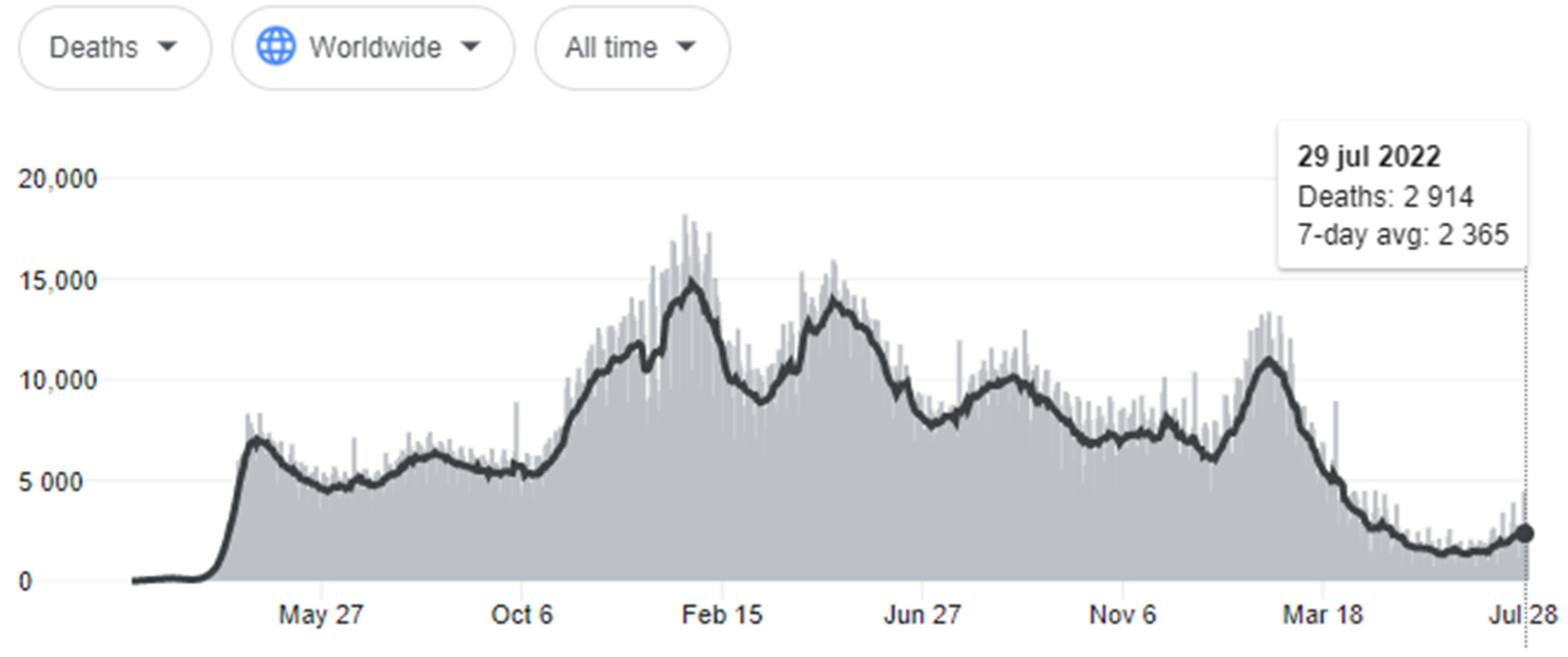
:1. Introduction
2. The Delta Variant
3. The Omicron Variant
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Genomic Sequencing of SARS-CoV-2: A Guide to Implementation for Maximum Impact on Public Health; World Health Organization: Geneva, Switzerland, 2021; Available online: https://www.who.int/publications/i/item/9789240018440 (accessed on 5 June 2022).
- Park, M.; Loverdo, C.; Schreiber, S.J.; Lloyd-Smith, J.O. Multiple scales of selection influence the evolutionary emergence of novel pathogens. Philos. Trans. R. Soc. Lond B Biol. Sci. 2013, 368, 20120333. [Google Scholar] [CrossRef] [Green Version]
- Wasik, B. On the biological success of viruses. Annu. Rev. Microbiol. 2013, 67, 519–541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Dorp, L.; Richard, D.; Tan, C.C.S.; Shaw, L.P.; Acman, M.; Balloux, F. No evidence for increased transmissibility from recurrent mutations in SARS-CoV-2. Nat. Commun. 2020, 11, 5986. [Google Scholar] [CrossRef] [PubMed]
- Thomson, E.C.; Rosen, L.E.; Shepherd, J.G.; Spreafico, R.; da Silva Filipe, A.; Wojcechowskyj, J.A.; Davis, C.; Piccoli, L.; Pascall, D.J.; Dillen, J.; et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell 2021, 184, 1171–1187.e1120. [Google Scholar] [CrossRef] [PubMed]
- Piccoli, L.; Park, Y.J.; Tortorici, M.A.; Czudnochowski, N.; Walls, A.C.; Beltramello, M.; Silacci-Fregni, C.; Pinto, D.; Rosen, L.E.; Bowen, J.E.; et al. Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology. Cell 2020, 183, 1024–1042.e1021. [Google Scholar] [CrossRef]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e819. [Google Scholar] [CrossRef]
- Yurkovetskiy, L.; Wang, X.; Pascal, K.E.; Tomkins-Tinch, C.; Nyalile, T.P.; Wang, Y.; Baum, A.; Diehl, W.E.; Dauphin, A.; Carbone, C.; et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell 2020, 183, 739–751.e738. [Google Scholar] [CrossRef]
- Greaney, A.J.; Loes, A.N.; Crawford, K.H.D.; Starr, T.N.; Malone, K.D.; Chu, H.Y.; Bloom, J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe 2021, 29, 463–476.e466. [Google Scholar] [CrossRef]
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D. Antibody resistance of SARS-CoV-2 variants B. 1.351 and B. 1.1. 7. Nature 2021, 593, 130–135. [Google Scholar] [CrossRef]
- Burnet, F.M.; White, D. Natural History of Infectious Disease; Cambridge University Press: Cambridge, UK, 1972. [Google Scholar]
- Levin, S.; Pimentel, D. Selection of intermediate rates of increase in parasite-host systems. Am. Nat. 1981, 117, 308–315. [Google Scholar] [CrossRef]
- Anderson, R.M.; May, R.M. Coevolution of hosts and parasites. Parasitology 1982, 85, 411–426. [Google Scholar] [CrossRef] [PubMed]
- May, R.M.; Anderson, R.M. Epidemiology and genetics in the coevolution of parasites and hosts. Proc. R. Soc. London. Ser. B Biol. Sci. 1983, 219, 281–313. [Google Scholar]
- Frank, S.A. Models of parasite virulence. Q. Rev. Biol. 1996, 71, 37–78. [Google Scholar] [CrossRef]
- Galvani, A.P. Epidemiology meets evolutionary ecology. Trends Ecol. Evol. 2003, 18, 132–139. [Google Scholar] [CrossRef]
- Grubaugh, N.D.; Petrone, M.E.; Holmes, E.C. We shouldn’t worry when a virus mutates during disease outbreaks. Nat. Microbiol. 2020, 5, 529–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Markov, P.V.; Katzourakis, A.; Stilianakis, N.I. Antigenic evolution will lead to new SARS-CoV-2 variants with unpredictable severity. Nat. Rev. Microbiol. 2022, 20, 251–252. [Google Scholar] [CrossRef] [PubMed]
- Yamasoba, D.; Kimura, I.; Nasser, H.; Morioka, Y.; Nao, N.; Ito, J.; Uriu, K.; Tsuda, M.; Zahradnik, J.; Shirakawa, K. Virological characteristics of SARS-CoV-2 BA. 2 variant. Cell 2022, 185, 2103–2115. [Google Scholar] [CrossRef]
- Qu, P.; Evans, J.P.; Faraone, J.N.; Zou, X.; Zheng, Y.M.; Carlin, C.; Bednash, J.S.; Lozanski, G.; Mallampalli, R.K.; Saif, L.J. Differential Evasion of Delta and Omicron Immunity and Enhanced Fusogenicity of SARS-CoV-2 Omicron BA. 4/5 and BA. 2.12. 1 Subvariants. bioRxiv 2022. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. Threat Assessment Brief: Emergence of SARS-CoV-2 B.1.617 Variants in India and Situation in the EU/EEA. Available online: https://www.ecdc.europa.eu/en/publications-data/threat-assessment-emergence-sars-cov-2-b1617-variants (accessed on 15 June 2022).
- Sheikh, A.; McMenamin, J.; Taylor, B.; Robertson, C. SARS-CoV-2 Delta VOC in Scotland: Demographics, risk of hospital admission, and vaccine effectiveness. Lancet 2021, 397, 2461–2462. [Google Scholar] [CrossRef]
- Twohig, K.A.; Nyberg, T.; Zaidi, A.; Thelwall, S.; Sinnathamby, M.A.; Aliabadi, S.; Seaman, S.R.; Harris, R.J.; Hope, R.; Lopez-Bernal, J. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B. 1.617. 2) compared with alpha (B. 1.1. 7) variants of concern: A cohort study. Lancet Infect. Dis. 2022, 22, 35–42. [Google Scholar] [CrossRef]
- Kim, S.; Liu, Y.; Lei, Z.; Dicker, J.; Cao, Y.; Zhang, X.F.; Im, W. Differential interactions between human ACE2 and Spike RBD of SARS-CoV-2 variants of concern. J. Chem. Theory Comput. 2021, 17, 7972–7979. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, J.; Johnson, B.A.; Xia, H.; Ku, Z.; Schindewolf, C.; Widen, S.G.; An, Z.; Weaver, S.C.; Menachery, V.D. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. Cell Rep. 2022, 39, 110829. [Google Scholar] [CrossRef]
- Saito, A.; Irie, T.; Suzuki, R.; Maemura, T.; Nasser, H.; Uriu, K.; Kosugi, Y.; Shirakawa, K.; Sadamasu, K.; Kimura, I. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature 2022, 602, 300–306. [Google Scholar] [CrossRef]
- Zhang, L.; Mann, M.; Syed, Z.A.; Reynolds, H.M.; Tian, E.; Samara, N.L.; Zeldin, D.C.; Tabak, L.A.; Ten Hagen, K.G. Furin cleavage of the SARS-CoV-2 spike is modulated by O-glycosylation. Proc. Natl. Acad. Sci. USA 2021, 118, e2109905118. [Google Scholar] [CrossRef]
- Cai, Y.; Zhang, J.; Xiao, T.; Lavine, C.L.; Rawson, S.; Peng, H.; Zhu, H.; Anand, K.; Tong, P.; Gautam, A. Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants. Science 2021, 373, 642–648. [Google Scholar] [CrossRef]
- Berger, I.; Schaffitzel, C. The SARS-CoV-2 spike protein: Balancing stability and infectivity. Cell Res. 2020, 30, 1059–1060. [Google Scholar] [CrossRef] [PubMed]
- Phan, T. Genetic diversity and evolution of SARS-CoV-2. Infect. Genet. Evol. 2020, 81, 104260. [Google Scholar] [CrossRef]
- Zhang, L.; Jackson, C.B.; Mou, H.; Ojha, A.; Peng, H.; Quinlan, B.D.; Rangarajan, E.S.; Pan, A.; Vanderheiden, A.; Suthar, M.S. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 2020, 11, 1–9. [Google Scholar] [CrossRef]
- Eaaswarkhanth, M.; Al Madhoun, A.; Al-Mulla, F. Could the D614G substitution in the SARS-CoV-2 spike (S) protein be associated with higher COVID-19 mortality? Int. J. Infect. Dis. 2020, 96, 459–460. [Google Scholar] [CrossRef]
- Jackson, C.B.; Zhang, L.; Farzan, M.; Choe, H. Functional importance of the D614G mutation in the SARS-CoV-2 spike protein. Biochem. Biophys. Res. Commun. 2021, 538, 108–115. [Google Scholar] [CrossRef]
- Mohammad, A.; Alshawaf, E.; Marafie, S.K.; Abu-Farha, M.; Abubaker, J.; Al-Mulla, F. Higher binding affinity of furin for SARS-CoV-2 spike (S) protein D614G mutant could be associated with higher SARS-CoV-2 infectivity. Int. J. Infect. Dis. 2021, 103, 611–616. [Google Scholar] [CrossRef] [PubMed]
- Becerra-Flores, M.; Cardozo, T. SARS-CoV-2 viral spike G614 mutation exhibits higher case fatality rate. Int. J. Clin. Pract. 2020, 74, e13525. [Google Scholar] [CrossRef] [PubMed]
- Fisman, D.N.; Tuite, A.R. Evaluation of the relative virulence of novel SARS-CoV-2 variants: A retrospective cohort study in Ontario, Canada. Cmaj 2021, 193, E1619–E1625. [Google Scholar] [CrossRef] [PubMed]
- WHO. Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern (26 November 2021). 2021. Available online: https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern (accessed on 10 June 2022).
- UKHSA. SARS-CoV-2 Variants of Concern and Variants under Investigation in England. Technical Briefing 35 (28 January 2022). Available online: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1050999/Technical-Briefing-35-28January2022.pdf (accessed on 12 June 2022).
- Roberts, D.S.; Mann, M.; Li, B.H.; Kim, D.; Brasier, A.R.; Jin, S.; Ge, Y. Distinct Core Glycan and O-Glycoform Utilization of SARS-CoV-2 Omicron Variant Spike Protein RBD Revealed by Top-Down Mass Spectrometry. bioRxiv 2022. [Google Scholar] [CrossRef]
- Aguilar, H.C.; Matreyek, K.A.; Filone, C.M.; Hashimi, S.T.; Levroney, E.L.; Negrete, O.A.; Bertolotti-Ciarlet, A.; Choi, D.Y.; McHardy, I.; Fulcher, J.A. N-glycans on Nipah virus fusion protein protect against neutralization but reduce membrane fusion and viral entry. J. Virol. 2006, 80, 4878–4889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, Y.; Bowden, T.A.; Wilson, I.A.; Crispin, M. Exploitation of glycosylation in enveloped virus pathobiology. Biochim. Et Biophys. Acta (BBA)-Gen. Subj. 2019, 1863, 1480–1497. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Hughes, T.A.; Kelkar, A.; Yu, X.; Cheng, K.; Park, S.; Huang, W.C.; Lovell, J.F.; Neelamegham, S. Inhibition of SARS-CoV-2 viral entry upon blocking N-and O-glycan elaboration. eLife 2020, 9, e61552. [Google Scholar] [CrossRef]
- Sikora, M.; von Bülow, S.; Blanc, F.E.; Gecht, M.; Covino, R.; Hummer, G. Computational epitope map of SARS-CoV-2 spike protein. PLoS Comput. Biol. 2021, 17, e1008790. [Google Scholar] [CrossRef]
- Suzuki, R.; Yamasoba, D.; Kimura, I.; Wang, L.; Kishimoto, M.; Ito, J.; Morioka, Y.; Nao, N.; Nasser, H.; Uriu, K. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature 2022, 603, 700–705. [Google Scholar] [CrossRef]
- Meng, B.; Abdullahi, A.; Ferreira, I.A.; Goonawardane, N.; Saito, A.; Kimura, I.; Yamasoba, D.; Gerber, P.P.; Fatihi, S.; Rathore, S. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 2022, 603, 706–714. [Google Scholar] [CrossRef]
- Cheng, Y.W.; Chao, T.L.; Li, C.L.; Wang, S.H.; Kao, H.C.; Tsai, Y.M.; Wang, H.Y.; Hsieh, C.L.; Lin, Y.Y.; Chen, P.J. D614G substitution of SARS-CoV-2 spike protein increases syncytium formation and virus titer via enhanced furin-mediated spike cleavage. Mbio 2021, 12, e00587-21. [Google Scholar] [CrossRef]
- Reading, P.C.; Pickett, D.L.; Tate, M.D.; Whitney, P.G.; Job, E.R.; Brooks, A.G. Loss of a single N-linked glycan from the hemagglutinin of influenza virus is associated with resistance to collectins and increased virulence in mice. Respir. Res. 2009, 10, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, C.J.; Boyington, J.C.; Dai, K.; Houser, K.V.; Pearce, M.B.; Kong, W.P.; Yang, Z.y.; Tumpey, T.M.; Nabel, G.J. Cross-neutralization of 1918 and 2009 influenza viruses: Role of glycans in viral evolution and vaccine design. Sci. Transl. Med. 2010, 2, ra21–ra24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Jayaraman, A.; Maniprasad, P.; Raman, R.; Houser, K.V.; Pappas, C.; Zeng, H.; Sasisekharan, R.; Katz, J.M.; Tumpey, T.M. N-linked glycosylation of the hemagglutinin protein influences virulence and antigenicity of the 1918 pandemic and seasonal H1N1 influenza A viruses. J. Virol. 2013, 87, 8756–8766. [Google Scholar] [CrossRef] [Green Version]
- Bentley, E.G.; Kirby, A.; Sharma, P.; Kipar, A.; Mega, D.F.; Bramwell, C.; Penrice-Randal, R.; Prince, T.; Brown, J.C.; Zhou, J. SARS-CoV-2 Omicron-B. 1.1. 529 Variant leads to less severe disease than Pango B and Delta variants strains in a mouse model of severe COVID-19. bioRxiv 2021. [Google Scholar] [CrossRef]
- McMahan, K.; Giffin, V.; Tostanosky, L.; Chung, B.; Siamatu, M.; Suthar, M.S.; Halfmann, P.; Kawaoka, Y.; Piedra-Mora, C.; Jain, N.; et al. Reduced pathogenicity of the SARS-CoV-2 Omicron variant in hamsters. Med 2022, 3, 262–268.e4. [Google Scholar] [CrossRef] [PubMed]
- Shuai, H.; Chan, J.F.W.; Hu, B.; Chai, Y.; Yuen, T.T.T.; Yin, F.; Huang, X.; Yoon, C.; Hu, J.C.; Liu, H. Attenuated replication and pathogenicity of SARS-CoV-2 B. 1.1. 529 Omicron. Nature 2022, 603, 693–699. [Google Scholar] [CrossRef] [PubMed]
- Wolter, N.; Jassat, W.; Walaza, S.; Welch, R.; Moultrie, H.; Groome, M.; Amoako, D.G.; Everatt, J.; Bhiman, J.N.; Scheepers, C. Early assessment of the clinical severity of the SARS-CoV-2 omicron variant in South Africa: A data linkage study. Lancet 2022, 399, 437–446. [Google Scholar] [CrossRef]
- Sievers, C.; Zacher, B.; Ullrich, A.; Huska, M.; Fuchs, S.; Buda, S.; Haas, W.; Diercke, M.; an der Heiden, M.; Kröger, S. SARS-CoV-2 Omicron variants BA. 1 and BA. 2 both show similarly reduced disease severity of COVID-19 compared to Delta, Germany, 2021 to 2022. Eurosurveillance 2022, 27, 2200396. [Google Scholar] [CrossRef] [PubMed]
- Fonager, J.; Bennedbæk, M.; Bager, P.; Wohlfahrt, J.; Ellegaard, K.M.; Ingham, A.C.; Edslev, S.M.; Stegger, M.; Sieber, R.N.; Lassauniere, R. Molecular epidemiology of the SARS-CoV-2 variant Omicron BA. 2 sub-lineage in Denmark, 29 November 2021 to 2 January 2022. Eurosurveillance 2022, 27, 2200181. [Google Scholar] [CrossRef]
- Wolter, N.; Jassat, W.; von Gottberg, A.; Cohen, C. Clinical severity of Omicron sub-lineage BA. 2 compared to BA. 1 in South Africa. Lancet 2022, 400, 93–96. [Google Scholar] [CrossRef]
- Gautret, P.; Hoang, V.T.; Jimeno, M.T.; Lagier, J.C.; Rossi, P.; Fournier, P.E.; Colson, P.; Raoult, D. The severity of the first 207 infections with the SARS-CoV-2 Omicron BA. 2 variant, in Marseille, France, December 2021–February 2022. J. Med. Virol. 2022, 94, 3494–3497. [Google Scholar] [CrossRef]
- Xia, B.; Wang, Y.; Pan, X.; Cheng, X.; Ji, H.; Zuo, X.; Jiang, H.; Li, J.; Gao, Z. Why is the SARS-CoV-2 Omicron variant milder? Innovation 2022, 3, 100251. [Google Scholar] [CrossRef]
- Ioannidis, J.P.A. The end of the COVID-19 pandemic. Eur. J. Clin. Investig. 2022, 52, e13782. [Google Scholar] [CrossRef] [PubMed]
- Lyngse, F.P.; Kirkeby, C.T.; Denwood, M.; Christiansen, L.E.; Mølbak, K.; Møller, C.H.; Skov, R.L.; Krause, T.G.; Rasmussen, M.; Sieber, R.N. Transmission of SARS-CoV-2 Omicron VOC subvariants BA. 1 and BA. 2: Evidence from Danish Households. MedRxiv 2022. [Google Scholar]
- Chen, J.; Wei, G.W. Omicron ba. 2 (b. 1.1. 529.2): High potential for becoming the next dominant variant. J. Phys. Chem. Lett. 2022, 13, 3840–3849. [Google Scholar] [CrossRef]
- Tegally, H.; Moir, M.; Everatt, J.; Giovanetti, M.; Scheepers, C.; Wilkinson, E.; Subramoney, K.; Moyo, S.; Amoako, D.G.; Althaus, C.L. Continued emergence and evolution of Omicron in South Africa: New BA. 4 and BA. 5 lineages. medRxiv 2022. [Google Scholar]
- WHO. Tracking SARS-CoV-2 Variants. Available online: who.int (accessed on 18 June 2022).
- Wymant, C.; Bezemer, D.; Blanquart, F.; Ferretti, L.; Gall, A.; Hall, M.; Golubchik, T.; Bakker, M.; Ong, S.H.; Zhao, L. A highly virulent variant of HIV-1 circulating in the Netherlands. Science 2022, 375, 540–545. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yap, Y.; Danchin, A. Testing the hypothesis of a recombinant origin of the SARS-associated coronavirus. Arch. Virol. 2005, 150, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, S.A.; Brown, J.; Pedersen, B.S.; Quinlan, A.R.; Elde, N.C. Extensive recombination-driven coronavirus diversification expands the pool of potential pandemic pathogens. BioRxiv 2021. [Google Scholar]
- Gribble, J.; Stevens, L.J.; Agostini, M.L.; Anderson-Daniels, J.; Chappell, J.D.; Lu, X.; Pruijssers, A.J.; Routh, A.L.; Denison, M.R. The coronavirus proofreading exoribonuclease mediates extensive viral recombination. PLoS Pathog. 2021, 17, e1009226. [Google Scholar] [CrossRef]
- Moisan, A.; Mastrovito, B.; De Oliveira, F.; Martel, M.; Hedin, H.; Leoz, M.; Nesi, N.; Schaeffer, J.; Ar Gouilh, M.; Plantier, J.C. Evidence of transmission and circulation of Deltacron XD recombinant SARS-CoV-2 in Northwest France. Clin. Infect. Dis. 2022, 10, ciac360. [Google Scholar] [CrossRef] [PubMed]
- Brinkac, L.; Diepold, S.; Mitchell, S.; Sarnese, S.; Kolakowski, L.F.; Nelson, W.M.; Jennings, K. SARS-CoV-2 Delta variant isolates from vaccinated individuals. BMC Genom. 2022, 23, 417. [Google Scholar] [CrossRef]
- Luo, R.; Delaunay-Moisan, A.; Timmis, K.; Danchin, A. SARS-CoV-2 biology and variants: Anticipation of viral evolution and what needs to be done. Environ. Microbiol. 2021, 23, 2339–2363. [Google Scholar] [CrossRef]
- Bouwman, K.M.; Parsons, L.M.; Berends, A.J.; De Vries, R.P.; Cipollo, J.F.; Verheije, M.H. Three amino acid changes in avian coronavirus spike protein allow binding to kidney tissue. J. Virol. 2020, 94, e01363-19. [Google Scholar] [CrossRef] [Green Version]
- Pasick, J.; Kalicharran, K.; Dales, S. Distribution and trafficking of JHM coronavirus structural proteins and virions in primary neurons and the OBL-21 neuronal cell line. J. Virol. 1994, 68, 2915–2928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Zhang, M.; Garcia Jr, G.; Tian, E.; Cui, Q.; Chen, X.; Sun, G.; Wang, J.; Arumugaswami, V.; Shi, Y. ApoE-isoform-dependent SARS-CoV-2 neurotropism and cellular response. Cell Stem Cell 2021, 28, 331–342.e5. [Google Scholar] [CrossRef] [PubMed]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Bullen, C.K.; Hogberg, H.T.; Bahadirli-Talbott, A.; Bishai, W.R.; Hartung, T.; Keuthan, C.; Looney, M.M.; Pekosz, A.; Romero, J.C.; Sillé, F.C. Infectability of human Brain Sphere neurons suggests neurotropism of SARS-CoV-2. ALTEX-Altern. Anim. Exp. 2020, 37, 665–671. [Google Scholar]
- Coutard, B.; Valle, C.; de Lamballerie, X.; Canard, B.; Seidah, N.G.; Decroly, E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020, 176, 104742. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Chen, W.; Zhang, Z.; Deng, Y.; Lian, J.Q.; Du, P.; Wei, D.; Zhang, Y.; Sun, X.X.; Gong, L. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 2020, 5, 283. [Google Scholar] [CrossRef] [PubMed]
- Merolli, A.; Kasaei, L.; Ramasamy, S.; Kolloli, A.; Kumar, R.; Subbian, S.; Feldman, L.C. An intra-cytoplasmic route for SARS-CoV-2 transmission unveiled by Helium-ion microscopy. Sci. Rep. 2022, 12, 3794. [Google Scholar] [CrossRef] [PubMed]
- Sardu, C.; D’Onofrio, N.; Balestrieri, M.L.; Barbieri, M.; Rizzo, M.R.; Messina, V.; Maggi, P.; Coppola, N.; Paolisso, G.; Marfella, R. Outcomes in patients with hyperglycemia affected by COVID-19: Can we do more on glycemic control? Diabetes Care 2020, 43, 1408–1415. [Google Scholar] [CrossRef] [PubMed]
- Sardu, C.; Gargiulo, G.; Esposito, G.; Paolisso, G.; Marfella, R. Impact of diabetes mellitus on clinical outcomes in patients affected by Covid-19. Cardiovasc. Diabetol. 2020, 19, 76. [Google Scholar] [CrossRef]
- Marfella, R.; D’Onofrio, N.; Sardu, C.; Scisciola, L.; Maggi, P.; Coppola, N.; Romano, C.; Messina, V.; Turriziani, F.; Siniscalchi, M.; et al. Does poor glycaemic control affect the immunogenicity of the COVID-19 vaccination in patients with type 2 diabetes: The CAVEAT study. Diabetes Obes. Metab. 2022, 24, 160–165. [Google Scholar] [CrossRef] [PubMed]
- International Diabetes Atlas. Available online: https://diabetesatlas.org/atlas/tenth-edition/ (accessed on 20 July 2022).
- Nordström, P.; Ballin, M.; Nordström, A. Risk of infection, hospitalisation, and death up to 9 months after a second dose of COVID-19 vaccine: A retrospective, total population cohort study in Sweden. Lancet 2022, 399, 814–823. [Google Scholar] [CrossRef]
- Reuschl, A.K.; Thorne, L.G.; Whelan, M.V.X.; Mesner, D.; Ragazzini, R.; Dowgier, G.; Bogoda, N.; Turner, J.L.E.; Furnon, W.; Cowton, V.M.; et al. Enhanced innate immune suppression by SARS-CoV-2 Omicron subvariants BA.4 and BA.5. Biorxiv 2022. [Google Scholar] [CrossRef]
- Rubio-Casillas, A.; Redwan, E.M.; Uversky, V.N. SARS-CoV-2: A Master of Immune Evasion. Biomedicines 2022, 10, 1339. [Google Scholar] [CrossRef] [PubMed]
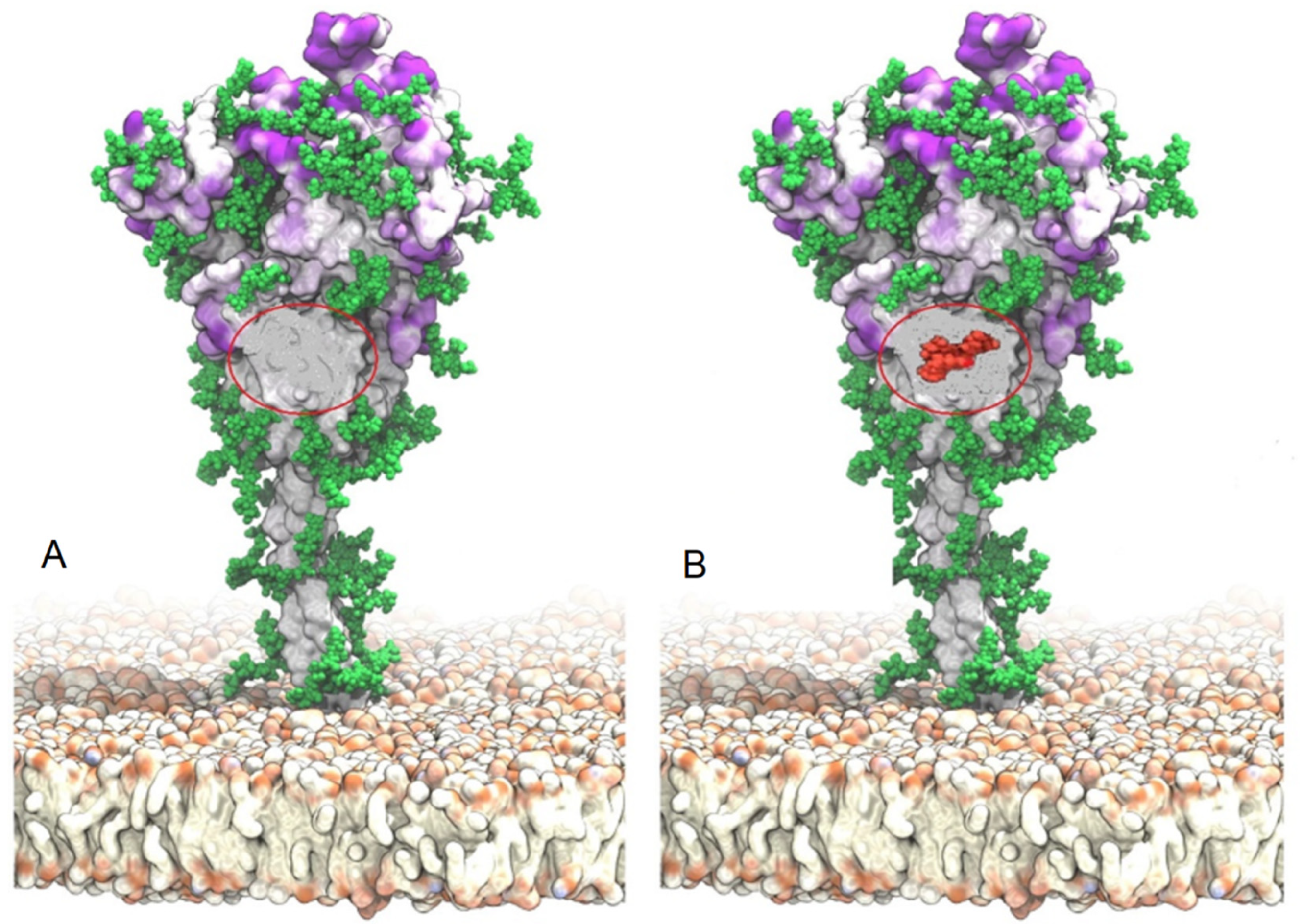
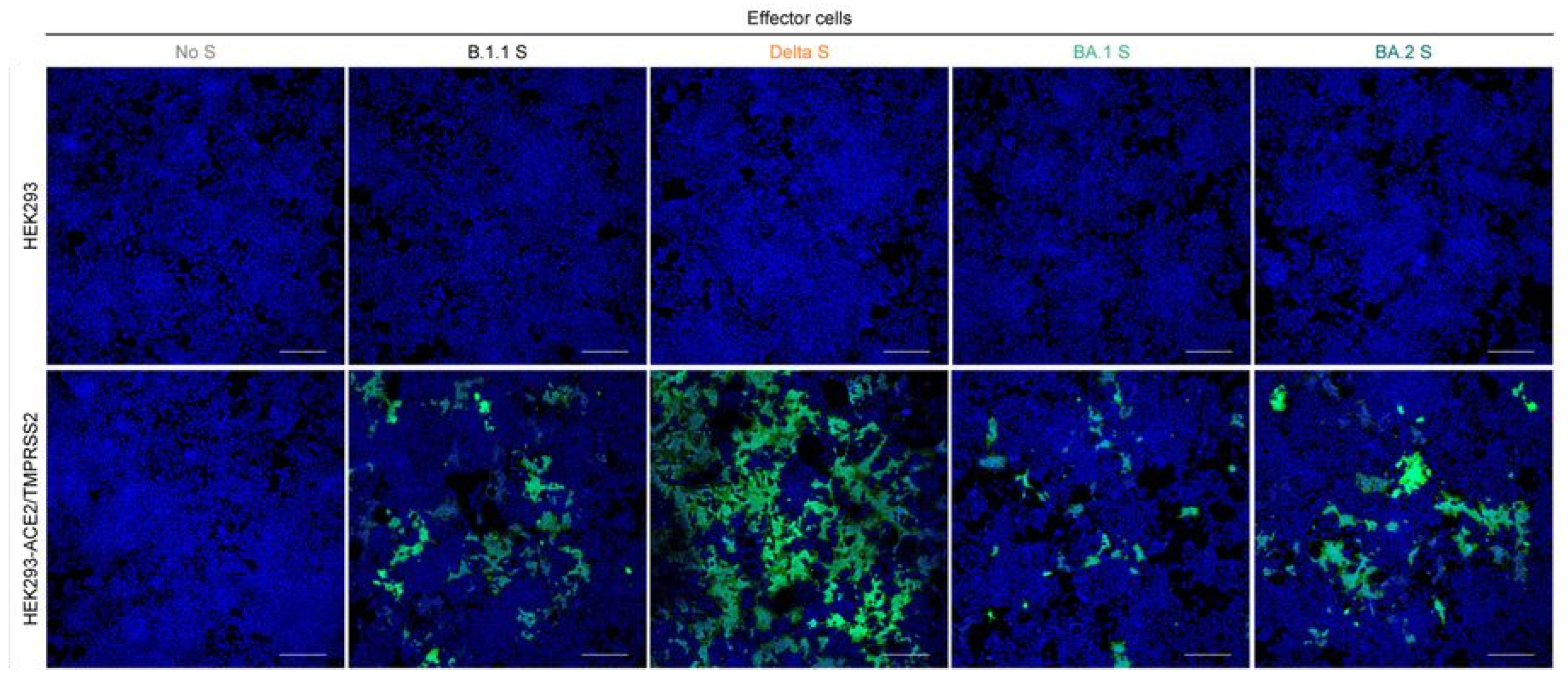
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rubio-Casillas, A.; Redwan, E.M.; Uversky, V.N. SARS-CoV-2 Intermittent Virulence as a Result of Natural Selection. COVID 2022, 2, 1089-1101. https://doi.org/10.3390/covid2080080
Rubio-Casillas A, Redwan EM, Uversky VN. SARS-CoV-2 Intermittent Virulence as a Result of Natural Selection. COVID. 2022; 2(8):1089-1101. https://doi.org/10.3390/covid2080080
Chicago/Turabian StyleRubio-Casillas, Alberto, Elrashdy M. Redwan, and Vladimir N. Uversky. 2022. "SARS-CoV-2 Intermittent Virulence as a Result of Natural Selection" COVID 2, no. 8: 1089-1101. https://doi.org/10.3390/covid2080080
APA StyleRubio-Casillas, A., Redwan, E. M., & Uversky, V. N. (2022). SARS-CoV-2 Intermittent Virulence as a Result of Natural Selection. COVID, 2(8), 1089-1101. https://doi.org/10.3390/covid2080080





