Risk Markers of COVID-19, a Study from South-Lebanon
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Setting
2.2. Sample Collection and Transportation
2.3. RNA Extraction and SARS-CoV-2 Detection by qRT-PCR
3. Statistical Analysis
4. Results
4.1. Demographic Data of All Patients
4.2. Prevalence of SARS-CoV-2 Infection and the Distribution of Positive Cases Regarding Different Parameters
| Test Result | ||||||
|---|---|---|---|---|---|---|
| Total | Negative | Positive * | p-Value | |||
| Age group | n (%) | 95% CI | n (%) | 95% CI | <0.001 | |
| Children 0–9 years | 3172 | 2723 (85.8) | (84.59–87.01) | 449 (14.2) | (12.99–15.41) | |
| Adolescent 10–19 | 5602 | 4276 (76.3) | (75.19–77.41) | 1326 (23.7) | (22.59–24.81) | |
| Adult 20–39 | 35,217 | 28,540 (81.0) | (80.59–81.41) | 6677 (19.0) | (18.59–19.41) | |
| Mature 40–69 | 21,392 | 17,168 (80.3) | (79.77–80.83) | 4224 (19.7) | (19.17–20.23) | |
| Elderly 70+ | 3621 | 3015 (83.3) | (82.08–84.52) | 606 (16.7) | (15.48–17.92) | |
| Total | 69,004 | 55,722 (80.8) | 13,282 (19.2) | |||
| Gender | n (%) | 95% CI | n (%) | 95% CI | 0.398 | |
| Male | 35,288 | 28,452 (80.6) | (80.19–81.01) | 6836 (19.4) | (18.99–19.81) | |
| Female | 33,727 | 27,279 (80.9) | (80.48–81.32) | 6448 (19.1) | (18.68–19.52) | |
| Total | 69,015 | 55,731 (80.8) | 13,284 (19.2) | |||
| Address | n (%) | 95% CI | n (%) | 95% CI | <0.001 | |
| Central area | 3065 | 2469 (80.6) | (79.20–82.00) | 596 (19.4) | (18.00–20.80) | |
| Close area | 19,167 | 15,206 (79.3) | (78.73–79.87) | 3961 (20.7) | (20.13–21.27) | |
| Remote area | 7828 | 6448 (82.4) | (81.56–83.24) | 1380 (17.6) | (16.76–18.44) | |
| Total | 30,060 | 24,123 (80.2) | 5937 (19.8) | |||
| ABO Blood group | n (%) | 95% CI | n (%) | 95% CI | 0.044 | |
| O | 6791 | 5752 (84.7) | (83.84–85.56) | 1039 (15.3) | (14.44–16.16) | |
| A | 6646 | 5529 (83.2) | (82.30–84.10) | 1117 (16.8) | (15.90–17.70) | |
| B | 2966 | 2469 (83.2) | (81.85–84.55) | 497 (16.8) | (15.45–18.15) | |
| AB | 1059 | 872 (82.3) | (80.00–84.60) | 187 (17.7) | (15.40–20.00) | |
| Total | 17,462 | 14,622 (83.7) | 2840 (16.3) | |||
| Rhesus Group | n (%) | 95% CI | n (%) | 95% CI | 0.831 | |
| Rh− | 1878 | 1569 (83.5) | (81.82–85.18) | 309 (16.5) | (14.82–18.18) | |
| Rh+ | 15,584 | 13,053 (83.8) | (83.22–84.38) | 2531 (16.2) | (15.62–16.78) | |
| Total | 17,462 | 14,622 (83.7) | 2840 (16.3) | |||
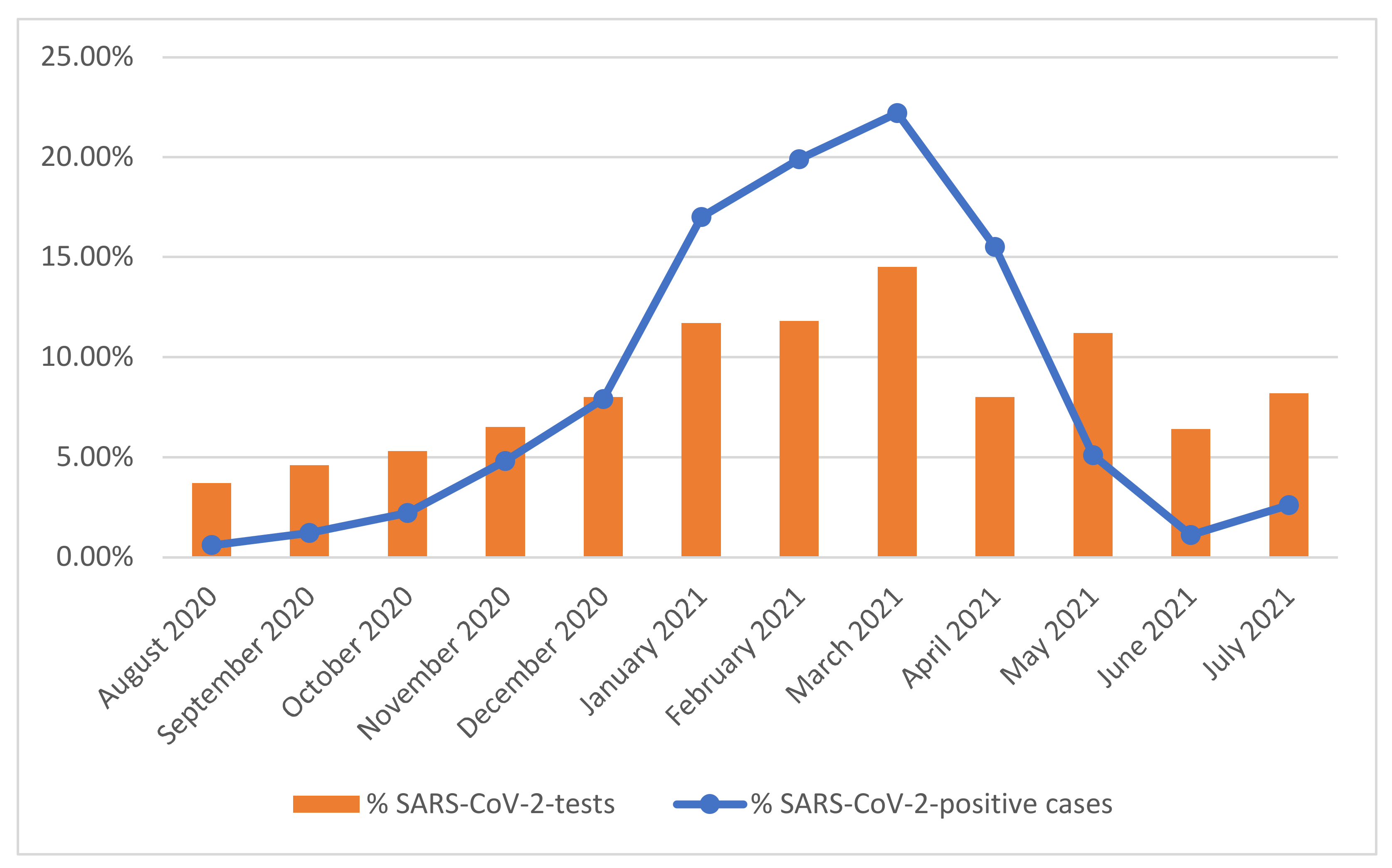
| Months | n (%) | 95% CI | n (%) | 95% CI | <0.001 | |
| August 2020 | 2571 | 2493 (97.0) | (96.34–97.66) | 78 (3.0) | (2.34–3.66) | |
| September 2020 | 3164 | 3007 (95.0) | (94.24–95.76) | 157 (5.0) | (4.24–5.76) | |
| October 2020 | 3670 | 3377 (92.0) | (91.12–92.88) | 293 (8.0) | (7.12–8.88) | |
| November 2020 | 4467 | 3833 (85.8) | (84.78–86.82) | 634 (14.2) | (13.18–15.22) | |
| December 2020 | 5528 | 4479 (81.0) | (79.97–82.03) | 1049 (19.0) | (17.97–20.03) | |
| January 2021 | 8085 | 5829 (72.1) | (71.12–73.08) | 2256 (27.9) | (26.92–28.88) | |
| February 2021 | 8163 | 5517 (67.6) | (66.58–68.62) | 2646 (32.4) | (31.38–33.42) | |
| March 2021 | 9993 | 7041 (70.5) | (69.61–71.39) | 2952 (29.5) | (28.61–30.39) | |
| April 2021 | 7744 | 5688 (73.5) | (72.52–74.48) | 2056 (26.5) | (25.52–27.48) | |
| May 2021 | 5556 | 4884 (87.9) | (87.04–88.76) | 672 (12.1) | (11.24–12.96) | |
| June 2021 | 4387 | 4238 (96.6) | (96.06–97.14) | 149 (3.4) | (2.86–3.94) | |
| July 2021 | 5688 | 5345 (94.0) | (93.38–94.62) | 343 (6.0) | (5.38–6.62) | |
| Total | 69,016 | 55,731 (80.8) | 13,285 (19.2) | |||
4.3. Risk Markers of COVID-19
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Noureddine, F.Y.; Chakkour, M.; El Roz, A.; Reda, J.; Al Sahily, R.; Assi, A.; Joma, M.; Salami, H.; Hashem, S.J.; Harb, B.; et al. The Emergence of SARS-CoV-2 Variant(s) and Its Impact on the Prevalence of COVID-19 Cases in the Nabatieh Region, Lebanon. Med. Sci. 2021, 9, 40. [Google Scholar] [CrossRef] [PubMed]
- WHO. Coronavirus Disease 2019 (COVID-19) Weekly Epidemiological Update 75. Weekly Epidemiological Update on COVID-19–18 January 2022. Available online: who.int (accessed on 18 January 2022).
- WHO. Lebanon: Who Coronavirus Disease (COVID-19) Dashboard with Vaccination Data. World Health Organization. Available online: https://covid19.who.int/region/emro/country/lb (accessed on 21 January 2022).
- Zheng, Z.; Peng, F.; Xu, B.; Zhao, J.; Liu, H.; Peng, J.; Li, Q.; Jiang, C.; Zhou, Y.; Liu, S.; et al. Risk factors of critical & mortal COVID-19 cases: A systematic literature review and meta-analysis. J. Infect. 2020, 81, e16–e25. [Google Scholar] [CrossRef]
- Cheng, Y.; Cheng, G.; Chui, C.H.; Lau, F.Y.; Chan, P.K.; Ng, M.H.; Sung, J.J.; Wong, R.S. ABO blood group and susceptibility to severe acute respiratory syndrome. JAMA 2005, 293, 1450–1451. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yang, Y.; Huang, H.; Li, D.; Gu, D.; Lu, X.; Zhang, Z.; Liu, L.; Liu, T.; Liu, Y.; et al. Relationship Between the ABO Blood Group and the Coronavirus Disease 2019 (COVID-19) Susceptibility. Clin. Infect. Dis. 2021, 73, 328–331. [Google Scholar] [CrossRef] [PubMed]
- Ellinghaus, D.; Degenhardt, F.; Bujanda, L.; Buti, M.; Albillos, A.; Invernizzi, P.; Fernandez, J.; Prati, D.; Baselli, G.; Asselta, R.; et al. Genomewide Association Study of Severe COVID-19 with Respiratory Failure. N. Engl. J. Med. 2020, 383, 1522–1534. [Google Scholar] [CrossRef] [PubMed]
- Zietz, M.; Zucker, J.; Tatonetti, N.P. Associations between blood type and COVID-19 infection, intubation, and death. Nat. Commun. 2020, 11, 5761. [Google Scholar] [CrossRef]
- Barnkob, M.B.; Pottegard, A.; Stovring, H.; Haunstrup, T.M.; Homburg, K.; Larsen, R.; Hansen, M.B.; Titlestad, K.; Aagaard, B.; Moller, B.K.; et al. Reduced prevalence of SARS-CoV-2 infection in ABO blood group O. Blood Adv. 2020, 4, 4990–4993. [Google Scholar] [CrossRef]
- Solmaz, I.; Arac, S. ABO blood groups in COVID-19 patients; Cross-sectional study. Int. J. Clin. Pract. 2021, 75, e13927. [Google Scholar] [CrossRef]
- Adhiah, A.; Abdullah, M.; Alsudani, M.; Shnawa, R.; Al-Saady, A.; Allami, R.; Mishaal, K.; Jassim, I.; Taqi, E. Association between ABO blood groups and susceptibility to COVID-19: Profile of age and gender in Iraqi patients. Egypt. J. Med. Hum. Genet. 2020, 21, 76. [Google Scholar] [CrossRef]
- Scott, R.A.; Scott, L.J.; Magi, R.; Marullo, L.; Gaulton, K.J.; Kaakinen, M.; Pervjakova, N.; Pers, T.H.; Johnson, A.D.; Eicher, J.D.; et al. An Expanded Genome-Wide Association Study of Type 2 Diabetes in Europeans. Diabetes 2017, 66, 2888–2902. [Google Scholar] [CrossRef] [Green Version]
- Nikpay, M.; Goel, A.; Won, H.H.; Hall, L.M.; Willenborg, C.; Kanoni, S.; Saleheen, D.; Kyriakou, T.; Nelson, C.P.; Hopewell, J.C.; et al. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015, 47, 1121–1130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, C.M.; Wang, R.Y.; Chen, J.W.; Fann, C.S.; Leu, H.B.; Ho, H.Y.; Ting, C.T.; Lin, T.H.; Sheu, S.H.; Tsai, W.C.; et al. A genome-wide association study identifies new loci for ACE activity: Potential implications for response to ACE inhibitor. Pharm. J. 2010, 10, 537–544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reilly, M.P.; Li, M.; He, J.; Ferguson, J.F.; Stylianou, I.M.; Mehta, N.N.; Burnett, M.S.; Devaney, J.M.; Knouff, C.W.; Thompson, J.R.; et al. Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: Two genome-wide association studies. Lancet 2011, 377, 383–392. [Google Scholar] [CrossRef] [Green Version]
- Klarin, D.; Busenkell, E.; Judy, R.; Lynch, J.; Levin, M.; Haessler, J.; Aragam, K.; Chaffin, M.; Haas, M.; Lindstrom, S.; et al. Genome-wide association analysis of venous thromboembolism identifies new risk loci and genetic overlap with arterial vascular disease. Nat. Genet. 2019, 51, 1574–1579. [Google Scholar] [CrossRef] [PubMed]
- Anstee, D.J. The relationship between blood groups and disease. Blood 2010, 115, 4635–4643. [Google Scholar] [CrossRef] [Green Version]
- Flegr, J. Influence of latent Toxoplasma infection on human personality, physiology and morphology: Pros and cons of the Toxoplasma-human model in studying the manipulation hypothesis. J. Exp. Biol. 2013, 216, 127–133. [Google Scholar] [CrossRef] [Green Version]
- Ray, J.G.; Schull, M.J.; Vermeulen, M.J.; Park, A.L. Association Between ABO and Rh Blood Groups and SARS-CoV-2 Infection or Severe COVID-19 Illness: A Population-Based Cohort Study. Ann. Intern. Med. 2021, 174, 308–315. [Google Scholar] [CrossRef]
- Kerbage, A.; Haddad, S.F.; Nasr, L.; Riachy, A.; Mekhael, E.; Nassim, N.; Hoyek, K.; Sleilaty, G.; Nasr, F.; Riachy, M. Impact of ABO and Rhesus blood groups on COVID-19 susceptibility and severity: A case-control study. J. Med. Virol. 2021, 94, 1162–1166. [Google Scholar] [CrossRef]
- Bakhos, J.J.; Khalife, M.; Teyrouz, Y.; Saliba, Y. Blood Donation in Lebanon: A Six-Year Retrospective Study of a Decentralized Fragmented Blood Management System. Cureus 2022, 14, e21858. [Google Scholar] [CrossRef]
- Shibeeb, S.; Khan, A. ABO blood group association and COVID-19. COVID-19 susceptibility and severity: A review. Hematol. Transfus. Cell Ther. 2022, 44, 70–75. [Google Scholar] [CrossRef]
- Guillon, P.; Clement, M.; Sebille, V.; Rivain, J.G.; Chou, C.F.; Ruvoen-Clouet, N.; Le Pendu, J. Inhibition of the interaction between the SARS-CoV spike protein and its cellular receptor by anti-histo-blood group antibodies. Glycobiology 2008, 18, 1085–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miotto, M.; Di Rienzo, L.; Gosti, G.; Milanetti, E.; Ruocco, G. Does blood type affect the COVID-19 infection pattern? PLoS ONE 2021, 16, e0251535. [Google Scholar] [CrossRef] [PubMed]
- Gerard, C.; Maggipinto, G.; Minon, J.M. COVID-19 and ABO blood group: Another viewpoint. Br. J. Haematol. 2020, 190, e93–e94. [Google Scholar] [CrossRef]
- Zhang, Y.; Garner, R.; Salehi, S.; La Rocca, M.; Duncan, D. Association between ABO blood types and coronavirus disease 2019 (COVID-19), genetic associations, and underlying molecular mechanisms: A literature review of 23 studies. Ann. Hematol. 2021, 100, 1123–1132. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, F.Z.; Zaidi, A.R.Z.; Abdullah, S.M.; Zaidi, S.Z.A. COVID-19 and the ABO blood group connection. Transfus. Apher. Sci. 2020, 59, 102838. [Google Scholar] [CrossRef]
- Yamamoto, F.; Yamamoto, M.; Muniz-Diaz, E. Blood group ABO polymorphism inhibits SARS-CoV-2 infection and affects COVID-19 progression. Vox Sang. 2021, 116, 15–17. [Google Scholar] [CrossRef]
- Malani, A.; Shah, D.; Kang, G.; Lobo, G.N.; Shastri, J.; Mohanan, M.; Jain, R.; Agrawal, S.; Juneja, S.; Imad, S.; et al. Seroprevalence of SARS-CoV-2 in slums versus non-slums in Mumbai, India. Lancet Glob. Health 2021, 9, e110–e111. [Google Scholar] [CrossRef]
- Lundkvist, A.; Hanson, S.; Olsen, B. Pronounced difference in COVID-19 antibody prevalence indicates cluster transmission in Stockholm, Sweden. Infect. Ecol. Epidemiol. 2020, 10, 1806505. [Google Scholar] [CrossRef]
- Franceschi, V.B.; Santos, A.S.; Glaeser, A.B.; Paiz, J.C.; Caldana, G.D.; Machado Lessa, C.L.; de Menezes Mayer, A.; Kuchle, J.G.; Gazzola Zen, P.R.; Vigo, A.; et al. Population-based prevalence surveys during the COVID-19 pandemic: A systematic review. Rev. Med. Virol. 2021, 31, e2200. [Google Scholar] [CrossRef]
- Ritchie, H.; Mathieu, E.; Rodés-Guirao, L.; Appel, C.; Giattino, C.; Ortiz-Ospina, E.; Hasell, J.; Macdonald, B.; Beltekian, D.; Roser, M. Lebanon: Coronavirus Pandemic Country Profile. Our World in Data. Available online: https://ourworldindata.org/coronavirus/country/lebanon (accessed on 27 March 2022).
- Mumtaz, G.; Jabbour, M.; Makhoul, M.; Harb, A.; El-Jardali, F. K2P COVID-19 Series: Modeling COVID-19 Vaccine Rollout in Lebanon for Better Impact (Full Version); Knowledge to Policy (K2P) Center: Beirut, Lebanon, 2021. [Google Scholar]
- Kemp, F.; Proverbio, D.; Aalto, A.; Mombaerts, L.; Fouquier d’Herouel, A.; Husch, A.; Ley, C.; Goncalves, J.; Skupin, A.; Magni, S. Modelling COVID-19 dynamics and potential for herd immunity by vaccination in Austria, Luxembourg and Sweden. J. Theor. Biol. 2021, 530, 110874. [Google Scholar] [CrossRef]
| Univariate Analysis | Multivariate Analysis | ||||
|---|---|---|---|---|---|
| Variable | Categories | OR (95% CI) | p-Value | OR (95% CI) | p-Value |
| Age group | Children 0–9 years | 1 | - | 1 | - |
| Adolescent 10–19 | 1.88 (1.67–2.12) | <0.001 | 1.94 (1.21–3.10) | 0.006 | |
| Adult 20–39 | 1.42 (1.28–1.57) | <0.001 | 2.03 (1.36–3.03) | 0.001 | |
| Mature 40–69 | 1.49 (1.34–1.66) | <0.001 | 1.90 (1.27–2.86) | 0.002 | |
| Elderly 70+ | 1.22 (1.07–1.39) | 0.003 | 1.58 (1.02–2.44) | 0.039 | |
| Gender | Male | 1 | - | ||
| Female | 0.984 (0.947–1.022) | 0.398 | |||
| ABO Blood Group | O | 1 | - | 1 | - |
| B | 1.11 (0.99–1.25) | 0.069 | |||
| A | 1.12 (1.02–1.23) | 0.017 | 1.09 (0.99–1.21) | 0.092 | |
| AB | 1.19 (1.00–1.41) | 0.049 | 1.15 (0.94–1.40) | 0.176 | |
| Rhesus Group | Rh+ | 1 | - | ||
| Rh− | 1.02 (0.89–1.16) | 0.814 | |||
| Address | Central Area | 1 | - | 1 | - |
| Close Area | 1.08 (0.98–1.19) | 0.120 | |||
| Remote Area | 0.89 (0.80–0.99) | 0.027 | 0.87 (0.78–0.98) | 0.018 | |
| Months | August 2020 | 0.19 (0.15–0.24) | <0.001 | 0.03 (0.01–0.12) | <0.001 |
| September 2020 | 0.32 (0.26–0.38) | <0.001 | 0.17 (0.10–0.29) | <0.001 | |
| October 2020 | 0.53 (0.45–0.61) | <0.001 | 0.49 (0.34–0.69) | <0.001 | |
| November 2020 | 1 | - | 1 | - | |
| December 2020 | 1.42 (1.27–1.58) | <0.001 | 1.25 (0.97–1.62) | 0.087 | |
| January 2021 | 2.34 (2.12–2.58) | <0.001 | 2.59 (2.06–3.25) | <0.001 | |
| February 2021 | 2.90 (2.63–3.19) | <0.001 | 3.39 (2.70–4.25) | <0.001 | |
| March 2021 | 2.54 (2.31–2.79) | <0.001 | 2.65 (2.12–3.32) | <0.001 | |
| April 2021 | 2.19 (1.98–2.41) | <0.001 | 2.23 (1.77–2.82) | <0.001 | |
| May 2021 | 0.83 (0.74–0.94) | <0.001 | 0.91 (0.69–1.20) | 0.495 | |
| June 2021 | 0.21 (0.18–0.26) | <0.001 | 0.18 (0.11–0.29) | <0.001 | |
| July 2021 | 0.39 (0.34–0.45) | <0.001 | 0.26 (0.17–0.39) | <0.001 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chakkour, M.; Salami, A.; Olleik, D.; Kamal, I.; Noureddine, F.Y.; Roz, A.E.; Ghssein, G. Risk Markers of COVID-19, a Study from South-Lebanon. COVID 2022, 2, 867-876. https://doi.org/10.3390/covid2070063
Chakkour M, Salami A, Olleik D, Kamal I, Noureddine FY, Roz AE, Ghssein G. Risk Markers of COVID-19, a Study from South-Lebanon. COVID. 2022; 2(7):867-876. https://doi.org/10.3390/covid2070063
Chicago/Turabian StyleChakkour, Mohamed, Ali Salami, Dana Olleik, Israa Kamal, Fatima Y. Noureddine, Ali El Roz, and Ghassan Ghssein. 2022. "Risk Markers of COVID-19, a Study from South-Lebanon" COVID 2, no. 7: 867-876. https://doi.org/10.3390/covid2070063
APA StyleChakkour, M., Salami, A., Olleik, D., Kamal, I., Noureddine, F. Y., Roz, A. E., & Ghssein, G. (2022). Risk Markers of COVID-19, a Study from South-Lebanon. COVID, 2(7), 867-876. https://doi.org/10.3390/covid2070063






