Abstract
The accurate detection of SARS-CoV-2 through respiratory sampling is critical for the prevention of further transmission and timely initiation of treatment. There is a diverse range of SARS-CoV-2 detection rates in reported studies, with uncertainty regarding the optimal sampling method for COVID-19 diagnosis and monitoring. Oropharyngeal sampling (OPS) is one of the most commonly used methods of respiratory sampling in Ghana and other parts of the world for the detection of SARS-CoV-2 viral RNA. However, this sampling technique has a number of drawbacks, which include difficulty in obtaining high-quality swab samples, increased risk of infection to healthcare workers, and increased cost from a regular supply of swabs, transport media, and personal protective equipment (PPE). This study, therefore, sought to evaluate the diagnostic performance of sputum specimens in the diagnosis of COVID-19. This was a cross-sectional analytical study conducted in two health facilities in Kumasi, Ghana, between April and September 2021. Paired samples (an oropharyngeal swab and sputum) were taken from each recruited patient and run concurrently for the detection of SARS-CoV-2 genes (the N and ORF1ab genes) using RT-qPCR. Of the 317 patients recruited, 50.8% were males, and 60.4% were young adults aged 20–39 years. A significant proportion (65.9%) of the patients did not have any co-morbidity, and the majority were with symptoms; predominantly cough (36.3%), headache (31.5%), general weakness (24.0%), fever (20.2%), and sore throat (16.1%). Being symptomatic (p = 0.003), having comorbidity (p = 0.001), and the reporting facility (p = 0.010) were significantly associated with the COVID-19 status. The sputum samples yielded more COVID-positive, 120/317 (37.9%), as compared to OPS, 83/317 (26.2%). The sputum samples were 85.5% (95% CI, 76.4–91.5) sensitive, 79.1% (95% CI, 73.4–83.7) specific, and with positive and negative predictive values of 59.2% and 93.9%, respectively, when compared with OPS. The overall median of the SARS-CoV-2 viral loads for sputum (3.70 × 103 copies/mL) were significantly higher than in OPS (1.18 × 102 copies/mL) (p = 0.003). Findings from the study suggest self-collected sputum as a useful alternative to OPS for the diagnosis of COVID-19, providing a comparable diagnostic performance and, thereby, easing the uncomfortable process and mitigating risk of aerosol transmission to healthcare workers.
1. Introduction
The emergence and spread of SARS-CoV-2 have caused major havoc to global health systems, and its economic impact cannot be underestimated. The WHO named it a pandemic in March 2020, owing to its global spread [1]. As of 31 January 2022, there have been 373,229,380 confirmed cases of COVID-19 worldwide, with over 5.5 million deaths. Europeans have the most confirmed cases (143,224,359), followed by the Americas (135,082,663 cases), South-East Asia (52,155,418 cases), the Eastern Mediterranean (18,933,395), the Western Pacific (15,767,915 cases), and Africa (8,064,866 cases) [2].
Relative to other continents, sub-Sahara Africa has escaped major mayhem in terms of a viral spread and death rate. Ghana, located in West Africa, has currently reported 156,920 cases with 1349 deaths as of January 2022 [3]. With the ongoing spread of the SARS-CoV-2 infection in Ghana and throughout the world, efficient laboratory testing is critical to prompt the detection of positive cases and management strategies, rather than solely depending on clinical empirical evidence [4].
COVID-19 has been diagnosed in the laboratory mostly through the detection of SARS-CoV-2 by the reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) in lower or upper respiratory samples, including sputum, nasopharyngeal wash/aspirate, oropharyngeal, and nasopharyngeal swab samples [5]. With the goal of enhancing the scalability of SARS-CoV-2 testing under heightened demand, several alternative sampling approaches have been explored. Notable among them include anal swabs, saliva, blood, urine, and even pooled nasal and oropharyngeal swabs [6,7]. According to the literature, these sample types have yielded varying diagnostic characteristics in terms of sensitivity and specificity [8]. Currently, nasopharyngeal and oropharyngeal swabs are widely used and recommended for the clinical detection of SARS-CoV-2 [9,10]. However, these swabs could pose a potential risk of causing acute injuries, such as bleeding and mucosal layer ulceration, particularly in patients with predisposing factors [11,12]. Additionally, health workers are exposed to the danger of infection as they collect such invasive samples [12]. There are also reports of decreased test sensitivity of OPS for the detection of SARS-CoV-2 [13]. This ultimately calls for a search for an alternative sampling approach(es) with invariably good to excellent clinical sensitivity. This study, therefore, sought to compare the diagnostic performance of self-produced sputum to an oropharyngeal swab for the diagnosis of COVID-19.
2. Materials and Methods
2.1. Ethics Statement
The study’s protocol was reviewed and approved by the Committee on Human Research Publication and Ethics (CHRPE) of the School of Medicine and Dentistry, Kwame Nkrumah University of Science and Technology (KNUST) Kumasi, Ghana (Approval number: CHRPE/AP/076/21).
2.2. Study Area
The Suntreso Government Hospital (SGH) and the Kwadaso Seventh-Day Adventist Hospital (KWA), both situated within the Kumasi, Ashanti region (Ghana), were the recruitment sites for this study. Kumasi is the second-largest and most populous city in the country, with a population of 3,353,850 people as of 2021 and harboring over 200 health facilities [14].
SGH is a district hospital under the Ghana Health Service located in the North Suntreso district of the Kumasi metropolis. The hospital provides secondary health care not only to its district but to all other neighbouring communities. KWA, on the other hand, is the largest Adventist hospital in the region with an affiliation with the Ghana Adventist Health Service (GAHS) and the Christian Health Association of Ghana (CHAG) under the Ministry of Health, Ghana. It is located in the Kwadaso district and provides quality healthcare to several communities within Kwadaso and beyond while also serving as a referral hospital for some government and private health centres in the Atwima Nwabiagya and Atwima Nkwahoma districts. Both recruitment sites are 2.6 km apart (an approximately 10min drive) and were part of the most visited hospitals in the metropolis at the peak of the COVID-19 pandemic.
The Kumasi Centre for Collaborative Research in Tropical Medicine (KCCR) of KNUST was the site where all of the laboratory procedures were conducted for this study. KCCR, also located in Kumasi, Ghana, was established as a joint venture between the stakeholders of the Ministry of Health of the Republic of Ghana, KNUST, and the Bernhard Nocht Institute of Tropical Medicine (BNITM) in Germany. It is equipped with various laboratories and is currently the second-largest COVID-19 testing site in the country, with approximately 800 tests per day [15].
2.3. Study Design and Sample Collection
This was a cross-sectional study that was conducted between April and September 2021. Samples were collected from patients aged 10 years and above and who had the ability to expectorate sputum. Prior to recruitment, written informed consent was obtained from all participants, including children (below 18 years) for whom parental consent was sought. Paired samples (sputum and OPS) were taken from each of the suspected patients recruited in this study. Oropharyngeal sampling (OPS) was taken with a sterile swab (BioTeke Corporation, ST9001-1 model) by first getting a clear view of the throat using a tongue depressor and swabbing the posterior oropharynx, tonsillar area, and other inflamed areas whilst rotating it several times. The swab was then kept in a sterile viral transport medium. Sputum was self-produced by expectorating into sterile screw-cap containers. The samples were transported in a cold box (+4 to +8 °C) within an hour of collection in a triple package to the Kumasi Centre for Collaborative Research in Tropical Medicine (KCCR) for laboratory analysis.
2.4. Laboratory Analysis
RNA was extracted from the sputum and OPS specimens using the LBP nucleic acid extraction and purification kit, series C (Guangzhou LBP Medical Science and Technology Co., Ltd., Guangzhou, China) and using the spin column method. Five microlitres (5 uL) of the extracted viral RNA were tested using a one-step reverse transcriptase quantitative polymerase chain reaction (RT-qPCR); technology targeting the open reading frame 1ab gene and N gene of SARS-CoV-2. RT-qPCR was performed with a LineGene 9600 Plus (FQD-96A, Bioer Technology, China) thermocycler using the following cycling conditions: 50 °C for 15 mins (1 cycle), denaturation 95 °C for 15 mins (1cycle), and 45 cycles each at 94 °C and 55 °C for 15 s and 45 s, respectively, (Da An gene PCR kit, DA-TF/nCoV-D/002, Guangzhou, China) for all tests. The viral load was extrapolated from a standard curve generated from plotting known viral concentrations against the cycle threshold (Ct) values of a single target. A positive and negative control were added to every PCR run for validation.
2.5. Statistical Analysis
Data were entered and cleaned in Microsoft Excel 2016. Statistical analysis was done using the Statistical Package for the Social Sciences (SPSS) version 26 and GraphPad Prism version 8 (GraphPad software, San Diego, CA, USA). The chi-square test was used to determine the association between socio-demographic, clinical parameters, and COVID-19 status. Contingency tables were used to determine the sensitivity, specificity, and predictive values of the sputum samples compared to the oropharyngeal samples. Comparative analysis of viral load genes was performed by the Mann–Whitney U and Wilcoxon matched-pairs signed-rank test. p-values of less than 0.05 were considered statistically significant for all analyses.
3. Results
3.1. Sociodemographic and Clinical Characteristics of Study Participants
Three hundred and seventeen (317) suspected COVID-19 patients were recruited into the study from the two study sites: KWA (166; 52.4%) and SGH (151; 47.6%). Most of the subjects were predominantly young persons aged 20 to 39 years (60.4%), with slightly more males (50.8%) than females (49.2%). More than half of the total number (65.9%) did not have any comorbidity. A total of N (70%) had some form of occupation, while the rest were either unemployed or retired. The majority of the patients (64%) presented with symptoms, and 31.9% were asymptomatic. The most prevalent symptoms among the patients were cough (36.3%), headache (31.5%), general weakness (24.0%), fever (20.2%), and sore throat (16.1%). Being symptomatic (p = 0.003) and having comorbidity (p = 0.001) were significantly associated with their COVID-19 status. Also, coughing (p = 0.006), runny nose (p = 0.011), fever (p < 0.0001), nausea or vomiting (p = 0.018), general weakness (p = 0.002), loss of smell or taste (p = 0.003), and diarrhea (p = 0.001) were significantly associated with their COVID-19 status (Table 1).

Table 1.
Factors associated with COVID-19 status.
3.2. Biodistribution and Diagnostic Accuracy of SARS-CoV-2 in Sputum and Oropharyngeal Swab Samples
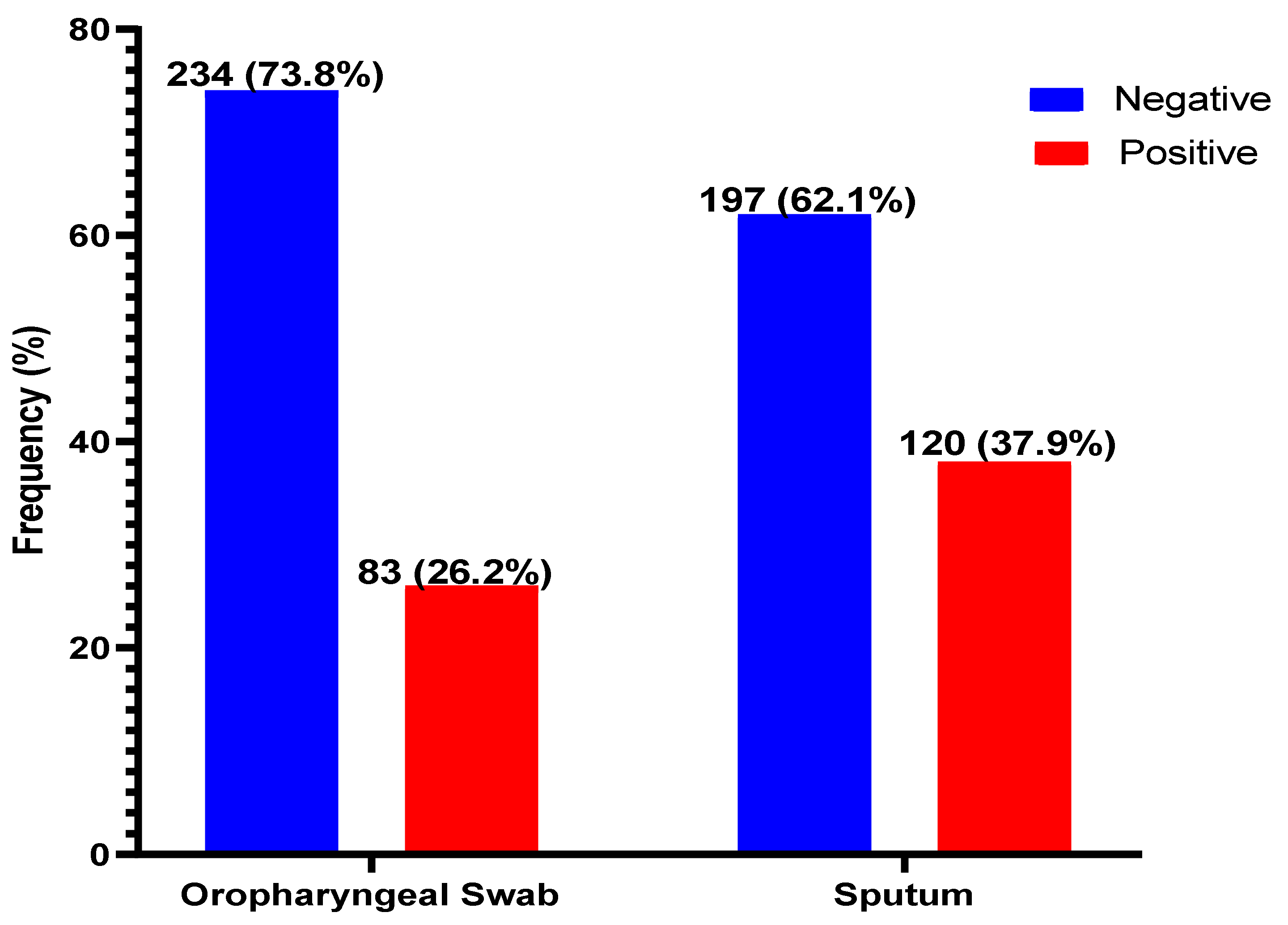
Of the 317 suspected patients sampled, sputum produced the most positives, 120/317 (37.9%), with oropharyngeal producing 83/317 (26.2%) positivity for COVID-19 (Figure 1). The sputum samples were 85.5% (95% CI, 76.4–91.5) sensitive, 79.1% (95% CI, 73.4–83.7) specific, and with positive and negative predictive values of 59.2% and 93.9%, respectively, when compared with OPS (Table 2).
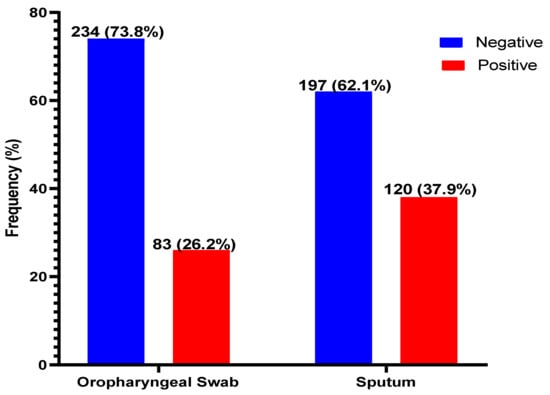
Figure 1.
Distribution of SARS-CoV-2 in oropharyngeal and sputum samples. SARS-CoV-2 was detected in both the sputum and OPS of 132 (41.6%) patients. However, the distribution of positives and negatives by either of the samples are shown.

Table 2.
Diagnostic comparison of sputum and oropharyngeal swab.
3.3. Comparison of Viral Load between Oropharyngeal and Sputum Samples
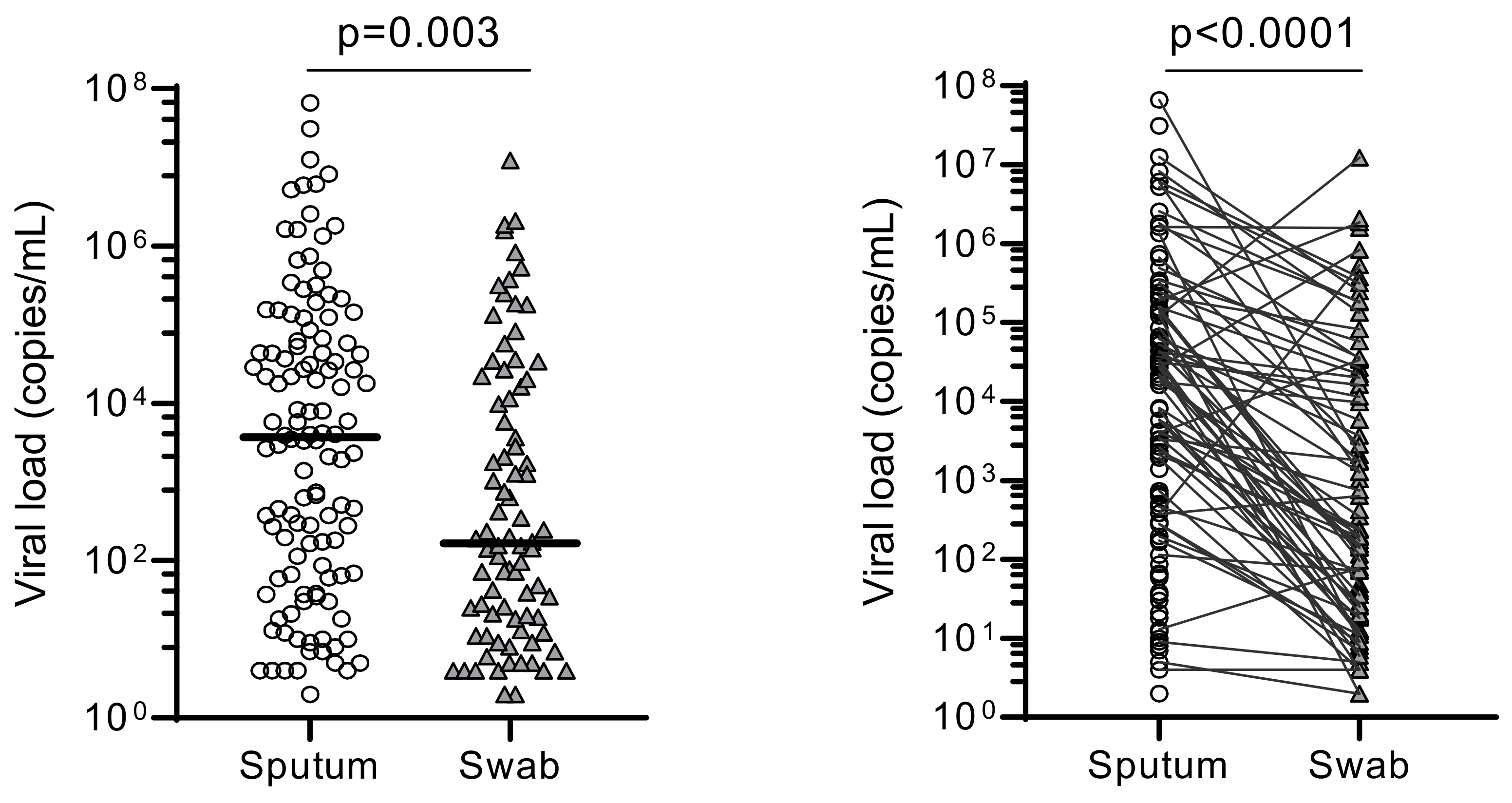
The overall median of the SARS-CoV-2 viral loads for sputum (3.70 × 103 copies/mL) were significantly higher than in OPS (1.18 × 102 copies/mL) (p = 0.003) (Figure 2).
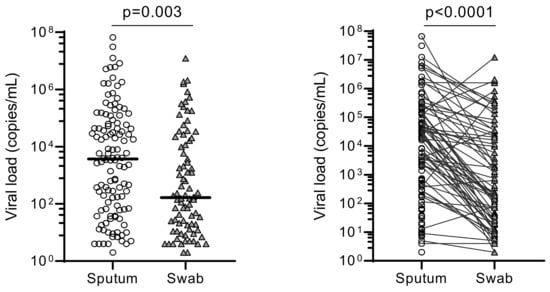
Figure 2.
Comparison of overall viral loads between sputum and oropharyngeal samples. The overall viral loads (“copies/mL”) obtained using sputum and swab (OPS) were statistically compared using their median viral load (p = 0.003). Pair-wise comparison was also made between them (p < 0.0001).
4. Discussion
This study showed that the presence of comorbidity and clinical symptoms have a direct association with disease status, as reported in several studies in Ghana [16,17]. The majority of the patients showed symptoms (64%), predominantly with cough, headache, general weakness, fever, and sore throat, among others; this is a finding similar to other studies [17,18]. It has been reported by Adjei et al. (2020) that fever is the third commonest symptom seen in COVID-19 patients in Greater Accra (Ghana); however, in our study in Kumasi (Ghana), it was the fourth most common symptom. Patients with cough, general weakness, runny nose, anosmia, fever, diarrhea, and nausea were also most likely to have COVID-19. Studies in other countries have also reported varying prevalent symptoms [19,20].
The literature has shown that COVID-19 affects the elderly [17,20,21,22] and, contrarily, the majority of suspected patients who tested positive in this study were in their 30 s and 20 s. This may be attributed to the design of the study, as most walk-in patients were young in age and/or because such age groups are more active and engage themselves in outdoor activities and social gatherings, making themselves more vulnerable to infection. This is in concordance with another study in Ghana, which attributed such an occurrence to increased outdoor engagement and decreased adherence to safety protocols by individuals of a younger age [16]. Even so, general reports worldwide show that the case fatality rate (CFR) is higher in the elderly and people with underlying medical conditions [23]. Despite the fact that there were more males than females in attendance, infection in females was slightly higher, which is contrary to reports from other studies where males were found to be more affected than females [18,19,20]. Although the majority of the study population (209; 65.9%) had no SARS-CoV-2 comorbid condition, there was a significant proportion with an unknown comorbidity history (89; 28.1%). This alarming rate of unknowns was reported due to the subjects’ low level of knowledge about their medical history. It is important for patients to be aware of their medical history, especially in the era of COVID-19, as comorbidities have regularly been cited as important risk factors for severe outcomes [24].
The sensitivity, specificity, and predictive values for sputum were compared to the oropharyngeal samples for the COVID-19 diagnosis in this study. Sputum was found to be sensitive and specific for SARS-CoV-2 detection, giving a “sensitivity + specificity” value of greater than 1.5 [25]. Suspected patients who tested negative with sputum can be appropriately identified as not having the disease with a 93.9 percent NPV, the type of predictive value required in diagnosis during an outbreak. Lower respiratory tract samples have been shown to produce the most positives when compared with other clinical samples used for COVID-19 diagnosis [22,26]. The same observation was seen in this study, as the sputum samples produced more positive results than the oropharyngeal samples when paired samples were taken. This is most likely due to the abundance of the angiotensin-converting enzyme-2 (ACE2) receptors in the pneumocytes and epithelial cells of the lower respiratory airway compared to the epithelial cells in the upper airway [27,28].
Sputum is better than oropharyngeal sampling not only in the detection of SARS-CoV-2, but it also outperforms it in the detection of non-COVID respiratory viruses [29,30]. These findings point to the possibility that just collecting oropharyngeal samples may lead to a misdiagnosis of COVID-19 and an incorrect assumption of the SARS-CoV-2 viral clearance, which influences how long a person is isolated from the rest of society and when they can return to work. There are also implications for clinical diagnosis and early treatment; hence, it is recommended that clinicians with a high index of suspicion for COVID-19 repeat negative OPS results or request sputum samples from suspected COVID-19 patients. This reaffirms the use of self-collected sputum as an alternative to oropharyngeal sampling in COVID-19 diagnosis. Sputum gives a comparative diagnostic accuracy in terms of positivity, sensitivity, predictive values, and viral loads. Also, sputum reduces discomfort to patients and the risk of HCWs to infectious aerosols. During the design phase of the study, ‘restriction’ was utilized to control and minimize the impact of ‘age’ as a confounding variable.
One potential drawback of using sputum is that not all COVID-19 patients can expectorate sputum, which may be reflected in the decreased number of examined sputum samples when compared to oropharyngeal samples worldwide. However, for those who are unable to produce sputum, induced sputum or a self-collected deep cough specimen could be explored for SARS-CoV-2 detection and may be comparable to that of the pharyngeal swabs [31,32,33,34]. Pharyngeal sampling may, nonetheless, continue to play an important role in the diagnosis or monitoring of COVID-19 patients as it is still considered the most preferred sampling method, especially in early diagnosis. Another limitation to the study was our inability to capture data at the time of onset symptoms; hence, the interpretation of results should be conducted with caution.
5. Conclusions
Findings from this study demonstrate that self-collected sputum is a useful alternative to OPS for the diagnosis of COVID-19. Sputum samples are easier to collect, have a comparable viral load, and produce the highest sample positivity than oropharyngeal sampling. Opting for sputum for COVID-19 diagnosis carries significant logistical and cost advantages, especially in the cost build-up per one SARS-CoV-2 RT-qPCR test being currently charged by the testing centres in Ghana. The methodology of self-collection, again, helps by alleviating the risk of aerosol transmission to healthcare workers and obviating the need for full protective suits. With high sputum reliability and a lower false-negative rate than pharyngeal sampling, we believe sputum is more useful for the confirmation of COVID-19. This study does not, however, completely eliminate the use of pharyngeal sampling but suggests an equivalent self-collected sample with obvious advantages.
Author Contributions
Conceptualization, E.A. and M.O.; methodology, E.A., A.A. and J.S.K.; formal analysis, E.A. and G.A. (Godfred Acheampong); investigation, E.A., G.A. (Godfred Acheampong) and M.O.; data curation, E.A. and G.A. (George Agyei); writing—original draft preparation, E.A.; writing—review and editing, G.A. (Godfred Acheampong), N.K.A.-B., F.O.A., D.O.O., B.N., M.M. and A.A.S.; visualization, M.O. and G.A. (Godfred Acheampong); supervision, M.O. All authors have read and agreed to the published version of the manuscript.
Funding
Laboratory support was provided by the Kumasi Centre for Collaborative Research in Tropical Medicine through support of the Ministry of Health, Ghana. The APC was funded by the Centre for Health System Strengthening, Kumasi, Ghana.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Committee on Human Research, Publication and Ethics (CHRPE) of the School of Medicine and Dentistry, Kwame Nkrumah University of Science and Technology (KNUST) Kumasi, Ghana (Approval number: CHRPE/AP/076/21).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
The authors are thankful to the participants of the study.
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this article.
References
- Khurshid, Z.; Asiri, F.Y.I.; Al Wadaani, H. Human Saliva: Non-Invasive Fluid for Detecting Novel Coronavirus (2019-nCoV). Int. J. Environ. Res. Public Health 2020, 17, 2225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WHO. WHO Coronavirus (COVID-19) Dashboard. 2022. Available online: https://covid19.who.int/ (accessed on 2 February 2022).
- COVID-19 Updates|Ghana. WHO Coronavirus (COVID-19) Dashboard-Ghana. 2022. Available online: https://covid19.who.int/region/afro/country/gh (accessed on 28 January 2022).
- Long, C.; Xu, H.; Shen, Q.; Zhang, X.; Fan, B.; Wang, C.; Zeng, B.; Li, Z.; Li, X.; Li, H. Diagnosis of the Coronavirus disease (COVID-19): rRT-PCR or CT? Eur. J. Radiol. 2020, 126, 108961. [Google Scholar] [CrossRef]
- CDC. Interim Guidelines for Collecting and Handling of Clinical Specimens for COVID-19 Testing. 2020. Available online: https://www.cdc.gov/coronavirus/2019-nCoV/lab/guidelines-clinical-specimens.html (accessed on 8 December 2021).
- Pinninti, S.; Trieu, C.; Pati, S.K.; Latting, M.; Cooper, J.; Seleme, M.C.; Boppana, S.; Arora, N.; Britt, W.J.; Boppana, S.B. Comparing Nasopharyngeal and Midturbinate Nasal Swab Testing for the Identification of Severe Acute Respiratory Syndrome Coronavirus 2. Clin. Infect. Dis. 2020, 72, 1253–1255. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Zhao, X.; Ma, X.; Wang, W.; Niu, P.; Xu, W.; Gao, G.F.; Wu, G. A Novel Coronavirus Genome Identified in a Cluster of Pneumonia Cases—Wuhan, China 2019−2020. China CDC Wkly. 2020, 2, 61–62. [Google Scholar] [CrossRef]
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in different types of clinical specimens. Jama 2020, 323, 1843–1844. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.; Xiang, J.; Yan, M.; Li, H.; Huang, S.; Shen, C. Comparison of throat swabs and sputum specimens for viral nucleic acid detection in 52 cases of novel coronavirus Presenting characteristics. Clin. Chem. Lab. Med. (CCLM) 2020, 58, 1089–1094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Yang, M.; Yuan, J.; Wang, F.; Wang, Z.; Li, J.; Zhang, M.; Xing, L.; Wei, J.; Peng, L.; et al. Laboratory Diagnosis and Monitoring the Viral Shedding of SARS-CoV-2 Infection Laboratory Diagnosis and Monitoring the Viral Shedding of SARS-CoV-2 Infection. Innovation 2020, 1, 100061. [Google Scholar] [CrossRef]
- Mawaddah, A.; Gendeh, H.S.; Lum, S.G.; Baki, M.M. Upper respiratory tract sampling in COVID-19. Malays. J. Pathol. 2020, 42, 23–35. [Google Scholar]
- Rao, S.N.; Manissero, D.; Steele, V.R.; Pareja, J. A Systematic Review of the Clinical Utility of Cycle Threshold Values in the Context of COVID-19. Infect. Dis. Ther. 2020, 9, 573–586. [Google Scholar] [CrossRef]
- Wang, H.; Liu, Q.; Hu, J.; Zhou, M.; Yu, M.Q.; Li, K.Y.; Xu, D.; Xiao, Y.; Yang, J.Y.; Lu, Y.J.; et al. Nasopharyngeal Swabs Are More Sensitive Than Oropharyngeal Swabs for COVID-19 Diagnosis and Monitoring the SARS-CoV-2 Load. Front. Med. 2020, 7, 334. [Google Scholar] [CrossRef]
- Ghana Statistical Service. Ghana 2021 population and housing census. Gen. Rep. 2021, 3A–3N, 104–105. [Google Scholar]
- Acheampong, G.; Owusu, M.; Nkrumah, B.; Obeng-Boadi, P.; Opare, D.A.; Sambian, D.J.; Angra, P.; Walker, C. Laboratory capacity in COVID-19 diagnosis and the need to enhance molecular testing in Ghana. Glob. Secur. Health Sci. Policy 2021, 6, 10–17. [Google Scholar] [CrossRef]
- Owusu, M.; Sylverken, A.A.; Ankrah, S.T.; El-Duah, P.; Ayisi-Boateng, N.K.; Yeboah, R.; Gorman, R.; Asamoah, J.; Binger, T.; Acheampong, G.; et al. Epidemiological profile of SARS-CoV-2 among selected regions in Ghana: A cross-sectional retrospective study. PLoS ONE 2020, 15, e0243711. [Google Scholar] [CrossRef]
- Adjei, P.; Afriyie-mensah, J.; Ganu, V.J.; Puplampu, P.; Opoku-Asare, B.; Dzefi-tettey, K.; Tachi, K.; Dey, D.; Akamah, J.; Akpalu, A.; et al. Clinical characteristics of COVID-19 patients admitted at the Korle-Bu Teaching Hospital, Accra, Ghana. Ghana Med. J. 2020, 54, 33–38. [Google Scholar] [CrossRef]
- Alsofayan, Y.M.; Althunayyan, S.M.; Khan, A.A.; Hakawi, A.M. Clinical characteristics of COVID-19 in Saudi Arabia: A national retrospective study. J. Infect. Public Health 2020, 13, 920–925. [Google Scholar] [CrossRef]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Guan, W.; Ni, Z.; Hu, Y.; Liang, W.; Ou, C.; He, J.; Liu, L.; Shan, H.; Lei, C.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Yokota, I.; Hattori, T.; Shane, P.Y.; Konno, S.; Nagasaka, A.; Takeyabu, K.; Fujisawa, S.; Nishida, M.; Teshima, T. Equivalent SARS-CoV-2 viral loads between nasopharyngeal swab and saliva in symptomatic patients. MedRxiv 2020, 11, 4500. [Google Scholar] [CrossRef]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA-J. Am. Med. Assoc. 2020, 323, 1061–1069. [Google Scholar] [CrossRef]
- Lamptey, J.; Oyelami, F.O.; Owusu, M.; Nkrumah, B.; Idowu, O.P.; Czika, A.; El-duah, P.; Yeboah, R.; Sylverken, A.; Olasunkanmi, I.O.; et al. Genomic and epidemiological characteristics of SARS-CoV-2 in Africa. PLoS Negl. Trop. Dis. 2021, 15, e0009335. [Google Scholar] [CrossRef]
- Ng, W.H.; Tipih, T.; Makoah, N.A.; Vermeulen, J.G.; Goedhals, D.; Sempa, J.B.; Burt, F.J.; Taylor, A.; Mahalingam, S. Comorbidities in SARS-CoV-2 Patients: A Systematic Review and Meta-Analysis. mBio 2021, 12, e03647-20. [Google Scholar] [CrossRef] [PubMed]
- Power, M.; Fell, G.; Wright, M. Principles for high-quality, high-value testing. BMJ Evid. Based Med. 2013, 18, 5–10. [Google Scholar] [CrossRef] [PubMed]
- To, K.K.W.; Tsang, O.T.Y.; Leung, W.S.; Tam, A.R.; Wu, T.C.; Lung, D.C.; Yip, C.C.Y.; Cai, J.P.; Chan, J.M.C.; Chik, T.S.H.; et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: An observational cohort study. Lancet Infect. Dis. 2020, 20, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Bosch, B.J.; Smits, S.L.; Haagmans, B.L. Membrane ectopeptidases targeted by human coronaviruses. Curr. Opin. Virol. 2014, 6, 55–60. [Google Scholar] [CrossRef]
- Hamming, I.; Timens, W.; Bulthuis, M.L.C.; Lely, A.T.; Navis, G.J.; Van Goor, H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004, 203, 631–637. [Google Scholar] [CrossRef]
- Branche, A.R.; Walsh, E.E.; Formica, M.A.; Falsey, A.R. Detection of respiratory viruses in sputum from adults by use of automated multiplex PCR. J. Clin. Microbiol. 2014, 52, 3590–3596. [Google Scholar] [CrossRef] [Green Version]
- Falsey, A.R.; Formica, M.A.; Walsh, E.E. Simple Method for Combining Sputum and Nasal Samples for Virus Detection by Reverse Transcriptase PCR. J. Clin. Microbiol. 2012, 50, 2835. [Google Scholar] [CrossRef] [Green Version]
- Butler-laporte, G.; Lawandi, A.; Schiller, I.; Yao, M. Comparison of Saliva and Nasopharyngeal Swab Nucleic Acid Amplification Testing for Detection of SARS-CoV-2 A Systematic Review and Meta-analysis. JAMA Intern. Med. 2021, 181, 353–360. [Google Scholar] [CrossRef]
- Pasomsub, E.; Watcharananan, S.P.; Boonyawat, K.; Janchompoo, P.; Wongtabtim, G.; Nalumansi, A.; Lutalo, T.; Kayiwa, J.; Watera, C.; Balinandi, S.; et al. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: A cross-sectional study. Clin. Microbiol. Infect. 2021, 27, 281–285. [Google Scholar] [CrossRef]
- Warsi, I.; Khurshid, Z.; Shazam, H.; Umer, M.F.; Imran, E.; Khan, M.O.; Slowey, P.D.; Goodson, J.M. Saliva Exhibits High Sensitivity and Specificity for the Detection of SARS-CoV-2. Diseases 2021, 9, 38. [Google Scholar] [CrossRef]
- Vaz, N.S.; Souza, D.; Santana, D.; Netto, E.M.; Pedroso, C.; Wang, W.; Deminco, F.; Santos, A.; Brites, C. Saliva is a reliable, non-invasive specimen for SARS-CoV-2 detection. Braz. J. Infect. Dis. 2020, 24, 422–427. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).