The Impact of Maternal SARS-CoV-2 Infection Next to Pre-Immunization with Gam-COVID-Vac (Sputnik V) Vaccine on the 1-Day-Neonate’s Blood Plasma Small Non-Coding RNA Profile: A Pilot Study
Abstract
1. Introduction
2. Materials and Methods
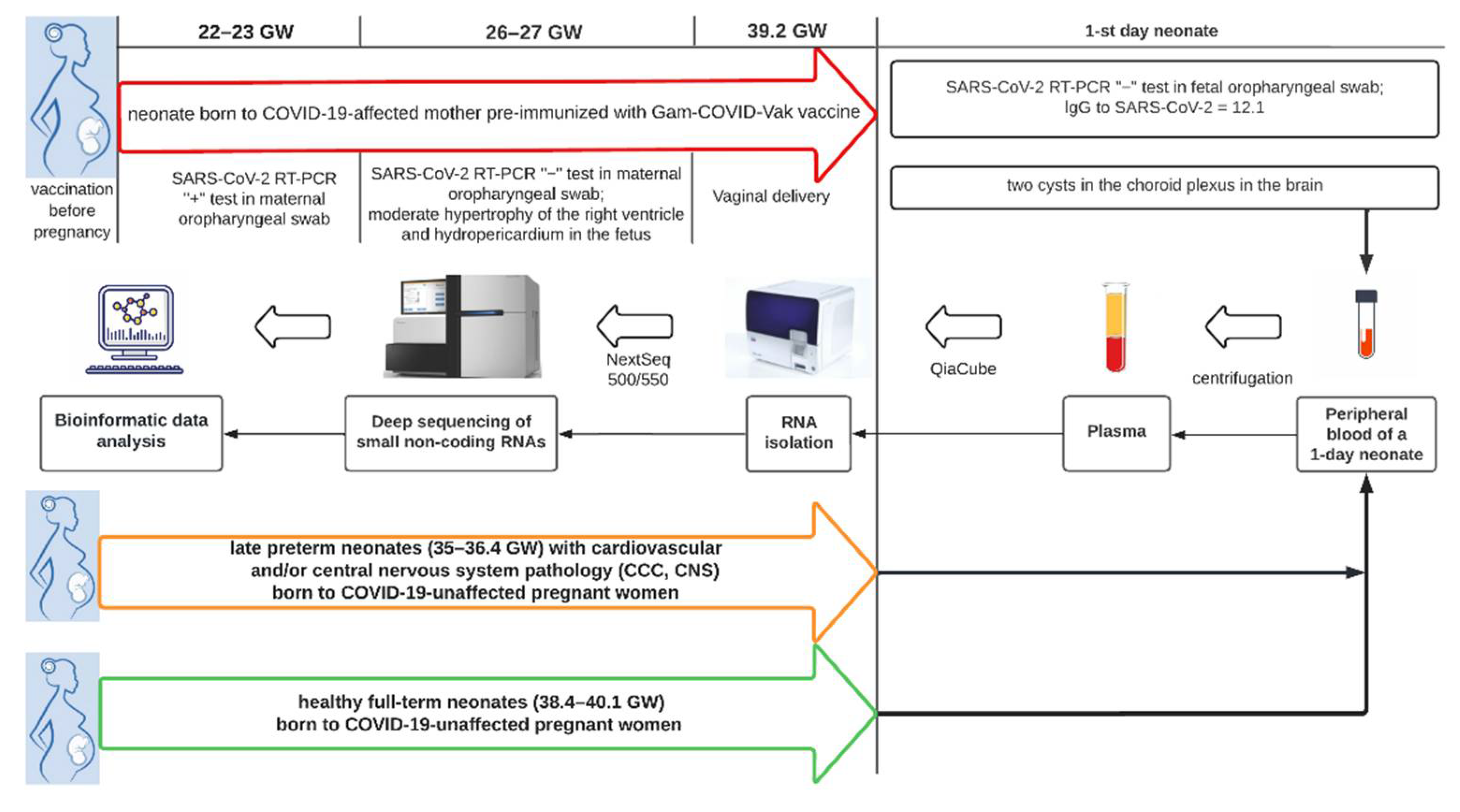
2.1. Patients
2.2. RNA Isolation from Peripheral Blood Plasma
2.3. cDNA Library Preparation and RNA Deep Sequencing
2.4. Statistical Analysis of the Obtained Data
3. Results
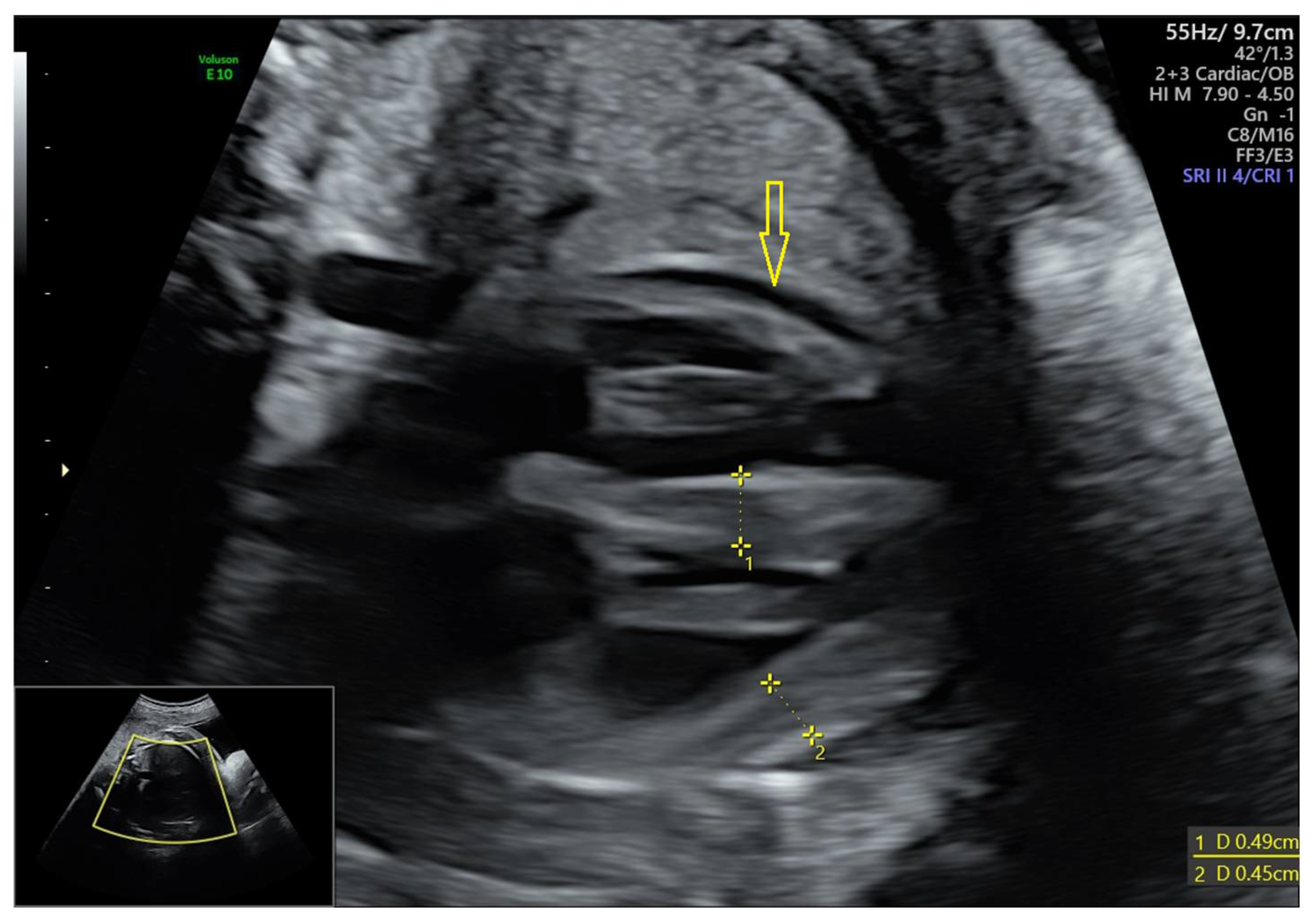
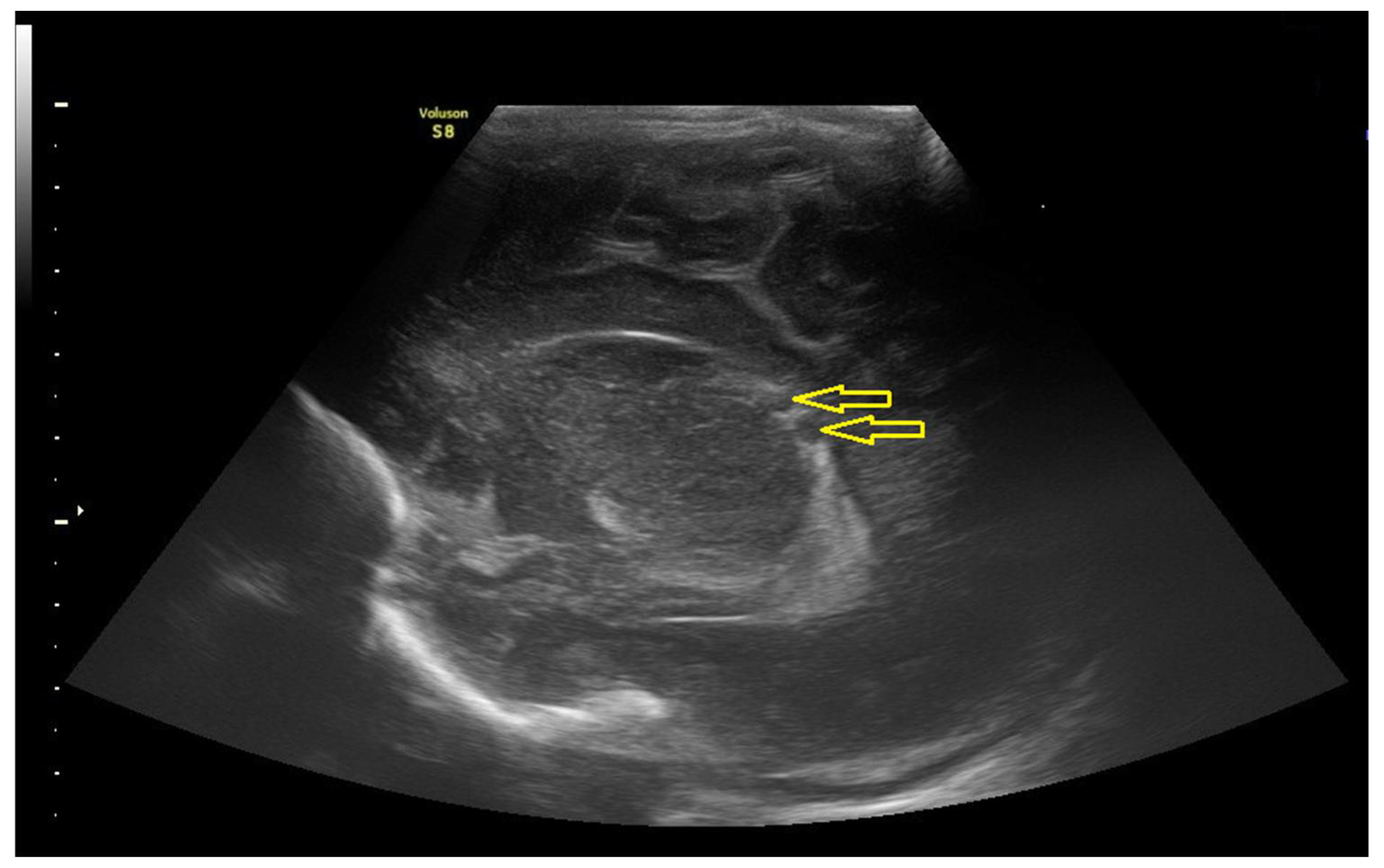
3.1. The Results of Clinical and Instrumental Methods of Examination of a Neonate Born to COVID-19-Affected Mother Pre-Immunized with Gam-COVID-Vac (Sputnik V) Vaccine
3.1.1. Laboratory Test Data of 1st Day Newborn
3.1.2. Data of the Instrumental Methods of Examination
3.2. Comparison of the Expression Profile of Small Non-Coding RNAs in Peripheral Blood of Neonates on the First Day of Life with Different Outcomes
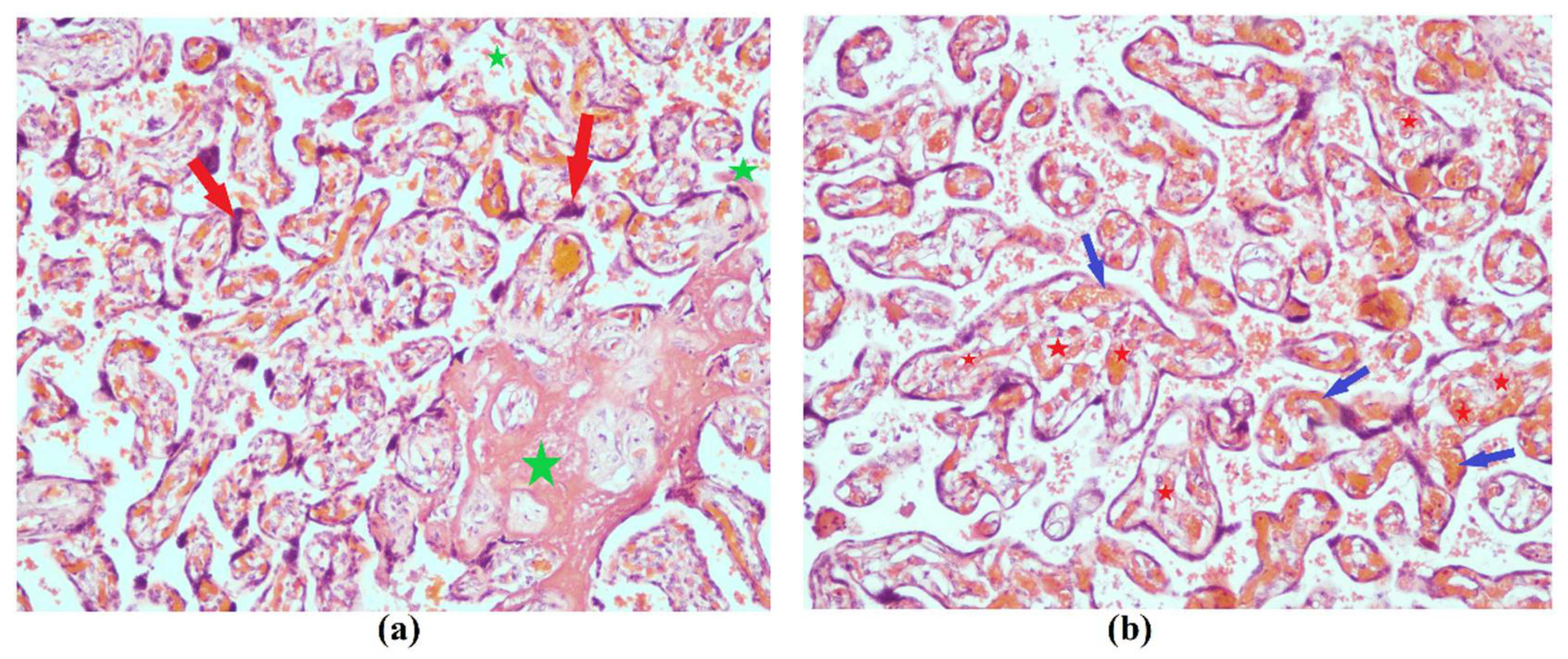
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cascella, M.; Rajnik, M.; Cuomo, A.; Dulebohn, S.C.; Di Napoli, R. Features, Evaluation and Treatment Coronavirus (COVID-19). In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Bahadur, G.; Bhat, M.; Acharya, S.; Janga, D.; Cambell, B.; Huirne, J.; Yoong, W.; Govind, A.; Pardo, J.; Homburg, R. Retrospective observational RT-PCR analyses on 688 babies born to 843 SARS-CoV-2 positive mothers, placental analyses and diagnostic analyses limitations suggest vertical transmission is possible. Facts Views Vis. ObGyn 2021, 13, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Gajbhiye, R.K.; Modi, D.N.; Mahale, S.D. Pregnancy outcomes, Newborn complications and Maternal-Fetal Transmission of SARS-CoV-2 in women with COVID-19: A systematic review of 441 cases. medRxiv 2020. medRxiv:2020.04.11.20062356. [Google Scholar] [CrossRef]
- Lamouroux, A.; Attie-Bitach, T.; Martinovic, J.; Leruez-Ville, M.; Ville, Y. Evidence for and against vertical transmission for severe acute respiratory syndrome coronavirus 2. Am. J. Obstet. Gynecol. 2020, 223, 91.e1–91.e4. [Google Scholar] [CrossRef] [PubMed]
- Kotlyar, A.M.; Grechukhina, O.; Chen, A.; Popkhadze, S.; Grimshaw, A.; Tal, O.; Taylor, H.S.; Tal, R. Vertical transmission of coronavirus disease 2019: A systematic review and meta-analysis. Am. J. Obstet. Gynecol. 2021, 224, 35–53.e3. [Google Scholar] [CrossRef]
- Wong, Y.P.; Khong, T.Y.; Tan, G.C. The Effects of COVID-19 on Placenta and Pregnancy: What Do We Know So Far? Diagnostics 2021, 11, 94. [Google Scholar] [CrossRef]
- Kreis, N.-N.; Ritter, A.; Louwen, F.; Yuan, J. A Message from the Human Placenta: Structural and Immunomodulatory Defense against SARS-CoV-2. Cells 2020, 9, 1777. [Google Scholar] [CrossRef]
- Burton, G.J.; Fowden, A.L. The placenta: A multifaceted, transient organ. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140066. [Google Scholar] [CrossRef]
- Celik, O.; Saglam, A.; Baysal, B.; Derwig, I.E.; Celik, N.; Ak, M.; Aslan, S.N.; Ulas, M.; Ersahin, A.; Tayyar, A.T.; et al. Factors preventing materno-fetal transmission of SARS-CoV-2. Placenta 2020, 97, 1–5. [Google Scholar] [CrossRef]
- Li, M.; Chen, L.; Zhang, J.; Xiong, C.; Li, X. The SARS-CoV-2 receptor ACE2 expression of maternal-fetal interface and fetal organs by single-cell transcriptome study. PLoS ONE 2020, 15, e0230295. [Google Scholar] [CrossRef]
- Roopenian, D.C.; Akilesh, S. FcRn: The neonatal Fc receptor comes of age. Nat. Rev. Immunol. 2007, 7, 715–725. [Google Scholar] [CrossRef]
- Maltepe, E.; Fisher, S.J. Placenta: The Forgotten Organ. Annu. Rev. Cell Dev. Biol. 2015, 31, 523–552. [Google Scholar] [CrossRef] [PubMed]
- Robbins, J.R.; Bakardjiev, A.I. Pathogens and the placental fortress. Curr. Opin. Microbiol. 2012, 15, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Heerema-McKenney, A. Defense and infection of the human placenta. APMIS 2018, 126, 570–588. [Google Scholar] [CrossRef] [PubMed]
- Pique-Regi, R.; Romero, R.; Tarca, A.L.; Luca, F.; Xu, Y.; Alazizi, A.; Leng, Y.; Hsu, C.-D.; Gomez-Lopez, N. Does the human placenta express the canonical cell entry mediators for SARS-CoV-2? Elife 2020, 9, e58716. [Google Scholar] [CrossRef]
- Tan, C.; Li, S.; Liang, Y.; Chen, M.; Liu, J. SARS-CoV-2 viremia may predict rapid deterioration of COVID-19 patients. Braz. J. Infect. Dis. 2020, 24, 565–569. [Google Scholar] [CrossRef]
- Sathiya, R.; Rajendran, J.; Sumathi, S. COVID-19 and Preeclampsia: Overlapping Features in Pregnancy. Rambam Maimonides Med. J. 2022, 13, e0007. [Google Scholar] [CrossRef]
- Liu, P.; Zheng, J.; Yang, P.; Wang, X.; Wei, C.; Zhang, S.; Feng, S.; Lan, J.; He, B.; Zhao, D.; et al. The immunologic status of newborns born to SARS-CoV-2-infected mothers in Wuhan, China. J. Allergy Clin. Immunol. 2020, 146, 101–109.e1. [Google Scholar] [CrossRef]
- Vimercati, A.; De Nola, R.; Trerotoli, P.; Metta, M.E.; Cazzato, G.; Resta, L.; Malvasi, A.; Lepera, A.; Ricci, I.; Capozza, M.; et al. COVID-19 Infection in Pregnancy: Obstetrical Risk Factors and Neonatal Outcomes-A Monocentric, Single-Cohort Study. Vaccines 2022, 10, 166. [Google Scholar] [CrossRef]
- Joma, M.; Fovet, C.-M.; Seddiki, N.; Gressens, P.; Laforge, M. COVID-19 and Pregnancy: Vertical Transmission and Inflammation Impact on Newborns. Vaccines 2021, 9, 391. [Google Scholar] [CrossRef]
- Schoenmakers, S.; Snijder, P.; Verdijk, R.M.; Kuiken, T.; Kamphuis, S.S.M.; Koopman, L.P.; Krasemann, T.B.; Rousian, M.; Broekhuizen, M.; Steegers, E.A.P.; et al. Severe Acute Respiratory Syndrome Coronavirus 2 Placental Infection and Inflammation Leading to Fetal Distress and Neonatal Multi-Organ Failure in an Asymptomatic Woman. J. Pediatr. Infect. Dis. Soc. 2021, 10, 556–561. [Google Scholar] [CrossRef]
- Correia, C.R.; Marçal, M.; Vieira, F.; Santos, E.; Novais, C.; Maria, A.T.; Malveiro, D.; Prior, A.R.; Aguiar, M.; Salazar, A.; et al. Congenital SARS-CoV-2 Infection in a Neonate With Severe Acute Respiratory Syndrome. Pediatr. Infect. Dis. J. 2020, 39, e439–e443. [Google Scholar] [CrossRef] [PubMed]
- Vivanti, A.J.; Vauloup-Fellous, C.; Prevot, S.; Zupan, V.; Suffee, C.; Do Cao, J.; Benachi, A.; De Luca, D. Transplacental transmission of SARS-CoV-2 infection. Nat. Commun. 2020, 11, 3572. [Google Scholar] [CrossRef] [PubMed]
- Di Nicola, P.; Ceratto, S.; Dalmazzo, C.; Roasio, L.; Castagnola, E.; Sannia, A. Concomitant SARS-CoV-2 infection and severe neurologic involvement in a late-preterm neonate. Neurology 2020, 95, 834–835. [Google Scholar] [CrossRef] [PubMed]
- Favre, G.; Mazzetti, S.; Gengler, C.; Bertelli, C.; Schneider, J.; Laubscher, B.; Capoccia, R.; Pakniyat, F.; Ben Jazia, I.; Eggel-Hort, B.; et al. Decreased Fetal Movements: A Sign of Placental SARS-CoV-2 Infection with Perinatal Brain Injury. Viruses 2021, 13, 2517. [Google Scholar] [CrossRef]
- Gale, C.; Quigley, M.A.; Placzek, A.; Knight, M.; Ladhani, S.; Draper, E.S.; Sharkey, D.; Doherty, C.; Mactier, H.; Kurinczuk, J.J. Characteristics and outcomes of neonatal SARS-CoV-2 infection in the UK: A prospective national cohort study using active surveillance. Lancet Child Adolesc. Healh 2021, 5, 113–121. [Google Scholar] [CrossRef]
- Yoon, S.H.; Kang, J.-M.; Ahn, J.G. Clinical outcomes of 201 neonates born to mothers with COVID-19: A systematic review. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 7804–7815. [Google Scholar] [CrossRef]
- Di Girolamo, R.; Khalil, A.; Alameddine, S.; D’Angelo, E.; Galliani, C.; Matarrelli, B.; Buca, D.; Liberati, M.; Rizzo, G.; D’Antonio, F. Placental histopathology after SARS-CoV-2 infection in pregnancy: A systematic review and meta-analysis. Am. J. Obstet. Gynecol. MFM 2021, 3, 100468. [Google Scholar] [CrossRef]
- Resta, L.; Vimercati, A.; Cazzato, G.; Mazzia, G.; Cicinelli, E.; Colagrande, A.; Fanelli, M.; Scarcella, S.V.; Ceci, O.; Rossi, R. SARS-CoV-2 and Placenta: New Insights and Perspectives. Viruses 2021, 13, 723. [Google Scholar] [CrossRef]
- Cribiù, F.M.; Erra, R.; Pugni, L.; Rubio-Perez, C.; Alonso, L.; Simonetti, S.; Croci, G.A.; Serna, G.; Ronchi, A.; Pietrasanta, C.; et al. Severe SARS-CoV-2 placenta infection can impact neonatal outcome in the absence of vertical transmission. J. Clin. Investig. 2021, 131, e145427. [Google Scholar] [CrossRef]
- Resta, L.; Vimercati, A.; Sablone, S.; Marzullo, A.; Cazzato, G.; Ingravallo, G.; Mazzia, G.; Arezzo, F.; Colagrande, A.; Rossi, R. Is the First of the Two Born Saved? A Rare and Dramatic Case of Double Placental Damage from SARS-CoV-2. Viruses 2021, 13, 995. [Google Scholar] [CrossRef]
- Kontou, A.; Virgiliou, C.; Mouskeftara, T.; Begou, O.; Meikopoulos, T.; Thomaidou, A.; Agakidou, E.; Gika, H.; Theodoridis, G.; Sarafidis, K. Plasma Lipidomic and Metabolomic Profiling after Birth in Neonates Born to SARS-CoV-19 Infected and Non-Infected Mothers at Delivery: Preliminary Results. Metabolites 2021, 11, 830. [Google Scholar] [CrossRef] [PubMed]
- Cai, A.; Hu, Y.; Zhou, Z.; Qi, Q.; Wu, Y.; Dong, P.; Chen, L.; Wang, F. PIWI-Interacting RNAs (piRNAs): Promising Applications as Emerging Biomarkers for Digestive System Cancer. Front. Mol. Biosci. 2022, 9, 848105. [Google Scholar] [CrossRef] [PubMed]
- Deogharia, M.; Gurha, P. The “guiding” principles of noncoding RNA function. Wiley Interdiscip. Rev. RNA 2021, e1704. [Google Scholar] [CrossRef]
- Zhu, Q.; Kirby, J.A.; Chu, C.; Gou, L.-T. Small Noncoding RNAs in Reproduction and Infertility. Biomed 2021, 9, 1884. [Google Scholar] [CrossRef] [PubMed]
- Santosh, B.; Varshney, A.; Yadava, P.K. Non-coding RNAs: Biological functions and applications. Cell Biochem. Funct. 2015, 33, 14–22. [Google Scholar] [CrossRef]
- Iwakawa, H.; Tomari, Y. Life of RISC: Formation, action, and degradation of RNA-induced silencing complex. Mol. Cell 2022, 82, 30–43. [Google Scholar] [CrossRef]
- Saulle, I.; Garziano, M.; Fenizia, C.; Cappelletti, G.; Parisi, F.; Clerici, M.; Cetin, I.; Savasi, V.; Biasin, M. MiRNA Profiling in Plasma and Placenta of SARS-CoV-2-Infected Pregnant Women. Cells 2021, 10, 1788. [Google Scholar] [CrossRef]
- Farr, R.J.; Rootes, C.L.; Rowntree, L.C.; Nguyen, T.H.O.; Hensen, L.; Kedzierski, L.; Cheng, A.C.; Kedzierska, K.; Au, G.G.; Marsh, G.A.; et al. Altered microRNA expression in COVID-19 patients enables identification of SARS-CoV-2 infection. PLoS Pathog. 2021, 17, e1009759. [Google Scholar] [CrossRef]
- Meidert, A.S.; Hermann, S.; Brandes, F.; Kirchner, B.; Buschmann, D.; Billaud, J.-N.; Klein, M.; Lindemann, A.; Aue, E.; Schelling, G.; et al. Extracellular Vesicle Associated miRNAs Regulate Signaling Pathways Involved in COVID-19 Pneumonia and the Progression to Severe Acute Respiratory Corona Virus-2 Syndrome. Front. Immunol. 2021, 12, 784028. [Google Scholar] [CrossRef]
- Male, V. SARS-CoV-2 infection and COVID-19 vaccination in pregnancy. Nat. Rev. Immunol. 2022, 22, 277–282. [Google Scholar] [CrossRef]
- Jamieson, D.J.; Rasmussen, S.A. An update on COVID-19 and pregnancy. Am. J. Obstet. Gynecol. 2022, 226, 177–186. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. A Language and Environment for Statistical Computing. In R Foundation for Statistical Computing; R Core Team: Vienna, Austria, 2020; Available online: https://www.r-project.org (accessed on 10 March 2021).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. RStudio: Integrated Development for R; RStudio: Vienna, Austria, 2021; Available online: http://www.rstudio.com/ (accessed on 23 March 2021).
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-regression: A basic tool of chemometrics. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Rajalahti, T.; Arneberg, R.; Kroksveen, A.C.; Berle, M.; Myhr, K.-M.; Kvalheim, O.M. Discriminating variable test and selectivity ratio plot: Quantitative tools for interpretation and variable (biomarker) selection in complex spectral or chromatographic profiles. Anal. Chem. 2009, 81, 2581–2590. [Google Scholar] [CrossRef]
- Li, M.; Chen, X.; Chen, L.; Chen, K.; Zhou, J.; Song, J. MiR-1-3p that correlates with left ventricular function of HCM can serve as a potential target and differentiate HCM from DCM. J. Transl. Med. 2018, 16, 161. [Google Scholar] [CrossRef]
- Kura, B.; Kalocayova, B.; Devaux, Y.; Bartekova, M. Potential Clinical Implications of miR-1 and miR-21 in Heart Disease and Cardioprotection. Int. J. Mol. Sci. 2020, 21, 700. [Google Scholar] [CrossRef]
- Schuchardt, E.L.; Miyamoto, S.D.; Crombleholme, T.; Karimpour-Fard, A.; Korst, A.; Neltner, B.; Howley, L.W.; Cuneo, B.; Sucharov, C.C. Amniotic Fluid microRNA in Severe Twin-Twin Transfusion Syndrome Cardiomyopathy-Identification of Differences and Predicting Demise. J. Cardiovasc. Dev. Dis. 2022, 9, 37. [Google Scholar] [CrossRef]
- Baulina, N.; Osmak, G.; Kiselev, I.; Matveeva, N.; Kukava, N.; Shakhnovich, R.; Kulakova, O.; Favorova, O. NGS-identified circulating miR-375 as a potential regulating component of myocardial infarction associated network. J. Mol. Cell. Cardiol. 2018, 121, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Cesarman, G.M.; Guevara, C.A.; Hajjar, K.A. An endothelial cell receptor for plasminogen/tissue plasminogen activator (t-PA). II. Annexin II-mediated enhancement of t-PA-dependent plasminogen activation. J. Biol. Chem. 1994, 269, 21198–21203. [Google Scholar] [CrossRef]
- Dassah, M.; Deora, A.B.; He, K.; Hajjar, K.A. The endothelial cell annexin A2 system and vascular fibrinolysis. Gen. Physiol. Biophys. 2009, 28, F20–F28. [Google Scholar] [PubMed]
- Ling, Q.; Jacovina, A.T.; Deora, A.; Febbraio, M.; Simantov, R.; Silverstein, R.L.; Hempstead, B.; Mark, W.H.; Hajjar, K.A. Annexin II regulates fibrin homeostasis and neoangiogenesis in vivo. J. Clin. Investig. 2004, 113, 38–48. [Google Scholar] [CrossRef]
- Timofeeva, A.V.; Fedorov, I.S.; Brzhozovskiy, A.G.; Bugrova, A.E.; Chagovets, V.V.; Volochaeva, M.V.; Starodubtseva, N.L.; Frankevich, V.E.; Nikolaev, E.N.; Shmakov, R.G.; et al. miRNAs and Their Gene Targets-A Clue to Differentiate Pregnancies with Small for Gestational Age Newborns, Intrauterine Growth Restriction, and Preeclampsia. Diagnostics 2021, 11, 729. [Google Scholar] [CrossRef]
- Cabrera-Garcia, D.; Miltiades, A.; Parsons, S.; Elisman, K.; Mansouri, M.T.; Wagener, G.; Harrison, N.L. High levels of plasminogen activator inhibitor-1, tissue plasminogen activator and fibrinogen in patients with severe COVID-19. medRxiv 2021. medRxiv:2020.12.29.20248869. [Google Scholar] [CrossRef]
- Rubina, K.; Shmakova, A.; Shabanov, A.; Andreev, Y.; Borovkova, N.; Kulabukhov, V.; Evseev, A.; Popugaev, K.; Petrikov, S.; Semina, E. Novel prognostic determinants of COVID-19-related mortality: A pilot study on severely-ill patients in Russia. PLoS ONE 2022, 17, e0264072. [Google Scholar] [CrossRef]
- Kagawa, T.; Shirai, Y.; Oda, S.; Yokoi, T. Identification of Specific MicroRNA Biomarkers in Early Stages of Hepatocellular Injury, Cholestasis, and Steatosis in Rats. Toxicol. Sci. 2018, 166, 228–239. [Google Scholar] [CrossRef]
- Timofeeva, A.; Drapkina, Y.; Fedorov, I.; Chagovets, V.; Makarova, N.; Shamina, M.; Kalinina, E.; Sukhikh, G. Small Noncoding RNA Signatures for Determining the Developmental Potential of an Embryo at the Morula Stage. Int. J. Mol. Sci. 2020, 21, 9399. [Google Scholar] [CrossRef]
- Flannery, D.D.; Gouma, S.; Dhudasia, M.B.; Mukhopadhyay, S.; Pfeifer, M.R.; Woodford, E.C.; Triebwasser, J.E.; Gerber, J.S.; Morris, J.S.; Weirick, M.E.; et al. Assessment of Maternal and Neonatal Cord Blood SARS-CoV-2 Antibodies and Placental Transfer Ratios. JAMA Pediatr. 2021, 175, 594–600. [Google Scholar] [CrossRef]
- Krechetova, L.V.; Inviyaeva, E.V.; Sadykov, V.F.; Vtorushina, V.V.; Ivanets, T.Y.; Silachev, D.N.; Pyregov, A.V.; Dolgushina, N.V.; Sukhikh, G.T. Immune status of covid-19 patients with different disease severity. Akusherstvo Ginekol. 2021, 2021, 75–88. [Google Scholar] [CrossRef]
- Laing, A.G.; Lorenc, A.; Del Molino Del Barrio, I.; Das, A.; Fish, M.; Monin, L.; Muñoz-Ruiz, M.; McKenzie, D.R.; Hayday, T.S.; Francos-Quijorna, I.; et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020, 26, 1623–1635. [Google Scholar] [CrossRef] [PubMed]
- Gee, S.; Chandiramani, M.; Seow, J.; Pollock, E.; Modestini, C.; Das, A.; Tree, T.; Doores, K.J.; Tribe, R.M.; Gibbons, D.L. The legacy of maternal SARS-CoV-2 infection on the immunology of the neonate. Nat. Immunol. 2021, 22, 1490–1502. [Google Scholar] [CrossRef] [PubMed]
- Raphael, I.; Joern, R.R.; Forsthuber, T.G. Memory CD4(+) T Cells in Immunity and Autoimmune Diseases. Cells 2020, 9, 531. [Google Scholar] [CrossRef]
- Kapustova, L.; Petrovicova, O.; Banovcin, P.; Antosova, M.; Bobcakova, A.; Urbancikova, I.; Rennerova, Z.; Jesenak, M. COVID-19 and the differences in physiological background between children and adults and their clinical consequences. Physiol. Res. 2021, 70, S209–S225. [Google Scholar] [CrossRef]
- Dhume, K.; McKinstry, K.K. Early programming and late-acting checkpoints governing the development of CD4 T-cell memory. Immunology 2018, 155, 53–62. [Google Scholar] [CrossRef]
- Belz, G.T.; Masson, F. Interleukin-2 tickles T cell memory. Immunity 2010, 32, 7–9. [Google Scholar] [CrossRef][Green Version]
- Raeber, M.E.; Zurbuchen, Y.; Impellizzieri, D.; Boyman, O. The role of cytokines in T-cell memory in health and disease. Immunol. Rev. 2018, 283, 176–193. [Google Scholar] [CrossRef]
- Kappanayil, M.; Balan, S.; Alawani, S.; Mohanty, S.; Leeladharan, S.P.; Gangadharan, S.; Jayashankar, J.P.; Jagadeesan, S.; Kumar, A.; Gupta, A.; et al. Multisystem inflammatory syndrome in a neonate, temporally associated with prenatal exposure to SARS-CoV-2: A case report. Lancet Child Adolesc. Healh 2021, 5, 304–308. [Google Scholar] [CrossRef]
- Arunachalam, P.S.; Wimmers, F.; Mok, C.K.P.; Perera, R.A.P.M.; Scott, M.; Hagan, T.; Sigal, N.; Feng, Y.; Bristow, L.; Tak-Yin Tsang, O.; et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science 2020, 369, 1210–1220. [Google Scholar] [CrossRef]
- Lu-Culligan, A.; Chavan, A.R.; Vijayakumar, P.; Irshaid, L.; Courchaine, E.M.; Milano, K.M.; Tang, Z.; Pope, S.D.; Song, E.; Vogels, C.B.F.; et al. Maternal respiratory SARS-CoV-2 infection in pregnancy is associated with a robust inflammatory response at the maternal-fetal interface. Med 2021, 2, 591–610.e10. [Google Scholar] [CrossRef] [PubMed]
- Brien, M.-E.; Baker, B.; Duval, C.; Gaudreault, V.; Jones, R.L.; Girard, S. Alarmins at the maternal-fetal interface: Involvement of inflammation in placental dysfunction and pregnancy complications (1). Can. J. Physiol. Pharmacol. 2019, 97, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Dubucs, C.; Groussolles, M.; Ousselin, J.; Sartor, A.; Van Acker, N.; Vayssière, C.; Pasquier, C.; Reyre, J.; Batlle, L.; Favarel Clinical Research Associate, S.; et al. Severe placental lesions due to maternal SARS-CoV-2 infection associated to intrauterine fetal death. Hum. Pathol. 2022, 121, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Mongula, J.E.; Frenken, M.W.E.; van Lijnschoten, G.; Arents, N.L.A.; de Wit-Zuurendonk, L.D.; Schimmel-de Kok, A.P.A.; van Runnard Heimel, P.J.; Porath, M.M.; Goossens, S.M.T.A. COVID-19 during pregnancy: Non-reassuring fetal heart rate, placental pathology and coagulopathy. Ultrasound Obstet. Gynecol. 2020, 56, 773–776. [Google Scholar] [CrossRef]
- Shchegolev, A.I.; Lyapin, V.M.; Tumanova, U.N.; Vodneva, D.N.; Shmakov, R.G. Histological changes in the placenta and vascularization of its villi in early-And lateonset Preeclampsia. Arkh. Patol. 2016, 78, 13–18. [Google Scholar] [CrossRef]
- Chen, D.-B.; Zheng, J. Regulation of placental angiogenesis. Microcirculation 2014, 21, 15–25. [Google Scholar] [CrossRef]
- Sukhikh, G.; Petrova, U.; Prikhodko, A.; Starodubtseva, N.; Chingin, K.; Chen, H.; Bugrova, A.; Kononikhin, A.; Bourmenskaya, O.; Brzhozovskiy, A.; et al. Vertical Transmission of SARS-CoV-2 in Second Trimester Associated with Severe Neonatal Pathology. Viruses 2021, 13, 447. [Google Scholar] [CrossRef]
- Di Gennaro, F.; Murri, R.; Segala, F.V.; Cerruti, L.; Abdulle, A.; Saracino, A.; Bavaro, D.F.; Fantoni, M. Attitudes towards Anti-SARS-CoV2 Vaccination among Healthcare Workers: Results from a National Survey in Italy. Viruses 2021, 13, 371. [Google Scholar] [CrossRef]


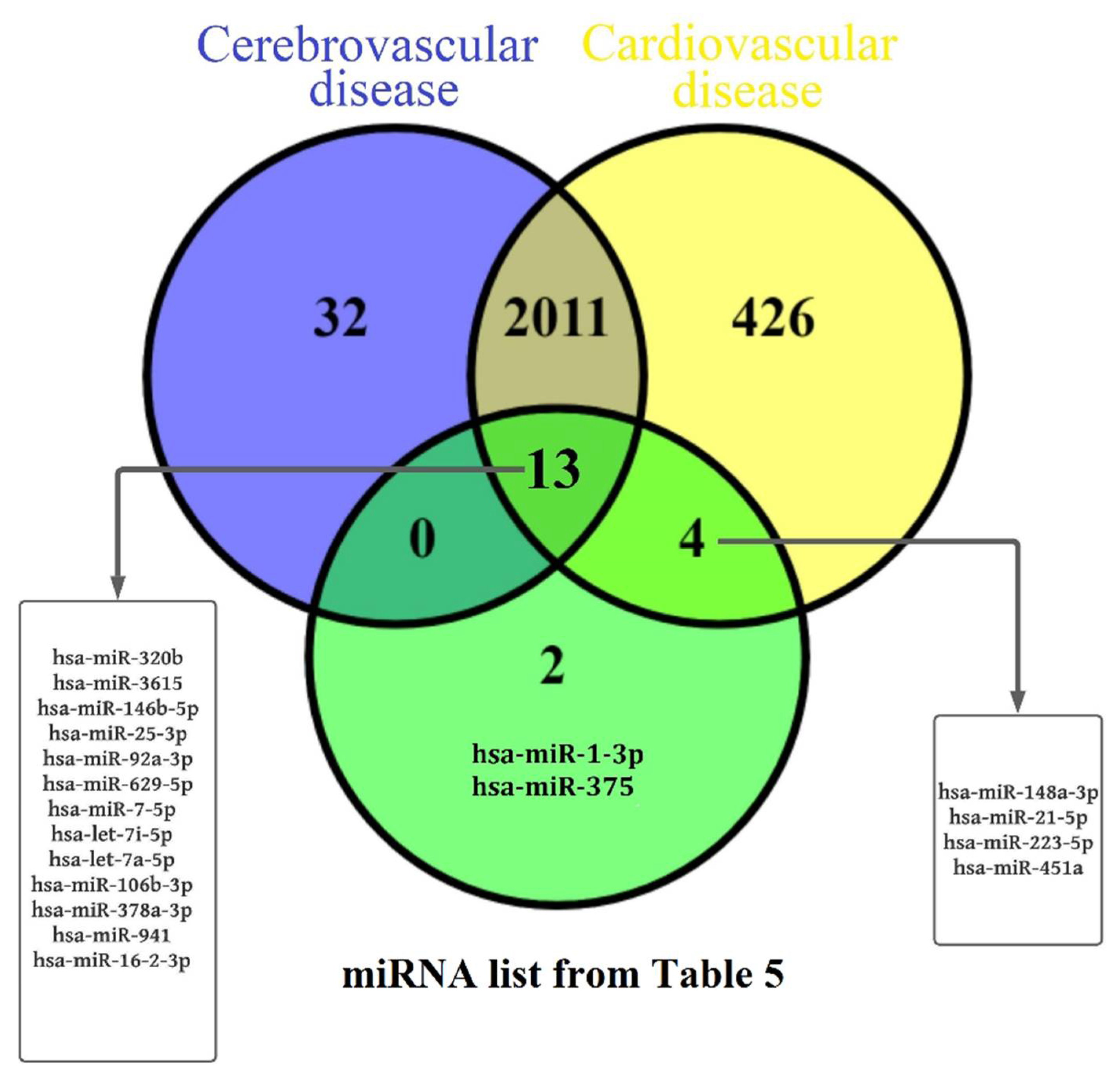
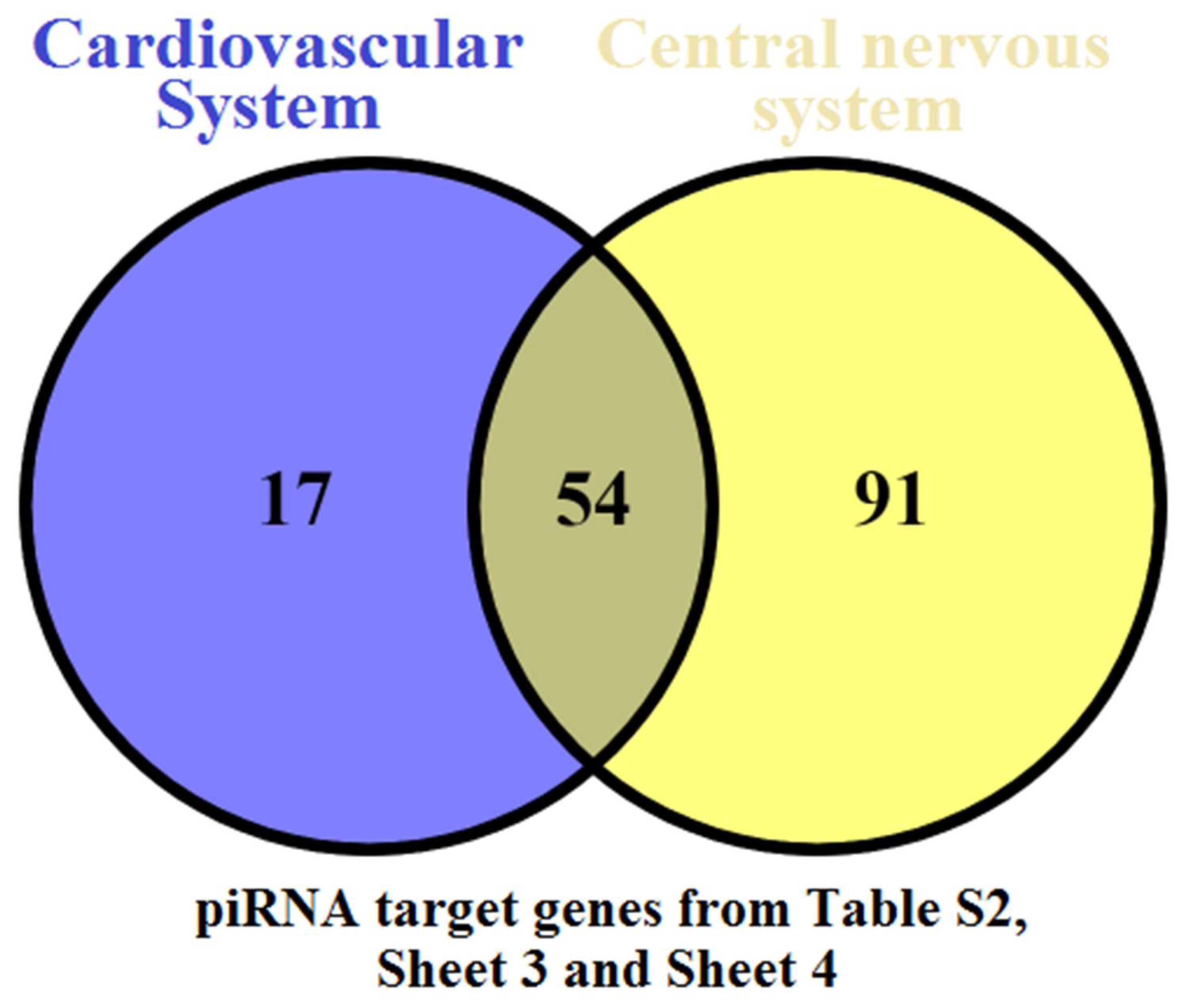
| 1st Day Newborn Blood Plasma Sample ID | Maternal Age | Mode of Delivery | Maternal Diagnosis | GW 1 | Newborn | APGAR Score at the 1st and 5th Minutes | CNS Disorder | Cardiovascular Disorder | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Gender | Weight | Growth | ||||||||
| #3 | 22 | Caesarean section | placenta previa, placenta increta | 35 | female | 2300 | 48 | 7 and 7 | Yes | Yes |
| #20 | 32 | Caesarean section | Gestational arterial hypertension. Placenta previa. Placenta accreta. | 35.1 | female | 2850 | 47 | 7 and 8 | Yes | No |
| #41 | 28 | Caesarean section | Low placentation. Preeclampsia. Chronic arterial hypertension. | 35.1 | male | 3144 | 51 | 8 and 9 | Yes | Yes |
| #151 | 36 | Caesarean section | Complete placenta previa. Placenta percreta. | 35.4 | female | 3062 | 50 | 8 and 9 | Yes | No |
| #52 | 27 | Caesarean section | Central placenta previa. Placenta accreta. | 36.4 | male | 2960 | 48 | 8 and 8 | Yes | Yes |
| #129 | 37 | Caesarean section | Placenta previa and placenta accreta | 36 | male | 2758 | 49 | 8 and 9 | Yes | Yes |
| #29 | 21 | Caesarean section | Placenta previa. Small uterine myoma. | 36 | male | 2780 | 52 | 8 and 8 | Yes | Yes |
| #26 | 36 | Caesarean section | Placenta previa. Placenta percreta. | 35 | female | 2480 | 49 | 7 and 7 | No | Yes |
| #152 | 30 | Caesarean section | Placenta previa. Placenta percreta | 36.1 | female | 2670 | 52 | 8 and 9 | Yes | Yes |
| #32 | 28 | spontaneous delivery | premature rupture of the foetal membrane | 36 | male | 2625 | 47 | 7 and 8 | Yes | Yes |
| #42 | 39 | Caesarean section | Chronic arterial hypertension. Primary antiphospholipid syndrome. | 35.3 | male | 2480 | 49 | 8 and 8 | No | No |
| #200 | 39 | physiological delivery | Timely spontaneous delivery | 40.1 | male | 3468 | 52 | 8 and 9 | No | No |
| #201 | 26 | Caesarean section | Chronic arterial hypertension. | 38.4 | male | 3344 | 53 | 8 and 9 | No | No |
| #202 | 38 | physiological delivery | Condition after COVID-19 infection. | 39.2 | male | 3402 | 52 | 8 and 9 | Yes | Yes (only at 26–27 GW) |
| Biochemical Indicator | Value | Reference Values |
|---|---|---|
| glucose, mmol/L | 4.0 | 3.9–6.4 |
| creatinin, µmol/L | 26.7 | 35–62 |
| direct bilirubin, µmol/L | 6.6 | 0–5.5 |
| alanine aminotransferase (ALT), U/L | 103.3 | 0–40 |
| aspartate aminotransferase (AST), U/L | 71.4 | 0–40 |
| alkaline phosphatase (ALP), U/L | 203.1 | 50–360 |
| gamma-glutamyl transferase (GGT), U/L | 121.7 | 0–250 |
| total bilirubin, µmol/L | 293.8 | 3.4–21 |
| lactate dehydrogenase (LDG), U/L | 1513.3 | 300–730 |
| uric acid, µmol/L | 217.3 | 202–416 |
| Indicator | Value | Reference Values |
|---|---|---|
| leukocytes/WBC, ×109/L | 10.51 | 5.9–17.5 |
| erythrocytes/RBC, ×1012/L | 5.00 | 3.9–5.9 |
| hemoglobin/HGB, g/L | 183 | 134–198 |
| hematocrit/HCT, L/L | 0.497 | 0.41–0.65 |
| mean corpuscular volume/MCV, fL | 99.4 | 88–140 |
| mean corpuscular haemoglobin/MCH, pg | 36.6 | 30–37 |
| mean corpuscular hemoglobin concentration/MCHC, g/dL | 36.8 | 28–36 |
| erythrocyte anisocytosis SD/RDW-SD, fL | 65.1 | 35.1–46.3 |
| erythrocyte anisocytosis CV/RDW-CV | 17.4 | 11.5–14.5 |
| platelets/PLT, ×109/L | 286 | 218–419 |
| platelet anisocytosis/PDW, fL | 10.7 | 5–30 |
| mean thrombocyte volume/MPV, fL | 9.6 | 9.4–12.3 |
| Platelet-Large Cell Ratio/P-LCR, % | 22.5 | 13–43 |
| thrombocrit/PCT, % | 0.27 | 0.1–0.4 |
| immature granulocytes (relative count)/IG% | 1.2 | 0–1.9 |
| neutrophilic leukocyte (relative count)/NEUT% | 41.0 | 20.2–66.1 |
| lymphocytes (relative count)/LYMPH% | 39.5 | 24.9–67.6 |
| monocytes (relative count)/MONO% | 11.8 | 6.7–19.9 |
| eosinophils (relative count)/EO% | 6.0 | 0.3–5.2 |
| basophils (relative count)/BASO% | 0.5 | 0–1 |
| immature granulocytes (absolute count)/IG, ×109/L | 0.13 | 0–0.28 |
| neutrophilic leukocyte (absolute count)/NEUT, ×109/L | 4.31 | 1.73–7.75 |
| lymphocytes (absolute count)/LYMPH, ×109/L | 4.15 | 1.75–7.53 |
| monocytes (absolute count)/MONO, ×109/L | 1.24 | 0.52–1.77 |
| eosinophils (absolute count)/EO, ×109/L | 0.63 | 0.12–0.66 |
| basophils (absolute count)/BASO, ×109/L | 0.05 | 0–0.15 |
| Indicator | Value | Reference Values |
|---|---|---|
| leukocytes ×109/L | 10.51 | 5.9–17.5 |
| lymphocytes (relative count) % | 39.5 | 24.9–67.6 |
| lymphocytes (absolute count) ×109/L | 4.15 | 1.75–7.53 |
| CD3+ (T lymphocytes) (relative count) % | 90.3 | 58–70 |
| CD3+ (T lymphocytes) (absolute count) ×109/L | 3.75 | 1–5.3 |
| CD3+CD4+ (T helper cells) (relative count) % | 71.3 | 38–50 |
| CD3+CD4+ (T helper cells) (absolute count) ×109/L | 2.96 | 0.7–3.8 |
| CD3+CD8+ (cytoxic T lymphocyte) (relative count) % | 20.5 | 15–25 |
| CD3+CD8+ (cytoxic T lymphocyte) (absolute count) ×109/L | 0.85 | 0.3–1.9 |
| CD3+CD4+/CD3+CD8+ | 3.5 | 1.5–2.9 |
| CD19+ (B lymphocytes) (relative count) % | 6.6 | 12–30 |
| CD19+ (B lymphocytes) (absolute count) ×109/L | 0.27 | 0.2–2.3 |
| CD3-CD16+CD56+ (NK cells) (relative count) % | 1.1 | 5–15 |
| CD3-CD16+CD56+ (NK cells) (absolute count) ×109/L | 0.05 | 0.09–1.12 |
| CD3+CD16+CD56+ (relative count) % | 0.1 | 0–10 |
| CD3+CD16+CD56+ (absolute count) ×109/L | 0.00 | 0–0.8 |
| CD19+CD5+ (relative count) % | 1.5 | 3–8 |
| CD19+CD5+ (absolute count) ×109/L | 0.06 | 0.05–0.6 |
| CD56+ (relative count) % | 0.9 | 5–15 |
| CD56+ (absolute count) ×109/L | 0.04 | 0.09–1.12 |
| CD3+HLA-Dr.+ (relative count) % | 0.2 | 2–10 |
| CD3+HLA-Dr.+ (absolute count) ×109/L | 0.01 | 0.02–0.4 |
| CD3+CD25+ (relative count) % | 5.9 | 0–7 |
| CD3+CD25+ (absolute count) ×109/L | 0.24 | 0–0.14 |
| CD3+CD56+ (relative count) % | 0.1 | 0–10 |
| CD3+CD56+ (absolute count) ×109/L | 0.00 | 0–0.4 |
| CD25+ (relative count) % | 6.2 | 1–10 |
| CD25+ (absolute count) ×109/L | 0.26 | 0.01–0.4 |
| Sample ID | #3 | #20 | #26 | #29 | #32 | #41 | #42 | #52 | #129 | #151 | #152 | #200 | #201 | #202 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| for plotting Figure 5A | ||||||||||||||
| hsa_piR_015150 | 27,6414 | 644,966 | 184,276 | 460,690 | 92,138 | 921,380 | 737,104 | 1,382,070 | 737,104 | 460,690 | 921,380 | 92,138 | 368,552 | 0 |
| hsa_piR_011187 | 70,008 | 233,360 | 560,064 | 280,032 | 280,032 | 350,040 | 256,696 | 373,376 | 466,720 | 140,016 | 280,032 | 23,336 | 93,344 | 0 |
| hsa-miR-106b-3p | 913 | 2813 | 2575 | 720 | 1658 | 804 | 482 | 51 | 55 | 94 | 91 | 63 | 84 | 20 |
| hsa-miR-16-2-3p | 1221 | 3140 | 2491 | 786 | 1312 | 874 | 541 | 70 | 99 | 196 | 142 | 113 | 110 | 34 |
| hsa-let-7a-5p | 3955 | 8260 | 5473 | 3807 | 7708 | 3504 | 2895 | 1307 | 744 | 1841 | 1306 | 849 | 1461 | 358 |
| hsa_piR_022017 | 666 | 6768 | 3078 | 3474 | 5652 | 2754 | 810 | 1404 | 5994 | 486 | 1512 | 126 | 144 | 72 |
| hsa-miR-3615 | 1942 | 3143 | 1861 | 679 | 2113 | 1670 | 674 | 118 | 99 | 235 | 136 | 111 | 140 | 48 |
| hsa-miR-451a | 14,247 | 23,142 | 21,306 | 6171 | 16,382 | 11,144 | 6930 | 2896 | 2411 | 5300 | 2263 | 2223 | 4132 | 1262 |
| hsa-miR-320b | 532 | 897 | 1415 | 848 | 2085 | 625 | 390 | 112 | 42 | 180 | 80 | 42 | 170 | 22 |
| hsa-miR-378a-3p | 400 | 692 | 738 | 220 | 604 | 358 | 246 | 129 | 62 | 201 | 122 | 79 | 75 | 19 |
| hsa-miR-375 | 495 | 1267 | 2000 | 1805 | 3813 | 249 | 114 | 202 | 26 | 162 | 55 | 26 | 376 | 4 |
| hsa-miR-148a-3p | 14,673 | 20,316 | 19,939 | 14,605 | 31,354 | 12,062 | 7634 | 1597 | 972 | 7224 | 2351 | 1033 | 1954 | 317 |
| hsa-miR-146b-5p | 204 | 146 | 221 | 172 | 281 | 113 | 78 | 56 | 34 | 85 | 47 | 39 | 56 | 5 |
| hsa-miR-21-5p | 1153 | 1573 | 2033 | 1765 | 2347 | 1043 | 640 | 225 | 157 | 335 | 235 | 202 | 252 | 51 |
| hsa-miR-25-3p | 5693 | 8269 | 11,923 | 4326 | 6966 | 4458 | 2669 | 680 | 595 | 1656 | 1399 | 621 | 844 | 248 |
| hsa-miR-92a-3p | 68,340 | 82,079 | 87,803 | 52,288 | 47,904 | 60,556 | 38,042 | 5127 | 5801 | 12,570 | 12,457 | 5309 | 10,097 | 2946 |
| hsa-miR-7-5p | 1020 | 1384 | 650 | 828 | 670 | 614 | 556 | 50 | 90 | 162 | 126 | 64 | 84 | 20 |
| hsa-let-7i-5p | 5480 | 12,349 | 4463 | 7444 | 7627 | 5115 | 4832 | 921 | 1176 | 1819 | 1942 | 1002 | 1665 | 544 |
| hsa-miR-223-5p | 78 | 92 | 183 | 66 | 216 | 125 | 99 | 42 | 23 | 62 | 46 | 47 | 54 | 5 |
| hsa-miR-941 | 3200 | 3025 | 1355 | 1530 | 2155 | 1095 | 660 | 100 | 75 | 255 | 185 | 95 | 95 | 45 |
| hsa-miR-629-5p | 446 | 944 | 1 | 256 | 610 | 178 | 217 | 55 | 29 | 58 | 74 | 58 | 65 | 13 |
| for plotting Figure 5B | ||||||||||||||
| hsa_piR_008983 | 63,472 | 222,152 | 142,812 | 174,548 | 174,548 | 190,416 | 31,736 | 79,340 | 142,812 | 31,736 | 0 | 0 | 0 | 15,868 |
| hsa_piR_008114 | 2122 | 3697 | 6787 | 4682 | 9581 | 4029 | 1451 | 2104 | 4361 | 1803 | 4891 | 507 | 1288 | 1199 |
| hsa_piR_022296 | 212,180 | 2,302,153 | 1,527,696 | 2,121,800 | 7,500,563 | 2,790,167 | 859,329 | 551,668 | 3,691,932 | 774,457 | 3,066,001 | 21,218 | 127,308 | 137,917 |
| hsa_piR_022421 | 210 | 6160 | 3955 | 3885 | 18,270 | 7630 | 2450 | 1470 | 9310 | 2065 | 7875 | 175 | 280 | 560 |
| for plotting Figure 5C | ||||||||||||||
| hsa_piR_020829 | 106 | 55 | 58 | 76 | 118 | 104 | 55 | 26 | 76 | 100 | 82 | 27 | 47 | 114 |
| hsa-miR-1-3p | 18 | 14 | 44 | 12 | 44 | 26 | 4 | 14 | 4 | 0 | 4 | 6 | 8 | 66 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Timofeeva, A.V.; Fedorov, I.S.; Chagovets, V.V.; Zubkov, V.V.; Makieva, M.I.; Sugak, A.B.; Frankevich, V.E.; Sukhikh, G.T. The Impact of Maternal SARS-CoV-2 Infection Next to Pre-Immunization with Gam-COVID-Vac (Sputnik V) Vaccine on the 1-Day-Neonate’s Blood Plasma Small Non-Coding RNA Profile: A Pilot Study. COVID 2022, 2, 837-857. https://doi.org/10.3390/covid2070061
Timofeeva AV, Fedorov IS, Chagovets VV, Zubkov VV, Makieva MI, Sugak AB, Frankevich VE, Sukhikh GT. The Impact of Maternal SARS-CoV-2 Infection Next to Pre-Immunization with Gam-COVID-Vac (Sputnik V) Vaccine on the 1-Day-Neonate’s Blood Plasma Small Non-Coding RNA Profile: A Pilot Study. COVID. 2022; 2(7):837-857. https://doi.org/10.3390/covid2070061
Chicago/Turabian StyleTimofeeva, Angelika V., Ivan S. Fedorov, Vitaliy V. Chagovets, Victor V. Zubkov, Mziya I. Makieva, Anna B. Sugak, Vladimir E. Frankevich, and Gennadiy T. Sukhikh. 2022. "The Impact of Maternal SARS-CoV-2 Infection Next to Pre-Immunization with Gam-COVID-Vac (Sputnik V) Vaccine on the 1-Day-Neonate’s Blood Plasma Small Non-Coding RNA Profile: A Pilot Study" COVID 2, no. 7: 837-857. https://doi.org/10.3390/covid2070061
APA StyleTimofeeva, A. V., Fedorov, I. S., Chagovets, V. V., Zubkov, V. V., Makieva, M. I., Sugak, A. B., Frankevich, V. E., & Sukhikh, G. T. (2022). The Impact of Maternal SARS-CoV-2 Infection Next to Pre-Immunization with Gam-COVID-Vac (Sputnik V) Vaccine on the 1-Day-Neonate’s Blood Plasma Small Non-Coding RNA Profile: A Pilot Study. COVID, 2(7), 837-857. https://doi.org/10.3390/covid2070061







