Single Cell Micro-Pillar-Based Characterization of Endothelial and Fibroblast Cell Mechanics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Immunostaining
2.3. Elastic Micropillar Arrays
2.4. Microscopy
2.5. Image Analysis
2.6. Statistics
3. Results
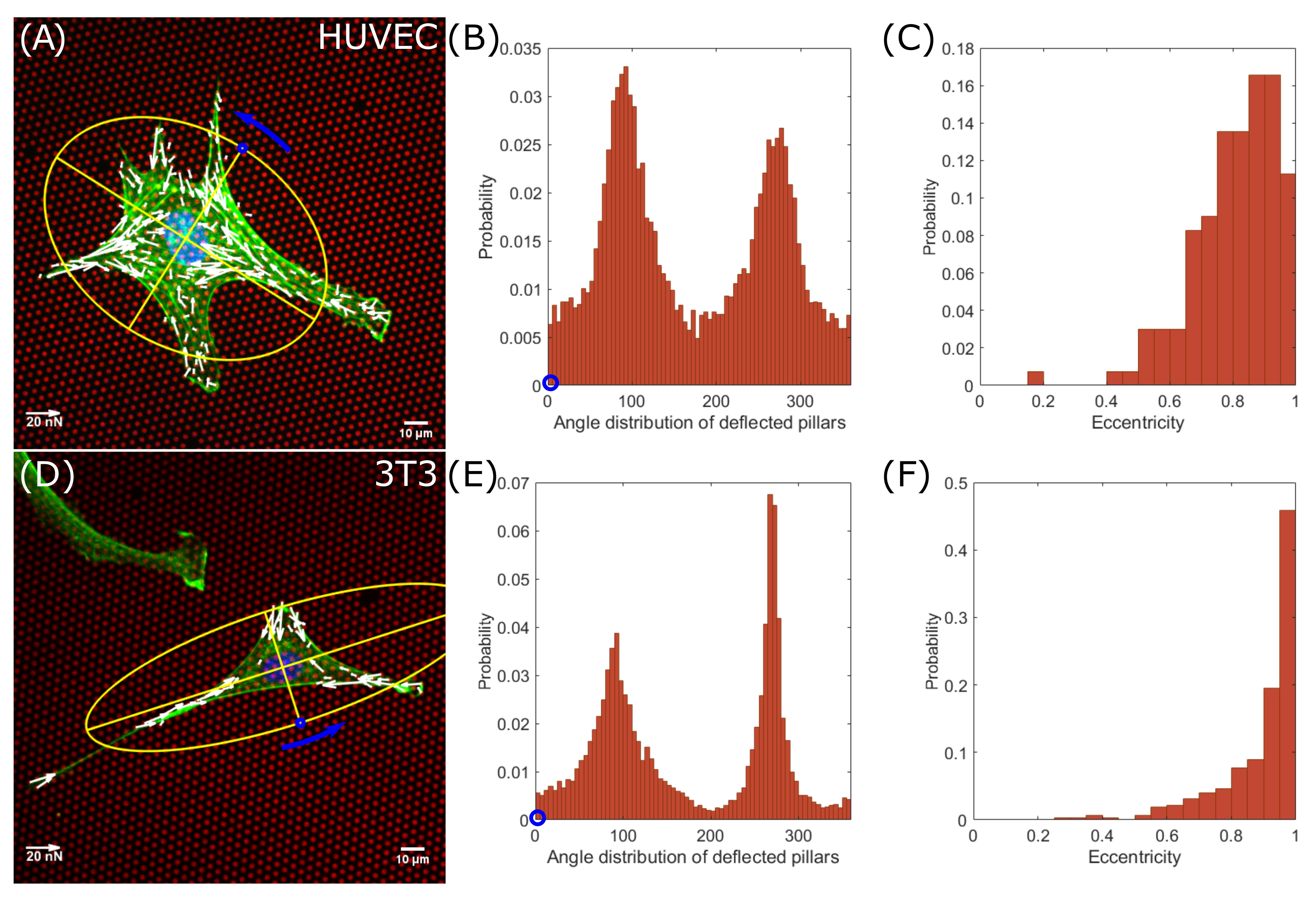
3.1. Endothelial Cells Apply Less Traction Forces Compared to Fibroblast Cells
3.2. Averaged Force Dipole Distribution Is Independent of Cell Type
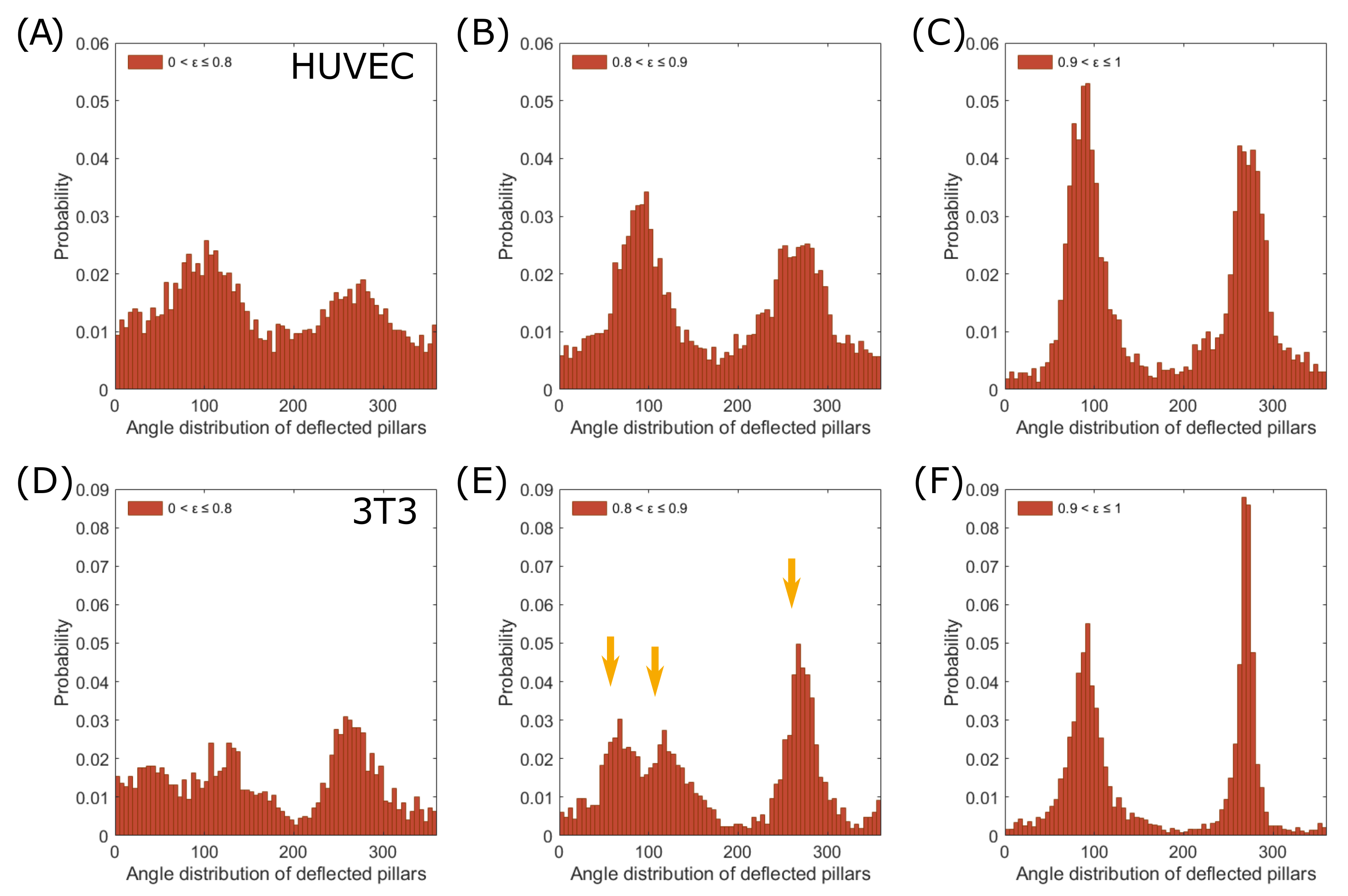
3.3. Force Pole Is Cell Morphology and Cell Type Dependent
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Evers, T.M.J.; Holt, L.J.; Alberti, S.; Mashaghi, A. Reciprocal regulation of cellular mechanics and metabolism. Nat. Metab. 2021, 3, 456–468. [Google Scholar] [CrossRef]
- Leslie, M.N.; Chou, J.; Young, P.M.; Traini, D.; Bradbury, P.; Ong, H.X. How Do Mechanics Guide Fibroblast Activity? Complex Disruptions during Emphysema Shape Cellular Responses and Limit Research. Bioengineering 2021, 8, 110. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Wang, J.H.C. Fibroblasts and myofibroblasts in wound healing: Force generation and measurement. J. Tissue Viability 2011, 20, 108–120. [Google Scholar] [CrossRef] [Green Version]
- Sorrell, J.M.; Baber, M.A.; Caplan, A.I. A Self-Assembled Fibroblast-Endothelial Cell Co-Culture System That Supports in vitro Vasculogenesis by both Human Umbilical Vein Endothelial Cells and Human Dermal Microvascular Endothelial Cells. Cells Tissues Organs 2007, 186, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, O.; Cooper-White, J.; Janmey, P.A.; Mooney, D.J.; Shenoy, V.B. Effects of extracellular matrix viscoelasticity on cellular behaviour. Nature 2020, 584, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Henderson, N.C.; Rieder, F.; Wynn, T.A. Fibrosis: From mechanisms to medicines. Nature 2020, 587, 555–566. [Google Scholar] [CrossRef]
- Kuehlmann, B.; Bonham, C.A.; Zucal, I.; Prantl, L.; Gurtner, G.C. Mechanotransduction in Wound Healing and Fibrosis. J. Clin. Med. 2020, 9, 1423. [Google Scholar] [CrossRef]
- Arslan, F.N.; Eckert, J.; Schmidt, T.; Heisenberg, C.P. Holding it together: When cadherin meets cadherin. Biophys. J. 2021, 120, 4182–4192. [Google Scholar] [CrossRef]
- Lemmon, C.A.; Sniadecki, N.J.; Ruiz, S.A.; Tan, J.L.; Romer, L.H.; Chen, C.S. Shear force at the cell-matrix interface: Enhanced analysis for microfabricated post array detectors. Mech. Chem. Biosyst. MCB 2005, 2, 1–16. [Google Scholar]
- Medina-Leyte, D.J.; Domínguez-Pérez, M.; Mercado, I.; Villarreal-Molina, M.T.; Jacobo-Albavera, L. Use of Human Umbilical Vein Endothelial Cells (HUVEC) as a Model to Study Cardiovascular Disease: A Review. Appl. Sci. 2020, 10, 938. [Google Scholar] [CrossRef] [Green Version]
- Van Hoorn, H.; Harkes, R.; Spiesz, E.M.; Storm, C.; van Noort, D.; Ladoux, B.; Schmidt, T. The Nanoscale Architecture of Force-Bearing Focal Adhesions. Nano Lett. 2014, 14, 4257–4262. [Google Scholar] [CrossRef] [PubMed]
- Eckert, J.; van Loon, J.J.A.; Eng, L.M.; Schmidt, T. Hypergravity affects cell traction forces of fibroblasts. Biophys. J. 2021, 120, 773–780. [Google Scholar] [CrossRef] [PubMed]
- Dembo, M.; Wang, Y.L. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys. J. 1999, 76, 2307–2316. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, U.S.; Safran, S.A. Elastic Interactions of Cells. Phys. Rev. Lett. 2002, 88, 048102. [Google Scholar] [CrossRef] [Green Version]
- Abuhattum, S.; Gefen, A.; Weihs, D. Ratio of total traction force to projected cell area is preserved in differentiating adipocytes. Integr. Biol. Quant. Biosci. Nano Macro 2015, 7, 1212–1217. [Google Scholar] [CrossRef]
- Abercrombie, M. Fibroblasts. J. Clin. Pathol. Suppl. R. Coll. Pathol. 1978, 12, 1–6. [Google Scholar]
- Li, X.; Das, A.; Bi, D. Mechanical Heterogeneity in Tissues Promotes Rigidity and Controls Cellular Invasion. Phys. Rev. Lett. 2019, 123, 058101. [Google Scholar] [CrossRef] [Green Version]
- Balaban, N.Q.; Schwarz, U.S.; Riveline, D.; Goichberg, P.; Tzur, G.; Sabanay, I.; Mahalu, D.; Safran, S.; Bershadsky, A.; Addadi, L.; et al. Force and focal adhesion assembly: A close relationship studied using elastic micropatterned substrates. Nat. Cell Biol. 2001, 3, 466–472. [Google Scholar] [CrossRef]
- Tan, J.L.; Tien, J.; Pirone, D.M.; Gray, D.S.; Bhadriraju, K.; Chen, C.S. Cells lying on a bed of microneedles: An approach to isolate mechanical force. Proc. Natl. Acad. Sci. USA 2003, 100, 1484–1489. [Google Scholar] [CrossRef] [Green Version]
- Shemesh, T.; Geiger, B.; Bershadsky, A.D.; Kozlov, M.M. Focal adhesions as mechanosensors: A physical mechanism. Proc. Natl. Acad. Sci. USA 2005, 102, 12383–12388. [Google Scholar] [CrossRef] [Green Version]
- Trichet, L.; Le Digabel, J.; Hawkins, R.J.; Vedula, S.R.K.; Gupta, M.; Ribrault, C.; Hersen, P.; Voituriez, R.; Ladoux, B. Evidence of a large-scale mechanosensing mechanism for cellular adaptation to substrate stiffness. Proc. Natl. Acad. Sci. USA 2012, 109, 6933–6938. [Google Scholar] [CrossRef] [Green Version]
- Balcioglu, H.E.; van Hoorn, H.; Donato, D.M.; Schmidt, T.; Danen, E.H.J. The integrin expression profile modulates orientation and dynamics of force transmission at cell–matrix adhesions. J. Cell Sci. 2015, 128, 1316–1326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Califano, J.P.; Reinhart-King, C.A. Exogenous and endogenous force regulation of endothelial cell behavior. J. Biomech. 2010, 43 1, 79–86. [Google Scholar] [CrossRef]
- Rhee, S.; Grinnell, F. Fibroblast mechanics in 3D collagen matrices. Adv. Drug Deliv. Rev. 2007, 59, 1299–1305. [Google Scholar] [CrossRef] [Green Version]
- Malandrino, A.; Mak, M.; Kamm, R.D.; Moeendarbary, E. Complex mechanics of the heterogeneous extracellular matrix in cancer. Extrem. Mech. Lett. 2018, 21, 25–34. [Google Scholar] [CrossRef]
- Mandal, K.; Wang, I.; Vitiello, E.; Orellana, L.A.C.; Balland, M. Cell dipole behaviour revealed by ECM sub-cellular geometry. Nat. Commun. 2014, 5, 5749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slany, A.; Bileck, A.; Kreutz, D.; Mayer, R.L.; Muqaku, B.; Gerner, C. Contribution of Human Fibroblasts and Endothelial Cells to the Hallmarks of Inflammation as Determined by Proteome Profiling. Mol. Cell. Proteom. 2016, 15, 1982–1997. [Google Scholar] [CrossRef] [Green Version]
- Tefft, J.B.; Chen, C.S.; Eyckmans, J. Reconstituting the dynamics of endothelial cells and fibroblasts in wound closure. APL Bioeng. 2021, 5, 016102. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eckert, J.; Abouleila, Y.; Schmidt, T.; Mashaghi, A. Single Cell Micro-Pillar-Based Characterization of Endothelial and Fibroblast Cell Mechanics. Micro 2021, 1, 242-249. https://doi.org/10.3390/micro1020018
Eckert J, Abouleila Y, Schmidt T, Mashaghi A. Single Cell Micro-Pillar-Based Characterization of Endothelial and Fibroblast Cell Mechanics. Micro. 2021; 1(2):242-249. https://doi.org/10.3390/micro1020018
Chicago/Turabian StyleEckert, Julia, Yasmine Abouleila, Thomas Schmidt, and Alireza Mashaghi. 2021. "Single Cell Micro-Pillar-Based Characterization of Endothelial and Fibroblast Cell Mechanics" Micro 1, no. 2: 242-249. https://doi.org/10.3390/micro1020018
APA StyleEckert, J., Abouleila, Y., Schmidt, T., & Mashaghi, A. (2021). Single Cell Micro-Pillar-Based Characterization of Endothelial and Fibroblast Cell Mechanics. Micro, 1(2), 242-249. https://doi.org/10.3390/micro1020018






