Abstract
Donkey milk is an underexplored biological niche with distinctive nutritional and microbiological properties, potentially harboring lactic acid bacteria (LAB) with technological or probiotic value. In this study, raw milk from the endangered Zamorano-Leonesa donkey breed was stored at 4 °C for 24 h to simulate realistic cold-chain conditions and favor the recovery of cold-tolerant microorganisms. Fourteen isolates were obtained, eight of which belonged to LAB or species with potential technological interest and were selected for functional evaluation. Phenotypic screening showed that most isolates tolerated acidic conditions (pH 2.5) and that four also resisted 0.3% bile salts. Acidification assays in pasteurized donkey milk revealed variable fermentation performance, with L. mesenteroides subsp. mesenteroides B8 and Lacticaseibacillus paracasei subsp. tolerans B19 displaying the most favorable profiles. These two strains were selected for genome sequencing. Genomic analysis revealed genes associated with acid and bile resistance, adhesion, cold and environmental stress responses, and carbohydrate metabolism. Both genomes also encoded biosynthetic gene clusters linked to secondary metabolites, including β-lactones, lincosamides, and RiPP-like compounds. No acquired antimicrobial resistance genes were detected. To our knowledge, this is the first study combining isolation, phenotypic screening, and genome-based characterization of cold-tolerant LAB from Zamorano-Leonesa donkey milk. Our findings highlight this milk as a valuable reservoir of safe, cold-adapted microorganisms with promising applications in functional dairy products and food biotechnology.
1. Introduction
Donkey milk has gained scientific and industrial interest due to its distinctive biochemical composition and hypoallergenic nature. It offers nutritional benefits for sensitive consumers, including infants, the elderly, and individuals allergic to bovine milk proteins [1]. Among European donkey breeds, the Zamorano-Leonesa—native to northwestern Spain—is a protected and endangered breed of notable genetic and cultural value [2]. Its milk, with low fat content, high lactose levels, and a protein profile that closely resembles human milk [3], is increasingly used for artisanal and functional dairy products [4]. However, donkey milk is a nutrient-rich biological fluid that is highly susceptible to microbial contamination and spoilage, which may compromise its sensory quality and safety [5]. Therefore, investigating its microbial composition and isolating beneficial strains is essential for quality preservation and biotechnological applications.
After milking, raw donkey milk is typically cooled and stored at 4 °C until further processing or transport. European and Codex Alimentarius regulations recommend refrigeration (≤6–8 °C) to maintain hygienic quality and inhibit the growth of pathogenic microorganisms [6,7]. This cold storage period, often lasts up to 24 h, represents realistic conditions within the dairy cold chain. Microorganisms capable of surviving or remaining metabolically active under refrigeration can influence product stability and possess technological relevance [8]. Thus, analyzing the culturable microbial fraction after 24 h at 4 °C provides an ecologically meaningful snapshot of cold-tolerant and psychrotrophic bacteria persisting under real storage conditions.
Lactic acid bacteria (LAB) are among the microorganisms capable of thriving at low temperatures and play a key role in food fermentation. They contribute to product preservation through lactic acid production and support sensory development by synthesizing flavor compounds and exopolysaccharides [9,10]. Some strains also exhibit probiotic traits, including acid and bile tolerance, mucosal adhesion, and antimicrobial activity against pathogens [11,12,13].
LAB have been well studied in bovine, goat, and ewe milk [14,15,16] but their occurrence and functional roles in donkey milk remain poorly characterized [17]. Recent studies indicate that donkey milk may contain LAB strains with relevant technological and health-promoting properties. However, systematic isolation and functional characterization remain limited [18].
Integrating phenotypic screening with genomic characterization provides a robust strategy to identify promising LAB candidates. Functional assays—including acidification kinetics and d—reflect fermentative performance and survivability under gastrointestinal-like conditions [19]. When combined with genomic data, these tests help identify strains with desirable functional traits.
This approach is especially relevant in the current food and health context. Antimicrobial resistance continues to rise. At the same time, consumers increasingly demand natural and minimally processed foods. Genomic exploration of LAB offers an effective way to identify strains with metabolic versatility, stress and antimicrobial tolerance, and biosynthetic potential [20]. Genome-mining analyses further highlight genetic determinants that support key technological functions and antimicrobial activity [21]. These genomic insights complement traditional microbiological assessments and support the rational selection of safe and functional LAB for technological and probiotic applications [22].
Within this framework, Zamorano-Leonesa donkey milk represents an unexplored microbial niche for isolating and characterizing cold-resistant LAB. Assessing their functional properties and genome-based traits expands current knowledge of donkey milk microbiota and supports their application in food biotechnology. To our knowledge, no previous study has combined phenotypic and genomic approaches to evaluate cold-tolerant LAB from this endangered donkey breed. Therefore, this study aimed to isolate and characterize cold-tolerant LAB from Zamorano-Leonesa donkey milk using functional assays and genome-based analyses to identify strains with technological and probiotic potential.
2. Materials and Methods
2.1. Sample Collection
Milk was obtained from twelve healthy lactating donkeys belonging to the BULEZA cooperative (Santa Croya de Tera, Zamora, Spain). Animals were machine-milked, and the milk was pumped into a refrigerated storage silo. A 50 mL composite milk aliquot, representative of the pooled milk from the 12 animals, was aseptically collected from the cooling silo into a sterile Falcon® tube and transported to the laboratory under refrigerated conditions (4 °C) for microbiological analysis.
2.2. Isolation of Lactic Acid Bacteria (LAB) Strains
LAB strains were isolated from unpasteurized donkey milk after 24 h of cold storage. For this purpose, 100 µL of milk were plated on MRS agar (pH 6.5 ± 0.2), a standard medium widely used for the primary recovery of lactic acid bacteria from dairy products. Although MRS is not fully selective and may allow the growth of other aerobic or psychrotrophic microorganisms, it effectively enriches LAB under aerobic incubation. Plates were incubated at 30 °C for 72 h, and colonies exhibiting LAB-like morphology were purified by successive streaking and subjected to taxonomic identification. Only isolates subsequently confirmed as LAB or strains showing probiotic potential were included in the phenotypic assays.
2.3. Identification of Bacterial Strains by 16S rRNA Amplification and Sequencing
Identification of bacterial isolates was performed by 16S rRNA gene amplification and sequencing following the procedure described by Menéndez-Cañamares et al. [23]. Briefly, genomic DNA was extracted from pure colonies, PCR products were sequenced by Macrogen Inc. (Seoul, Republic of Korea), and the resulting sequences were assembled using BioEdit v9. Taxonomic identification was achieved by comparing reference sequences of type strains in the EZBioCloud database (https://www.ezbiocloud.net/identify, accessed on 20 October 2025) [24].
2.4. Fresh Donkey Milk Fermentation
Fresh donkey milk from the BULEZA cooperative was used to evaluate the ability of LAB isolates to ferment donkey milk. The milk was heat-treated at 90 °C for 10 min to inactivate native microbiota while minimizing protein denaturation. Only isolates taxonomically identified as LAB or showing potential probiotic or technological relevance were included in this assay. Fermentation ability was determined by monitoring pH decrease over 72 h. A commercial Lactobacillus acidophilus starter (Genesis® Acidophilus, Genesis Laboratories Ltd., Sofia, Bulgaria) served as the control. Each 50 mL flask of heat-treated milk was inoculated with 100 µL of bacterial suspension (~107 CFU mL−1) and incubated at 30 °C for 72 h. pH was measured daily under aseptic conditions using a calibrated pH meter. All assays were performed in triplicate.
2.5. Qualitative Screening of Acid and Bile Tolerance
Acid and bile tolerance assays were performed as a qualitative primary screening to assess the ability of isolates to withstand acidic and bile conditions. Acid and bile tolerance were assessed following the method of Vinderola and Reinheimer [25], with minor modifications. Only isolates identified as LAB or showing probiotic or technological potential were tested, while strains associated with pathogenicity or spoilage were excluded.
For acid tolerance, 24 h MRS cultures were washed with phosphate-buffered saline (PBS, pH 7.0), adjusted to an optical density of OD600 = 1.0, and mixed 1:1 (v/v) with sterile PBS adjusted to pH 2.5 using 1 M HCl. Suspensions were incubated at 37 °C for 4 h; PBS (pH 7.0) served as the control.
For bile tolerance, cultures were inoculated (1% v/v) into MRS broth supplemented with 0.3% (w/v) ox bile (Difco Laboraoties™, Detroit, MI, USA) or bile-free MRS (control) and incubated under the same conditions. Tolerance was evaluated qualitatively based on turbidity compared to the control and confirmed by growth recovery on MRS agar. Growth intensity—applied to both the acid and bile tolerance assays—was classified as positive (+) when abundant colony growth covered most of the plate surface; weak (w) when only a few discrete colonies were scattered across the plate; or negative (−) when no colonies were recovered.
2.6. Genome Analysis, Metabolic Pathways and Secondary Metabolites
The strains B8 and B19 were cultivated on MRS agar and harvested after 48 h of incubation at 30 °C. Genomic DNA was extracted using the ZR Fungal/Bacterial DNA MiniPrep Kit (Zymo Research catalog # D6005) according to the manufacturer’s instructions. Draft genome sequencing was performed by shotgun sequencing on an Illumina MiSeq platform using a paired-end run (2 × 251 bp). Sequence reads were assembled with Velvet v1.2.10, and gene prediction and annotation were conducted with RAST 2.0 (Rapid Annotation using Subsystem Technology) and the SEED Viewer framework [26].
Secondary metabolite biosynthetic gene clusters were identified using antiSMASH v7.0 [27]. Carbohydrate-active enzymes were classified through the dbCAN2 server based on the CAZy database (dbCAN3) [28]. Genomic visualization, gene neighborhood inspection, and structural organization were performed using Prokka 1.14.6 [29]. To assess the presence of acquired antimicrobial resistance (AMR) genes, whole-genome sequences were screened with the ResFinder 4.0 database and pipeline [30]. Default parameters were used (90% minimum identity, 60% minimum length). Only AMR genes meeting these criteria and associated with known transferable determinants were considered [30].
2.7. Data/Statistical Analysis
Data were statistically analyzed using one-way analysis of variance (ANOVA). Mean differences were assessed with Fisher’s protected least significant difference (LSD) test at a significance level of p < 0.05. Statistical analyses were performed using StatView software version 5.0.
2.8. Antibiotic Susceptibility Testing
Antibiotic susceptibility was assessed using the disk diffusion method as a qualitative screening assay. Cultures were spread on MRS agar plates, antibiotic disks were applied, and incubated. Antibiotic disks included ampicillin (10 µg), streptomycin (10 µg), and gentamicin (120 µg). After incubation, inhibition zones were visually examined, and the presence of a clear inhibition halo was considered indicative of susceptibility, whereas the absence of a halo indicated resistance. This approach provided a rapid qualitative assessment of the isolates’ response to the tested antibiotics.
3. Results
3.1. Isolation and Identification of Bacterial Strains from Storage Zamorano-Leonese Donkey Milk
After 72 h of incubation on MRS agar plates, a total of 14 bacterial strains were isolated from refrigerated (4 °C) Zamorano-Leonese donkey milk. All isolates were identified by partial sequencing of their 16S rRNA gene, yielding fragments of approximately 1500 bp. Sequence analysis revealed overall nucleotide identities greater than 99% when compared with reference sequences in the EZBioCloud database (Table 1).

Table 1.
Identification and 16S rRNA sequence similarity of bacterial isolates from Zamorano-Leonese donkey milk after 24 h of cold storage.
The isolates were distributed across two major bacterial phyla: Firmicutes (64.3%) and Proteobacteria (35.7%). Within Firmicutes, nine isolates were assigned to the genera Leuconostoc (six isolates), Bacillus (one isolate), Enterococcus (one isolate), and Lacticaseibacillus (one isolate). The remaining five isolates, belonging to the phylum Proteobacteria, were identified as Enterobacter (four isolates) and Citrobacter (one isolate).
These results indicate that, after 24 h of refrigerated storage, cold-tolerant and mesophilic lactic acid bacteria predominate in Zamorano-Leonese donkey milk, with Leuconostoc mesenteroides subsp. mesenteroides being the most frequently recovered species.
3.2. Donkey Milk Fermentation Assay
The fermentation assay evaluated the acidification capacity of donkey milk by the LAB isolates, using the commercial Lactobacillus acidophilus starter Genesis® as a control. The initial pH of pasteurized donkey milk was 7.16. As shown in Table 2, after 24 h of incubation at 30 °C, the control showed the strongest acidification, reaching pH 4.67. In contrast, the LAB isolates displayed slower acidification, with values ranging from 5.03 (B8) to 6.65 (B9), while B4 also remained above 6.0 (6.40). Most isolates continued to acidify the milk over time. After 48 h, strain B19 reached the lowest pH (4.11), followed by B13 (4.44). This trend persisted at 72 h, where B19 remained the most acidifying isolate (4.04), again followed by B13 (4.34). Other isolates, such as B5 and B12, maintained comparatively high pH values throughout fermentation (5.62 and 5.94 at 72 h, respectively), indicating limited acidification activity. Overall, the commercial starter exhibited the fastest and strongest acidification, while isolates B19 and B13 showed the greatest acidification capacity among the LAB strains tested.

Table 2.
Evolution of pH during fermentation of pasteurized donkey milk by selected bacterial isolates.
3.3. Preliminary Screening for Acid and Bile Resistance
The acid and bile tolerance assays revealed notable variability among isolates (Table 3). At pH 2.5, strains B5, B8, B9, and B19 showed positive growth, indicating good acid tolerance, while B3, B4, and B12 displayed only weak growth and B13 showed no survival. Under exposure to 0.3% (w/v) bile salts, strains B5, B8, B12, and B19 showed positive growth, whereas B4 and B9 exhibited weak tolerance and B3 and B13 did not grow. Overall, strains B5, B8, and B19 demonstrated the most consistent performance, showing positive growth under both acidic and bile conditions.

Table 3.
Qualitative screening of bacteria isolates from donkey milk for tolerance to acidic (pH 2.5) and bile (0.3% ox bile) conditions after 4 h incubation at 37 °C.
3.4. Genome Analysis of L. mesenteroides B8 and L. paracasei subsp. tolerans B19 from Donkey Milk
Based on their acid and bile tolerance profiles and fermentation performance, two isolates—L. B8 and B19—were selected for whole-genome sequencing. These strains were chosen for their robust acid tolerance, distinctive fermentation behavior in donkey milk, and taxonomic classification as LAB, commonly known for their safety and technological importance. Their inclusion represents two functionally distinct LAB groups with complementary characteristics: a heterofermentative Leuconostoc and a technologically versatile Lacticaseibacillus.
The main genomic features of both strains are summarized in Table 4. The genome of L. mesenteroides subsp. mesenteroides B8 comprises 27 contigs with a total length of 2,119,290 base pairs (bp) and an average GC content of 37.7%. A total of 2169 coding sequences (CDSs) and 69 RNA genes were predicted. In contrast, the genome of L. paracasei subsp. tolerans B19 consists of 81 contigs totaling 3,154,893 bp, with a GC content of 46.2%, 3275 predicted CDSs, and 68 RNA genes.

Table 4.
Main features of the draft genome sequence of L. mesenteroides subsp. mesenteroides B8 and L. paracasei subsp. tolerans B19.
The draft genome sequences have been deposited in the NCBI database. L. paracasei subsp. tolerans B19 is associated with BioSample SAMN52826719 (accession number JBSHYA000000000), and L. mesenteroides subsp. mesenteroides B8 with accession number JBSHXZ000000000.
According to RAST annotation, approximately 29% of the L. mesenteroides subsp. mesenteroides B8 genome and 21% of the L. paracasei subsp. tolerans B19 genome were assigned to functional subsystems (Figure 1 and Figure 2, respectively), while the remaining genes corresponded to hypothetical or unclassified functions.
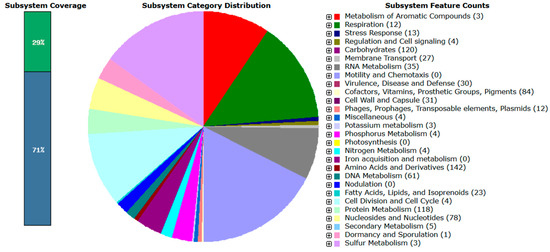
Figure 1.
Overview of RAST subsystem analysis for L. mesenteroides subsp. mesenteroides B8. The pie chart represents the presented subsystem by cellular process, and protein-coding genes (in parentheses) that are predicted to be involved in that cellular process are indicated.
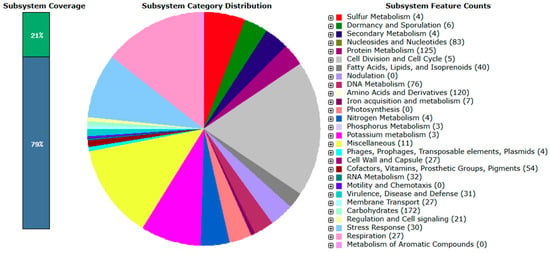
Figure 2.
Overview of RAST subsystem analysis for L. paracasei subsp. tolerans B19. The pie chart represents the presented subsystem by cellular process, and protein-coding genes (in parentheses) that are predicted to be involved in that cellular process are indicated.
In L. mesenteroides subsp. mesenteroides B8, the most abundant gene categories included amino acids and derivatives (142), carbohydrates (120), protein metabolism (118), cofactors, vitamins, prosthetic groups, and pigments (84), and nucleosides and nucleotides (78). Additional subsystems were related to DNA metabolism (61), RNA metabolism (35), capsule and cell wall synthesis (31), virulence, disease, and defense (30), membrane transport (27), fatty acids, lipids, and isoprenoids (23), and stress response (23). Minor categories included respiration (12), mobile genetic elements (12), and secondary metabolites (5).
In L. paracasei subsp. tolerans B19, the predominant functional categories were related to carbohydrate metabolism (172), protein metabolism (125), amino acids and derivatives (120), nucleosides and nucleotides (83), and DNA metabolism (76). Additional functional subsystems included cofactors, vitamins, prosthetic groups, and pigments (54), fatty acids, lipids, and isoprenoids (40), RNA metabolism (32), virulence, disease, and defense (31), stress response (30), capsule and cell wall synthesis (27), and membrane transport (27). Other minor categories were associated with respiration, signaling, iron acquisition, and secondary metabolite biosynthesis.
These findings indicate that both genomes encode extensive metabolic versatility, stress-response systems, and pathways relevant to probiotic and technological performance, consistent with their observed acid and bile tolerance and fermentation capacities.
3.4.1. CAZymes
Enzymes involved in the degradation, synthesis, and modification of carbohydrates, collectively known as carbohydrate-active enzymes (CAZymes), were analyzed in L. mesenteroides subsp. mesenteroides B8 and L. paracasei subsp. tolerans B19.
In L. mesenteroides subsp. mesenteroides B8, a total of 116 CAZymes were identified using the CAZy database (Figure 3). These included 66 glycoside hydrolases (GHs), 34 glycosyltransferases (GTs), 12 carbohydrate-binding modules (CBMs)—four of which were classified as both CBMs and GHs—and three carbohydrate esterases (CEs). Among the predicted CAZymes, RAST and Prokka annotations revealed genes encoding β-galactosidase, riboflavin synthase, and aromatic amino acid aminotransferase α, the latter associated with aromatic amino acid biosynthesis.
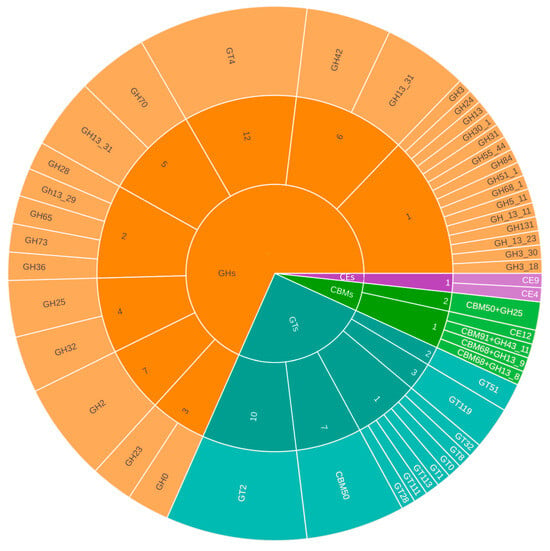
Figure 3.
Distribution of CAZy enzyme families in L. mesenteroides subsp. mesenteroides B8. Colors represent the major CAZy classes: orange shades correspond to Glycoside Hydrolases (GHs), teal/blue-green shades to Glycosyltransferases (GTs), green shades to Carbohydrate-Binding Modules (CBMs), and purple shades to Carbohydrate Esterases (CEs). Lighter or darker tones within each color group indicate the different families identified within each CAZy class.
In L. paracasei subsp. tolerans B19, 149 CAZymes were identified (Figure 4), comprising 68 GHs, 51 GTs, 15 CBMs, four CEs, two polysaccharide lyases (PLs), and one auxiliary activity enzyme (AA). Annotation with RAST and Prokka also identified genes related to maltodextrin glucosidase, acetyl-CoA biotin carboxylase, and thymidylate synthase, which are potentially linked to carbohydrate metabolism, lipid biosynthesis, and nucleotide synthesis, respectively.
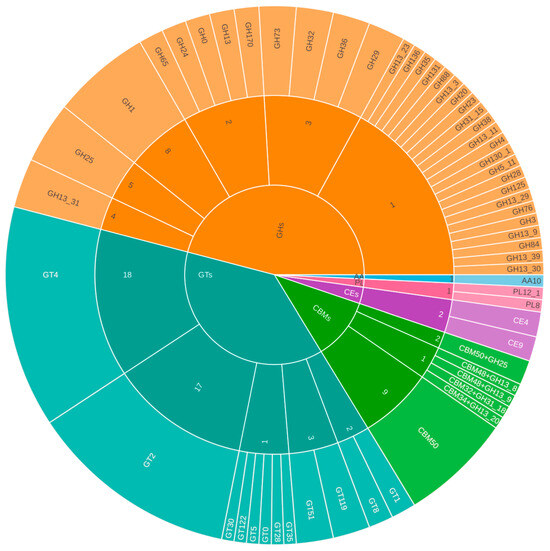
Figure 4.
Distribution of CAZy enzyme families in L. paracasei subsp. tolerans B19. Colors represent the major CAZy classes: orange shades correspond to Glycoside Hydrolases (GHs), teal/blue-green shades to Glycosyltransferases (GTs), green shades to Carbohydrate-Binding Modules (CBMs), and purple shades to Carbohydrate Esterases (CEs). Lighter or darker tones within each color group indicate the different families identified within each CAZy class.
These results indicate that both genomes possess a wide array of CAZymes, reflecting their metabolic versatility and potential ability to use diverse carbohydrate sources, a feature desirable for fermentation and probiotic functionality.
3.4.2. antiSMASH Analysis
AntiSMASH analysis identified several biosynthetic gene clusters (BGCs) in both isolates (Table 5 and Table 6). In B8, four clusters were detected: a cytokinin/terpenoid precursor cluster, a T3PKS cluster, and two clusters associated with β-lactones and lincosamide-like compounds.

Table 5.
Predicted biosynthetic gene clusters (BGCs) associated with secondary metabolite synthesis in L. mesenteroides subsp. mesenteroides B8 identified by antiSMASH.

Table 6.
Predicted biosynthetic gene clusters (BGCs) associated with secondary metabolite synthesis in L. paracasei subsp. tolerans B19 identified by antiSMASH.
The cytokinin/terpenoid cluster (359,483–406,232 bp) included genes encoding (2E,6E)-farnesyl diphosphate synthase (EC 2.5.1.10) and tRNA dimethylallyl transferase (EC 2.5.1.75), both linked to isoprenoid biosynthesis. The T3PKS cluster (64,089–105,246 bp) contained the hydroxymethylglutaryl-CoA synthase gene (EC 2.3.3.10). The β-lactone and lincosamide clusters encoded O-succinylbenzoic acid-CoA ligase (EC 6.2.1.26), 2-isopropylmalate synthase (EC 2.3.3.13), and several oxidoreductases, indicating the potential to synthesize diverse bioactive compounds.
In B19, four BGCs were also predicted. Three corresponded to RiPP clusters and one to a terpenoid precursor cluster. RiPPs include well-known antimicrobial peptides such as nisin, microcin, and plantazolicin. In B19, antiSMASH identified two short open reading frames (6038–6555 bp). BLAST+ 2.13.0 analysis showed 98–100% similarity to class Ib bacteriocins from Lactobacillus paracasei. The terpenoid cluster (93,206–114,057 bp) also contained the (2E,6E)-farnesyl diphosphate synthase gene, suggesting a role in isoprenoid metabolism.
Overall, both genomes contain genes associated with the production of antimicrobial and bioactive metabolites, consistent with the biosynthetic potential often observed in LAB.
3.4.3. Prokka
In addition to secondary metabolite biosynthesis, Prokka annotation identified multiple genes associated with stress adaptation, survival, and probiotic functionality in both isolates.
In B8 (Table 7), relevant genes were linked to cold stress (cspLA_1, cspLA_2, cspG), acid resistance (aroE, npr, murE, atp operon), and adhesion (dltA, dltC, dltD). Several genes related to biosynthesis and redox metabolism (coaBC, gshAB, ribD, ribBA) were also present. Carbohydrate-metabolism genes such as lacZ, bglB, bgaA and were detected as well, indicating metabolic versatility and environmental adaptability.

Table 7.
Functional genes predicted by Prokka annotation in L. mesenteroides subsp. mesenteroides B8 related to stress response, adhesion, and metabolism.
In B19, Prokka annotation revealed genes associated with gastrointestinal survival (uspA, msrA, msrB), heat and osmotic stress response (hrcA, ctsR, dnaK, dnaJ, clpB), and cell wall biosynthesis (murT, lysM, pilO). Additional genes involved in adhesion and capsular polysaccharide formation (cpsH, gtrA), protein folding (grpE, hslV), and bacteriocin export (abpD) were identified (Table 8).

Table 8.
Functional genes predicted by Prokka annotation in in L. paracasei subsp. tolerans B19 related to stress response, adhesion, and metabolism.
These genomic traits are consistent with the observed physiological tolerance to acid and bile, suggesting a robust genetic basis for probiotic functionality and technological potential.
In addition, KEGG pathway mapping confirmed the presence of core metabolic routes involved in amino acid biosynthesis, carbohydrate metabolism, and cofactor/vitamin synthesis, consistent with the fermentative and adaptive metabolism typical of LAB.
3.5. Antibiotic Susceptibility and Genome-Based AMR Assessment
Most isolates were susceptible to ampicillin and showed the expected intrinsic resistance to the aminoglycosides streptomycin and gentamicin. B8 and B19 displayed clear inhibition halos for ampicillin (25 ± 0.5 mm and 28 ± 0.6 mm, respectively), confirming their susceptibility to β-lactams.
These phenotypic results align with the genome-based AMR screening, which identified no acquired antimicrobial resistance genes in either strain.
4. Discussion
4.1. Ecological and Technological Relevance
Interest in the microbiota of donkey milk has increased notably in recent years, and recent metagenomic studies have contributed to a deeper understanding of its microbial ecology. Ruso et al. reported that, under cold storage conditions, raw donkey milk tends to shift towards psychrotrophic spoilage-associated taxa, such as Pseudomonas spp., while lactic acid bacteria (LAB) progressively decline. This observation is consistent with the storage conditions used in our study and may explain the limited number of LAB isolates recovered.
Despite this reduction, the LAB species isolated in our work agree well with previous reports. Lacticaseibacillus paracasei subsp. tolerans—recovered in our study—was also detected by Ruso et al. [31] and has recently been isolated from bulk donkey milk tanks [31]. This species is also widely distributed in raw cow and goat milk [18], suggesting that it is a common inhabitant of raw milk environments. Likewise, Leuconostoc mesenteroides subsp. mesenteroides, also isolated here, has been previously reported in cow’s and sheep’s milk as well as kefir grains [32] and in raw donkey milk [17]. These observations indicate that this species is naturally associated with raw dairy matrices and support our findings.
MRS medium was selected for LAB cultivation, as it is one of the most widely used media for this purpose [33]. However, MRS is not fully selective, and the recovery of non-LAB microorganisms—including species belonging to the genus Bacillus—has been reported [23]. Previous studies have identified potentially probiotic Bacillus species, such as Bacillus paramycoides in raw donkey milk [34]. In this study, we report Bacillus tequilensis in raw donkey milk for the first time. Collectively, these findings indicate that raw donkey milk may constitute a niche for the isolation of Bacillus species with potential probiotic properties.
Overall, the microbiota of Zamorano-Leonesa donkey milk represents an underexplored reservoir of functional biodiversity. This study expands the range of cultivable microorganisms identified in donkey milk and provides new evidence of its value as a source of technologically and functionally relevant strains. These findings highlight the potential of donkey milk microbiota for applications in food biotechnology and for the development of new functional dairy products.
4.2. Functional Performance and Stress Tolerance
Functional screening showed that several isolates produced strong acidification in pasteurized donkey milk and maintained growth under acidic conditions (pH 2.5) and in the presence of bile (0.3% ox bile). These traits are essential for technological performance, enabling rapid milk acidification, and for potential probiotic functionality, supporting survival under gastrointestinal-like stress [35].
Among the isolates, B8 and B19 were selected as representative strains due to their combined performance. B8 exhibited efficient growth and acid tolerance, consistent with the behavior of heterofermentative Leuconostoc species. In contrast, B19 showed strong resistance to both low pH and bile salts, a trait commonly observed in Lacticaseibacillus strains [36]. These characteristics align with the ecological origin of the isolates, as LAB from raw donkey milk are naturally exposed to fluctuating nutrient availability and temperature, which may enhance their stress resilience [17].
4.3. Genome-Based Functional Insights
The genomic profiles of B19 and B8 provide insights into their ecological adaptation and functional roles in dairy environments. Both strains encode numerous carbohydrate-metabolism genes, including phosphotransferase systems, permeases, and glycosyl hydrolases, reflecting specialization for milk-derived substrates. B19 contains a larger CAZyme repertoire (149 vs. 116 in B8), indicating an enhanced ability to use complex oligosaccharides from donkey milk, which may contribute to its faster acidification and growth during fermentation.
Both genomes also encode β-galactosidases (lacZ, bgaA) and α-glucosidases, enzymes involved in lactose hydrolysis and flavor development [37]. These features are consistent with desirable starter functionality. Stress-related operons such as dltA/dltC/dltD, associated with teichoic acid modification, suggest robust tolerance to acid and bile, similar to other probiotic lactobacilli [38]. Additional genes, including npr and murE, reinforce resilience to oxidative and osmotic stress, traits relevant for gastrointestinal survival and industrial fermentation [39].
The two strains showed distinct adaptive patterns. B8 encodes several cold-shock proteins, consistent with its isolation from refrigerated milk and indicative of psychrotrophic adaptation [40]. In contrast, B19 carries a more complex stress regulon (dnaK, dnaJ, hrcA, ctsR, clpB, msrA/msrB), supporting broader thermal and oxidative tolerance [41]. Such genomic differentiation supports niche specialization—B8 thriving under refrigeration, while B19 remains stable under variable or stress conditions.
Genes related to adhesion and biofilm formation (lysM, pilO, murT, cpsH, gtrA) were more abundant in B19, suggesting stronger mucosal adherence and persistence—key probiotic traits [42]. Their coexistence with the dlt operon may also indicate surface charge modulation, favoring colonization and competitive exclusion of pathogens.
AntiSMASH analysis identified several BGCs in both strains, including RiPP and terpenoid precursor clusters commonly associated with bacteriocin synthesis. The presence of abpD in B19 supports active bacteriocin export, suggesting antimicrobial capacity and natural biopreservation potential [43]. In contrast, B8 encoded several redox-related enzymes (ribD, ribBA, gshAB), which may contribute to oxidative stress regulation and cellular redox balance, enhancing resilience [44].
Overall, the two strains appear to play complementary roles. B8 is specialized for cold adaptation and flavor development, whereas B19 shows greater metabolic flexibility, stress tolerance, adhesion capacity, and antimicrobial potential. Together, these attributes support their use as a synergistic consortium in fermented or functional donkey milk, in line with current probiotic design approaches.
4.4. Biotechnological Implication
Combined phenotypic and genomic evidence supports the complementary functional roles of B8 and B19 as technologically relevant LAB. B8 shows key traits of heterofermentative starter cultures, including moderate acidification, aromatic compound production, and adaptation to low temperatures [45,46]. In contrast, B19 displays strong acid and bile tolerance, a broad enzymatic repertoire, and genomic markers associated with bacteriocin synthesis. Together, these features indicate that B8 and B19 could function as a complementary consortium for producing fermented or functional donkey milk products. They also represent promising probiotic candidates for nutraceutical applications.
More broadly, this study provides one of the first integrated analyses combining isolation, phenotypic screening, and genome-based functional annotation of LAB from donkey milk. These findings establish a foundation for future work aimed at validating probiotic traits, evaluating antimicrobial activity, and clarifying the metabolic contributions of these strains during milk fermentation.
4.5. Antibiotic Susceptibility and AMR Genomic Assessment
The disk diffusion results showed that B8 and B19 were susceptible to ampicillin and displayed the expected intrinsic resistance to aminoglycosides. This phenotype is consistent with the well-documented, non-transferable aminoglycoside resistance typical of lactic acid bacteria, which stems from the natural impermeability of their cell envelope [47].
Genomic screening using ResFinder did not detect any acquired antimicrobial resistance genes in either strain. The absence of AMR determinants supports the phenotypic findings and indicates that the susceptibility to β-lactams reflects a genuine lack of acquired resistance mechanisms. Likewise, the intrinsic aminoglycoside resistance observed phenotypically was not associated with any acquired AMR genes.
Overall, the agreement between phenotypic and genomic data supports the safe AMR profile of both strains, fully in line with current EFSA requirements for starter cultures and probiotics.
5. Conclusions
This study highlights donkey milk from the endangered Zamorano-Leonesa breed as a valuable ecological niche for recovering functionally relevant lactic acid bacteria. Among the isolates, L. mesenteroides subsp. mesenteroides B8 and L. paracasei subsp. tolerans B19 stood out for their complementary technological and probiotic traits, combining robust stress tolerance with desirable acidification capacity. Genomic analysis supported these phenotypes by revealing genes linked to adhesion, environmental adaptation, and beneficial metabolic functions.
Notably, the concordance between phenotypic antibiotic susceptibility and in silico AMR assessment confirmed the absence of acquired antimicrobial resistance genes, supporting the safety of both strains for food and biotechnological applications in line with current EFSA recommendations.
Overall, this integrative phenotypic–genomic approach expands current knowledge of donkey-milk microbiota and identifies two promising LAB strains with potential synergistic value in fermented dairy products and functional foods. Future work should include in vitro assays on antimicrobial activity, adhesion to intestinal epithelial cells, sensory evaluation of fermented products, and transcriptomic analyses under cold and stress conditions to validate in silico predictions and further substantiate their technological suitability.
Author Contributions
Conceptualization, A.D.-M.; methodology, A.D.-M.; validation, A.D.-M.; formal analysis, D.B. and I.A.; investigation, A.D.-M.; resources, A.D.-M.; data curation, D.B.; writing—original draft preparation, D.B. and A.D.-M.; writing—review and editing, A.D.-M.; visualization, A.D.-M.; supervision, A.D.-M.; review of the final manuscript, I.A., J.-M.J. and M.J.C.-A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The draft genome sequences generated in this study have been deposited in the NCBI database. Lactobacillus paracasei B19 is associated with BioSample SAMN52826719 (submission ID SUB15713980), and Leuconostoc mesenteroides B8 with submission ID SUB15714241.
Acknowledgments
The authors would like to thank the Cooperativa BULEZA for kindly donating the milk used in this research.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| AMR | Antimicrobial Resistance |
| CAZy | Carbohydrate active enzyme |
| CARD | Comprehensive Antibiotic Resistance Database |
| CDSs | Coding Sequences |
| EFSA | European Food Safety Authority |
| HPP | High pressure process |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| RiPPs | Ribosomally synthesized and post-translationally modified proteins |
References
- Sarti, L.; Martini, M.; Brajon, G.; Barni, S.; Salari, F.; Altomonte, I.; Ragona, G.; Mori, F.; Pucci, N.; Muscas, G.; et al. Donkey’s Milk in the Management of Children with Cow’s Milk Protein Allergy: Nutritional and Hygienic Aspects. Ital. J. Pediatr. 2019, 45, 102. [Google Scholar] [CrossRef]
- Albertos, I.; López, M.; Jiménez, J.M.; Cao, M.J.; Corell, A.; Castro-Alija, M.J. Characterisation of Zamorano-Leonese donkey milk as an alternative sustainably produced protein food. Front. Nutr. 2022, 9, 872409. [Google Scholar] [CrossRef]
- Martini, M.; Altomonte, I.; Licitra, R.; Salari, F. Nutritional and nutraceutical quality of donkey milk. J. Equine Vet. Sci. 2018, 65, 33–37. [Google Scholar] [CrossRef]
- Singh, M.P.; Vashisht, P.; Singh, L.; Awasti, N.; Sharma, S.; Mohan, C.; Charles, A.P.R. Donkey milk as a non-bovine alternative: A review of its nutri-functional properties, applications, and challenges. J. Food Sci. Technol. 2024, 61, 1652–1661. [Google Scholar] [CrossRef]
- Plotuna, A.M.; Hotea, I.; Ban-Cucerzan, A.; Badea, C.; Mladin, A.; Tîrziu, E. The microbial landscape of donkey milk: A systematic review. Rom. J. Vet. Sci. 2025, 58, 2. [Google Scholar] [CrossRef]
- European Commission. Regulation (EC) No 853/2004 of the European Parliament and of the Council of 29 April 2004 on the Hygiene of Food of Animal Origin. Off. J. Eur. Union 2004, L 139, 55–205. [Google Scholar]
- Codex Alimentarius. General Principles of Food Hygiene CXC 1-1969; 2020 Revision; FAO: Rome, Italy, 2020. [Google Scholar]
- Papademas, P.; Kamilari, E.; Aspri, M.; Anagnostopoulos, D.A.; Mousikos, P.; Kamilaris, A.; Tsaltas, D. Investigation of donkey milk bacterial diversity by 16S rDNA high-throughput sequencing on a Cyprus donkey farm. J. Dairy Sci. 2021, 104, 167–178. [Google Scholar] [CrossRef]
- Axelsson, L.; Fontana, A.; Morelli, L.; von Wright, A. Lactic Acid Bacteria: An Introduction to Taxonomy, Physiology, and Molecular Biology. In Lactic Acid Bacteria; CRC Press: Boca Raton, FL, USA, 2024; pp. 3–27. [Google Scholar]
- Hernández-Figueroa, R.H.; López-Malo, A.; Mani-López, E. Lactic acid bacteria-derived exopolysaccharides: Dual roles as functional ingredients and fermentation agents in food applications. Fermentation 2025, 11, 538. [Google Scholar] [CrossRef]
- Cirat, R.; Capozzi, V.; Benmechernene, Z.; Spano, G.; Grieco, F.; Fragasso, M. LAB antagonistic activities and their significance in food biotechnology: Molecular mechanisms, food targets, and other related traits of interest. Fermentation 2024, 10, 222. [Google Scholar] [CrossRef]
- Farid, W.; Masud, T.; Sohail, A.; Ahmad, N.; Naqvi, S.S.; Khan, S.; Manzoor, M.F. Gastrointestinal transit tolerance, cell surface hydrophobicity, and functional attributes of Lactobacillus acidophilus strains isolated from Indigenous Dahi. Food Sci. Nutr. 2021, 9, 5092–5102. [Google Scholar] [CrossRef] [PubMed]
- Mani-López, E.; Arrioja-Bretón, D.; López-Malo, A. The impacts of antimicrobial and antifungal activity of cell-free supernatants from lactic acid bacteria in vitro and foods. Compr. Rev. Food Sci. Food Saf. 2022, 21, 604–641. [Google Scholar] [CrossRef]
- Pellegrino, M.S.; Frola, I.D.; Natanael, B.; Gobelli, D.; Nader-Macias, M.E.; Bogni, C.I. In vitro characterization of lactic acid bacteria isolated from bovine milk as potential probiotic strains to prevent bovine mastitis. Probiotics Antimicrob. Proteins 2019, 11, 74–84. [Google Scholar] [CrossRef]
- Islam, M.Z.; Uddin, M.E.; Rahman, M.T.; Islam, M.A.; Harun-ur-Rashid, M. Isolation and characterization of dominant lactic acid bacteria from raw goat milk: Assessment of probiotic potential and technological properties. Small Rumin. Res. 2021, 205, 106532. [Google Scholar] [CrossRef]
- Patil, A.; Disouza, J.; Pawar, S. Shelf life stability of encapsulated lactic acid bacteria isolated from sheep milk thrived in different milk as natural media. Small Rumin. Res. 2019, 170, 19–25. [Google Scholar] [CrossRef]
- Massouras, T.; Bitsi, N.; Paramithiotis, S.; Manolopoulou, E.; Drosinos, E.H.; Triantaphyllopoulos, K.A. Microbial profile, antibacterial properties and chemical composition of raw donkey milk. Animals 2020, 10, 2001. [Google Scholar] [CrossRef] [PubMed]
- Greifová, G.; Drobná, E.; Olejníková, P.; Greif, G.; Greifová, M. Isolation, characterisation and technological properties of raw donkey’s milk isolate, Lacticaseibacillus paracasei, compared to raw goat’s and cow’s milk isolates. Czech J. Food Sci. 2025, 43, 118–128. [Google Scholar] [CrossRef]
- Negm El-Dein, A.; Noor El-Deen, A.; Tolba, S.; El-Shatoury, E.; Awad, G.; Ibrahim, M.; Farid, M. Probiotic properties and bile salt hydrolase activity of some isolated lactic acid bacteria. Egypt. J. Microbiol. 2017, 52, 87–100. [Google Scholar] [CrossRef]
- Mejía-Caballero, A.; López-Sánchez, R.; Ramos-Cerrillo, B.; Garciarrubio, A.; Segovia, L. Genomic insights into habitat adaptation of Lactobacillus species. World J. Microbiol. Biotechnol. 2025, 41, 61. [Google Scholar] [CrossRef]
- Huang, Z.; Zhou, X.; Stanton, C.; Ross, R.P.; Zhao, J.; Zhang, H.; Chen, W. Comparative genomics and specific functional characteristics analysis of Lactobacillus acidophilus. Microorganisms 2021, 9, 1992. [Google Scholar] [CrossRef]
- Wels, M.; Siezen, R.; van Hijum, S.; Kelly, W.J.; Bachmann, H. Comparative genome analysis of Lactococcus lactis indicates niche adaptation and resolves genotype/phenotype disparity. Front. Microbiol. 2019, 10, 4. [Google Scholar] [CrossRef]
- Menéndez-Cañamares, S.; Blázquez, A.; Albertos, I.; Poveda, J.; Díez, A. Probiotic Bacillus subtilis SB8 and edible coatings for sustainable fungal disease management in strawberry. Biol. Control 2024, 196, 105572. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Vinderola, G.; Gueimonde, M.; Gomez-Gallego, C.; Delfederico, L.; Salminen, S. Correlation between in vitro and in vivo assays in selection of probiotics from traditional species of bacteria. Trends Food Sci. Technol. 2017, 68, 83–90. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Medema, M.H.; Weber, T. The antiSMASH database version 4: Additional genomes and BGCs, new sequence-based searches and more. Nucleic Acids Res. 2024, 52, D586–D589. [Google Scholar] [CrossRef]
- Zheng, J.; Ge, Q.; Yan, Y.; Zhang, X.; Huang, L.; Yin, Y. dbCAN3: Automated carbohydrate-active enzyme and substrate annotation. Nucleic Acids Res. 2023, 51, W115–W121. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Aarestrup, F.M. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Russo, P.; Fiocco, D.; Albenzio, M.; Spano, G.; Capozzi, V. Microbial populations of fresh and cold-stored donkey milk by high-throughput sequencing provide indication for a correct management of this high-value product. Appl. Sci. 2020, 10, 2314. [Google Scholar] [CrossRef]
- Akpinar, A.; Yerlikaya, O. Some potential beneficial properties of Lacticaseibacillus paracasei subsp. paracasei and Leuconostoc mesenteroides strains originating from raw milk and kefir grains. J. Food Process. Preserv. 2021, 45, e15986. [Google Scholar] [CrossRef]
- Hayek, S.A.; Gyawali, R.; Aljaloud, S.O.; Krastanov, A.; Ibrahim, S.A. Cultivation media for lactic acid bacteria used in dairy products. J. Dairy Res. 2019, 86, 490–502. [Google Scholar] [CrossRef]
- Djerrab, L.; Chekroud, Z.; Rouabhia, A.; Dems, M.A.; Attailia, I.; Garcia, L.I.R.; Smadi, M.A. Potential use of Bacillus paramycoides for the production of the biopolymer polyhydroxybutyrate from leftover carob fruit agro-waste. AIMS Microbiol. 2022, 8, 318. [Google Scholar] [CrossRef]
- Singhal, N.; Singh, N.S.; Mohanty, S.; Kumar, M.; Virdi, J.S. Rhizospheric Lactobacillus plantarum (Lactiplantibacillus plantarum) strains exhibit bile salt hydrolysis, hypocholesterolemic and probiotic capabilities in vitro. Sci. Rep. 2021, 11, 15288. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Yu, L.; Tian, F.; Zhao, J.; Zhai, Q. Identification of Novel Bile Salt-Tolerant Genes in Lactobacillus Using Comparative Genomics and Its Application in the Rapid Screening of Tolerant Strains. Microorganisms 2022, 10, 2371. [Google Scholar] [CrossRef] [PubMed]
- Costa, T.J.N.; Costa, I.M.; Rocha, W.C.D.; Silva, J.F.; Sousa, L.F.; Pereira, A.L.F.; Gomes, M.M.; Albuquerque, R.F.; Nascimento, J.S.; Oliveira, M.E.G.; et al. Lactococcus lactis GV103, potentially probiotic, applied in the development of lactose-free fermented milk. Braz. J. Microbiol. 2025, 56, 1669–1679. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhang, Y.; Li, L.; Li, Y.; Yuan, L.; E, Y.; Qiao, J. Positive Regulation of the DLT Operon by TCSR7 Enhances Acid Tolerance of Lactococcus lactis F44. J. Dairy Sci. 2022, 105, 7940–7950. [Google Scholar] [CrossRef]
- Kandasamy, S.; Lee, K.H.; Yoo, J.; Yun, J.; Kang, H.B.; Kim, J.E.; Ham, J.S. Whole genome sequencing of Lacticaseibacillus casei KACC92338 strain with strong antioxidant activity reveals genes and gene clusters of probiotic and antimicrobial potential. Front. Microbiol. 2024, 15, 1458221. [Google Scholar] [CrossRef]
- Samodra, E.M.A.; Suroto, D.; Utami, T.; Wikandari, R.; Rahayu, E.S. Cold Stress Response Genes of Lactiplantibacillus plantarum subsp. plantarum Mut-3 and Lactiplantibacillus plantarum subsp. plantarum Mut-7 Support the Ability to Survive in Low-Temperature Conditions. HAYATI J. Biosci. 2023, 30, 65–70. [Google Scholar] [CrossRef]
- Bucka-Kolendo, J.; Sokołowska, B. Impact of High Hydrostatic Pressure on the Single Nucleotide Polymorphism of Stress-Related dnaK, hrcA, and ctsR in Lactobacillus Strains. Qual. Assur. Saf. Crops Foods 2022, 14, 54–66. [Google Scholar] [CrossRef]
- Nöldeke, E.R.; Stehle, T. Unraveling the Mechanism of Peptidoglycan Amidation by the Bifunctional Enzyme Complex GatD/MurT: A Comparative Structural Approach. Int. J. Med. Microbiol. 2019, 309, 151334. [Google Scholar] [CrossRef]
- Han, S.; Elnar, A.G.; Lim, C.; Kim, G.B. Complete Genome Sequence of Bacteriocin-Producing Ligilactobacillus salivarius B4311 Isolated from Fecal Samples of Broiler Chicken with Anti-Listeria Activity. J. Anim. Sci. Technol. 2024, 66, 232–236. [Google Scholar] [CrossRef]
- Averina, O.V.; Poluektova, E.U.; Marsova, M.V.; Danilenko, V.N. Biomarkers and Utility of the Antioxidant Potential of Probiotic Lactobacilli and Bifidobacteria as Representatives of the Human Gut Microbiota. Biomedicines 2021, 9, 1340. [Google Scholar] [CrossRef]
- Goyal, D.; Swaroop, S.; Prakash, O.; Pandey, J. Survival Strategies in Cold-Adapted Microorganisms. In Survival Strategies in Cold-Adapted Microorganisms; Springer: Singapore, 2021; pp. 173–186. [Google Scholar] [CrossRef]
- Martínez-Miranda, J.G.; Chairez, I.; Durán-Páramo, E. Mannitol Production by Heterofermentative Lactic Acid Bacteria: A Review. Appl. Biochem. Biotechnol. 2022, 194, 2762–2795. [Google Scholar] [CrossRef] [PubMed]
- Darsanaki, R.K.; Aliabadi, M.A.; Chakoosari, M.M.D. Antibiotic resistance of lactic acid bacteria. Sci. J. Microbiol. 2013, 2, 201–206. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).