Differential Rumen Microbial Taxa in Charolais Bulls with Divergent Residual Feed Intake
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals, Diet, RFI Determination, and Sampling
2.2. DNA Extraction and 16s rRNA Gene Sequencing
2.3. Data and Statistical Analysis
3. Results
3.1. Growth Performance
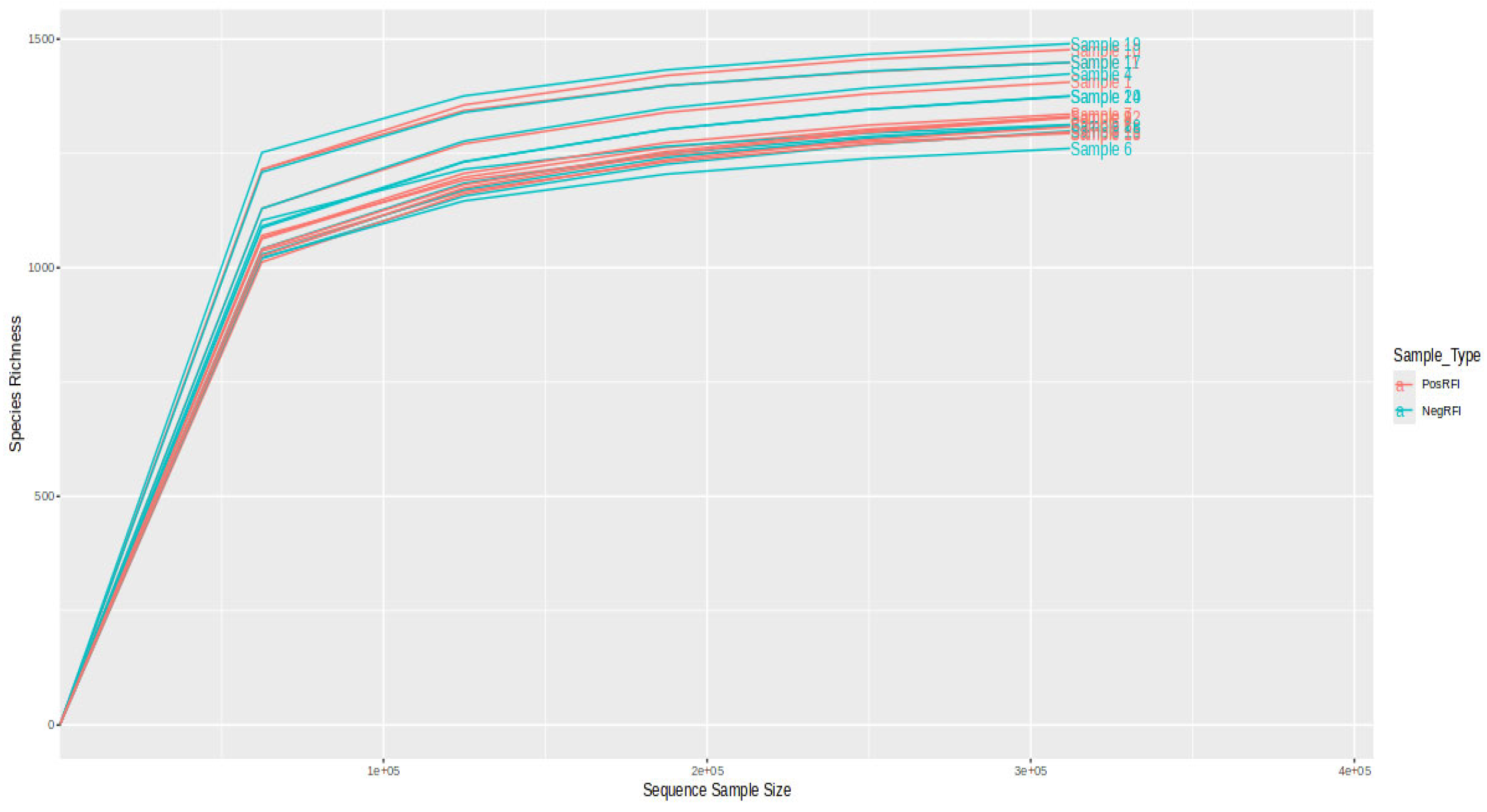
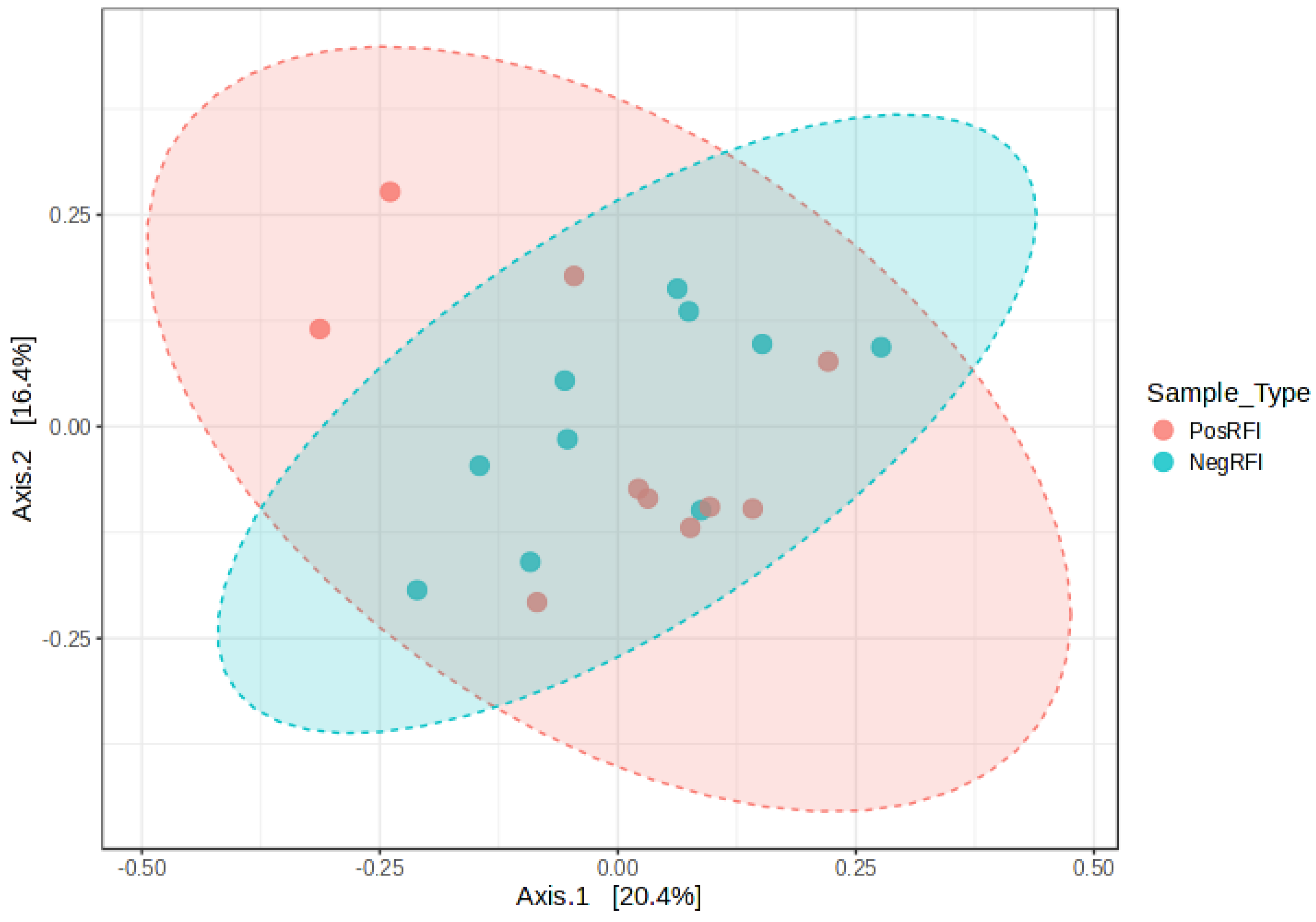
3.2. Rumen Bacterial Community
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Archer, J.A.; Richardson, E.C.; Herd, R.M.; Arthur, P.F. Potential for selection to improve efficiency of feed use in beef cattle: A review. Aust. J. Agric. Res. 1999, 50, 147–162. [Google Scholar] [CrossRef]
- Knight, R.; Hahn, W.; Taylor, H.; Terán, A.; Haley, M.; Grossen, G.; Valcu-Lisman, A.; Cornelius, M.; Collins, L.A. Livestock, Dairy, and Poultry: July 2022; USDA: Washington, DC, USA, 2022. [Google Scholar]
- Arthur, J.P.F.; Herd, R.M. Residual feed intake in beef cattle. Rev. Bras. Zootec. 2008, 37, 269–279. [Google Scholar] [CrossRef]
- Santiago, K.G.; Lopez, B.I.; Kim, S.-H.; Lee, D.-H.; Cho, Y.-G.; Song, Y.-N.; Seo, K. Genetic Parameters for Different Measures of Feed Efficiency and Their Relationship to Production Traits in Three Purebred Pigs. Life 2021, 11, 830. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Liu, J.; Yu, F.; Shen, Y.; Gong, S.; Lu, Y.; Peng, W.; Wang, Y.; Gan, Y.; Xiao, Q.; et al. Heritabilityand genetic correlation for residual feed intake of Pacific abalone Haliotis discus hannai. Aquaculture 2022, 553, 738060. [Google Scholar] [CrossRef]
- VandeHaar, M.J.; Armentano, L.E.; Weigel, K.; Spurlock, D.M.; Tempelman, R.J.; Veerkamp, R. Harnessing the geneticsof the modern dairy cow to continue improvements in feed efficiency. J. Dairy Sci. 2016, 99, 4941–4954. [Google Scholar] [CrossRef]
- Vytelle On Farm Genetic Selection for Feed Efficiency. 2021. Available online: https://vytelle.com/tools/on-farm-genetic-selection-for-feed-efficiency/ (accessed on 18 December 2024).
- McDonald, T.J.; Brester, G.W.; Bekkerman, A.; Paterson, J.A. CASE STUDY: Searching for the Ultimate Cow: The Economic Value of Residual Feed Intake at Bull Sales. Prof. Anim. Sci. 2010, 26, 655–660. [Google Scholar] [CrossRef]
- Vytelle Feed Savings Associated with Using a Low RFI Bull. 2021. Available online: https://vytelle.com/tools/feed-savings-associated-with-using-a-low-rfi-bull/ (accessed on 19 December 2024).
- Owens, F.N.; Basalan, M. Ruminal Fermentation. In Rumenology; Millen, D.D., De Beni Arrigoni, M., Pacheco, R.D.L., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 63–102. [Google Scholar] [CrossRef]
- Li, F.; Li, C.; Chen, Y.; Liu, J.; Zhang, C.; Irving, B.; Fitzsimmons, C.; Plastow, G.; Guan, L.L. Host genetics influence the rumen microbiota and heritable rumen microbial features associate with feed efficiency in cattle. Microbiome 2019, 7, 92. [Google Scholar] [CrossRef]
- Auffret, M.D.; Dewhurst, R.J.; Duthie, C.-A.; Rooke, J.A.; John Wallace, R.; Freeman, T.C.; Stewart, R.; Watson, M.; Roehe, R. The rumen microbiome as a reservoir of antimicrobial resistance and pathogenicity genes is directly affected by diet in beef cattle. Microbiome 2017, 5, 159. [Google Scholar] [CrossRef]
- Clemmons, B.A.; Voy, B.H.; Myer, P.R. Altering the Gut Microbiome of Cattle: Considerations of Host-Microbiome Interactions for Persistent Microbiome Manipulation. Microb. Ecol. 2019, 77, 523–536. [Google Scholar] [CrossRef]
- Virgínio Júnior, G.F.; da Silva, A.P.; de Toledo, A.F.; Poczynek, M.; Cezar, A.M.; Montenegro, H.; Coutinho, L.L.; Bittar, C.M.M. Ruminal and Fecal Bacteriome of Dairy Calves Fed Different Levels and Sources of NDF. Animals 2021, 11, 2705. [Google Scholar] [CrossRef]
- Gonzalez-Recio, O.; Zubiria, I.; García-Rodríguez, A.; Hurtado, A.; Atxaerandio, R. Short communication: Signs of host genetic regulation in the microbiome composition in 2 dairy breeds: Holstein and Brown Swiss. J. Dairy Sci. 2018, 101, 2285–2292. [Google Scholar] [CrossRef] [PubMed]
- Treon, E.; Sidney, T.; Taiwo, G.; Idowu, M.; Leal, Y.; Ologunagba, D.; Ogunade, I.M. Effects of dietary supplementation of a blend of Saccharomyces cerevisiae, multiple live probiotic bacteria, and their fermentation products onperformance, health, and rumen bacterial community of newly weaned beef steers during a 56-d receiving period. Transl. Anim. Sci. 2024, 8, txad143. [Google Scholar] [CrossRef] [PubMed]
- Idowu, M.; Taiwo, G.; Sidney, T.; Morenikeji, O.B.; Pech Cervantes, A.; Estrada-Reyes, Z.M.; Wilson, M.; Ogunade, I.M. The differential plasma and ruminal metabolic pathways and ruminal bacterial taxa associated with divergent residual body weight gain phenotype in crossbred beef steers. Transl. Anim. Sci. 2023, 7, txad054. [Google Scholar] [CrossRef]
- MacNeil, M.D.; Berry, D.P.; Clark, S.A.; Crowley, J.J.; Scholtz, M.M. Evaluation of partial body weight for predicting bodyweight and average daily gain in growing beef cattle. Transl. Anim. Sci. 2021, 5, txab126. [Google Scholar] [CrossRef]
- Taiwo, G.; Idowu, M.D.; Wilson, M.; Pech-Cervantes, A.; Estrada-Reyes, Z.M.; Ogunade, I.M. Residual Feed Intake in Beef Cattle Is Associated With Differences in Hepatic mRNA Expression of Fatty Acid, Amino Acid, and Mitochondrial Energy Metabolism Genes. Front. Anim. Sci. 2022, 3, 828591. [Google Scholar] [CrossRef]
- Durunna, O.N.; Mujibi, F.D.N.; Goonewardene, L.; Okine, E.K.; Basarab, J.A.; Wang, Z.; Moore, S.S. Feed efficiencydifferences and reranking in beef steers fed grower and finisher diets. J. Anim. Sci. 2011, 89, 158–167. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Chong, J.; Liu, P.; Zhou, G.; Xia, J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat Protoc. 2020, 15, 799–821. [Google Scholar] [CrossRef]
- Taiwo, G.; Morenikeji, O.B.; Idowu, M.; Sidney, T.; Adekunle, A.; Cervantes, A.P.; Peters, S.; Ogunade, I.M. Characterization of rumen microbiome and immune genes expression of crossbred beef steers with divergent residual feed intake phenotypes. BMC Genomics 2024, 25, 245. [Google Scholar] [CrossRef]
- Abbas, W.; Howard, J.; Paz, H.; Hales, K.; Wells, J.; Keuhn, L.; Erickson, G.; Spangler, M.; Fernando, S. Influence of host genetics in shaping the rumen bacterial community in beef cattle. Sci Rep. 2020, 10, 15101. [Google Scholar] [CrossRef]
- Rosselli, R.; Romoli, O.; Vitulo, N.; Vezzi, A.; Campanaro, S.; de Pascale, F.; Schiavon, R.; Tiarca, M.; Poletto, F.; Concheri, G.; et al. Direct 16S rRNA-seq from bacterial communities: A PCR-independent approach to simultaneously assess microbial diversity and functional activity potential of each taxon. Sci Rep. 2016, 6, 32165. [Google Scholar] [CrossRef]
- Commichaux, S.; Luan, T.; Muralidharan, H.S.; Pop, M. Database size positively correlates with the loss of species-leveltaxonomic resolution for the 16S rRNA and other prokaryotic marker genes. Biorxiv 2023. [Google Scholar] [CrossRef]
- Nguyen, N.P.; Warnow, T.; Pop, M.; White, B. A perspective on 16S rRNA operational taxonomic unit clustering usingsequence similarity. Npj Biofilms Microbiomes 2016, 2, 16004. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, R.; Rani, A.; Metwally, A.; McGee, H.; Perkins, D.L. Analysis of the microbiome: Advantages of whole genome shotgun versus 16s amplicon sequencing. Sci. Dir. 2016, 469, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.N.; Méndez–García, C.; Geier, R.R.; Iakiviak, M.; Chang, J.; Cann, I.; Mackie, R.I. Metabolic networks for nitrogen utilization in Prevotella ruminicola 23. Sci. Rep. 2017, 7, 7851. [Google Scholar] [CrossRef]
- Kovatcheva-Datchary, P.; Nilsson, A.; Akrami, R.; Lee, Y.S.; De Vadder, F.; Arora, T.; Hallen, A.; Martens, E.; Björck, I.; Bäckhed, F. Dietary Fiber-Induced Improvement in Glucose Metabolism Is Associated with Increased Abundance of Prevotella. Cell Metab. 2015, 22, 971–982. [Google Scholar] [CrossRef]
- Cruz-Morales, P.; Orellana, C.A.; Moutafis, G.; Moonen, G.; Rincon, G.; Nielsen, L.K.; Marcellin, E. Revisiting the Evolution and Taxonomy of Clostridia, a Phylogenomic Update. Genome Biol. Evol. 2019, 11, 2035–2044. [Google Scholar] [CrossRef]
- Betancur-Murillo, C.L.; SBAguilar-Marín Jovel, J. Prevotella: A Key Player in Ruminal Metabolism. Microorganisms 2022, 11, 1. [Google Scholar] [CrossRef]
- Kou, X.; Ma, Q.; Liu, Y.; Khan, M.Z.; Wu, B.; Chen, W.; Liu, X.; Wang, C.; Li, Y. Exploring the Effect of Gastrointestinal Prevotellaon Growth Performance Traits in Livestock Animals. Animals 2024, 14, 1965. [Google Scholar] [CrossRef]
- Shinkai, T.; Takizawa, S.; Enishi, O.; Higuchi, K.; Ohmori, H.; Mitsumori, M. Characteristics of rumen microbiota and Prevotella isolates found in high propionate and low methane-producing dairy cows. Front. Microbiol. 2024, 15, 1404991. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Chen, B.; Zhang, X.; Akbar, M.T.; Wu, T.; Zhang, Y.; Zhi, L.; Shen, Q. Exploration of the Muribaculaceae Family in the Gut Microbiota: Diversity, Metabolism, and Function. Nutrients 2024, 16, 2660. [Google Scholar] [CrossRef] [PubMed]
- Ormerod, K.L.; Wood, D.L.A.; Lachner, N.; Gellatly, S.L.; Daly, J.N.; Parsons, J.D.; Dal’Molin, C.G.O.; Palfreyman, R.W.; Nielsen, L.K.; Cooper, M.A.; et al. Genomic characterization of the uncultured Bacteroidales family S24-7 inhabiting the guts of homeothermic animals. Microbiome 2016, 4, 36. [Google Scholar] [CrossRef]
- Tong, J.; Zhang, H.; Yang, D.; Xiong, B.; Jiang, L. Illumina sequencing analysis of the ruminal microbiota in high-yield and low-yield lactating dairy cows. bioRxiv 2018. [Google Scholar] [CrossRef]
- Jiang, Y.; Ogunade, I.M.; Pech-Cervantes, A.A.; Fan, P.X.; Li, X.; Kim, D.H.; Arriola, K.G.; Poindexter, M.B.; Jeong, K.C.; Vyas, D.; et al. Effect of sequestering agents based on a Saccharomyces cerevisiae fermentation product and clay on the ruminal bacterial community of lactating dairy cows challenged with dietary aflatoxin B1. J. Dairy Sci. 2020, 103, 1431–1447. [Google Scholar] [CrossRef]
- Gaffney, J.; Embree, J.; Gilmore, S.; Embree, M. Ruminococcus bovis sp. nov., a novel species of amylolytic Ruminococcus isolated from the rumen of a dairy cow. Int. J. Syst. Evol. Microbiol. 2021, 71, 004924. [Google Scholar] [CrossRef]
- Christopherson, M.R.; Dawson, J.A.; Stevenson, D.M.; Cunningham, A.C.; Bramhacharya, S.; Weimer, P.J.; Kendziorski, C.; Suen, G. Unique aspects of fiber degradation by the ruminal methanologen Ruminococcus albus 7 revealed by physiological and transcriptomic analysis. BMC Genomics 2014, 15, 1066. [Google Scholar] [CrossRef]
- Myer, P.R.; Seay, T.B.; Rhinehart, J. Cattle Gut Microbe Series: Ruminococcus Species; University of Tennessee Institute of Agriculture: Knoxville, TN, USA, 2020; Available online: https://utbeef.tennessee.edu/wp-content/uploads/sites/127/2020/11/W938.pdf (accessed on 2 January 2025).
- Karri, S.; Vadela, M.B.; Gundi, V.A.K.B. Fiber degradation strategies of bacteria in rumen ecosystem. In Recent Developments in Applied Microbiology and Biochemistry; Academic Press: Cambridge, MA, USA, 2021; Volume 2, pp. 153–159. [Google Scholar] [CrossRef]
- McLoughlin, S.; Spillane, C.; Claffey, N.; Smith, P.E.; O’Rourke, T.; Diskin, M.G.; Waters, S.M. Rumen Microbiome Composition Is Altered in Sheep Divergent in Feed Efficiency. Front. Microbiol. 2020, 11, 1981. [Google Scholar] [CrossRef]
- Monteiro, H.F.; Zhou, Z.; Gomes, M.S.; Peixoto, P.M.G.; Bonsaglia, E.C.R.; Canisso, I.F.; Weimer, B.C.; Lima, F.S. Rumen and lower gut microbiomes relationship with feed efficiency and production traits throughout the lactation of Holstein dairy cows. Sci. Rep. 2022, 12, 4904. [Google Scholar] [CrossRef]
- Kyawt, Y.Y.; Aung, M.; Xu, Y.; Sun, Z.; Zhou, Y.; Zhu, W.; Padmakumar, V.; Tan, Z.; Cheng, Y. Dynamic changes of rumen microbiota and serum metabolome revealed increases in meat quality and growth performances of sheep fed bio-fermented rice straw. J. Anim. Sci. Biotechnol. 2024, 15, 34. [Google Scholar] [CrossRef]
- Konikoff, T.; Gophna, U. Oscillospira: A Central, Enigmatic Component of the Human Gut Microbiota. Trends Microbiol. 2016, 24, 523–524. [Google Scholar] [CrossRef] [PubMed]
- Niwińska, B.; Hanczakowska, E.; Arciszewski, M.B.; Klebaniuk, R. Review: Exogenous butyrate: Implications for the functional development of ruminal epithelium and calf performance. Animal 2017, 11, 1522–1530. [Google Scholar] [CrossRef] [PubMed]
- Gophna, U.; Konikoff, T.; Nielsen, H.B. Oscillospira and related bacteria—From metagenomic species to metabolic features. Environ Microbiol. 2017, 19, 835–841. [Google Scholar] [CrossRef] [PubMed]
- Difford, G.F.; Plichta, D.R.; Løvendahl, P.; Lassen, J.; Noel, S.J.; Højberg, O.; Wright, A.-D.G.; Zhu, Z.; Kristensen, L.; Nielsen, H.B.; et al. Host genetics and the rumen microbiome jointly associate with methane emissions in dairy cows. PLoS Genet. 2018, 14, e1007580. [Google Scholar] [CrossRef]
- Miettinen, H.; Huhtanen, P. Effects of the Ratio of Ruminal Propionate to Butyrate on Milk Yield and Blood Metabolites in Dairy Cows. J. Dairy Sci. 1996, 79, 851–861. [Google Scholar] [CrossRef]
- Zhuang, Y.; Abdelsattar, M.M.; Fu, Y.; Zhang, N.; Chai, J. Butyrate metabolism in rumen epithelium affected by host and diet regime through regulating microbiota in a goat model. Anim. Nutr. 2024, 19, 41–55. [Google Scholar] [CrossRef]
- Fan, P. Microbiome-Guided Strategies to Improve Cattle Production; Mississippi State University. 2023. Available online: https://www.adsa.org/Portals/0/SiteContent/Docs/Meetings/2023ADSA/PowerPoints/2303.pdf (accessed on 13 January 2025).
- Arrazuria, R.; Pérez, V.; Molina, E.; Juste, R.A.; Khafipour, E.; Elguezabal, N. Diet induced changes in the microbiota and cell composition of rabbit gut associated lymphoid tissue (GALT). Sci. Rep. 2018, 8, 14103. [Google Scholar] [CrossRef]

| Ingredients (%DM) | Inclusion (% of Dietary DM) |
|---|---|
| Corn silage | 65.61 |
| Hay a | 14.93 |
| Cracked corn | 14.50 |
| Concentrate/vitamin supplement b | 4.69 |
| Mineral supplement c | 0.27 |
| Nutrient Analysis | Value |
| DM, % | 48.3 |
| Crude Protein, % | 11.6 |
| NDF, % | 38.5 |
| NFC, % | 42.0 |
| Fat, % | 3.59 |
| Calcium, % | 0.57 |
| Phosphorus, % | 0.37 |
| Potassium, % | 1.28 |
| Magnesium, % | 0.15 |
| Item | PosRFI | NegRFI | SEM | p-Value |
|---|---|---|---|---|
| RFI-EPD, kg/d | 0.18 | −0.11 | 0.02 | <0.0001 |
| RFI, kg/d | 0.71 | −0.82 | 0.46 | 0.004 |
| Initial Weight, kg | 427 | 448 | 26.47 | 0.539 |
| Final Weight, kg 1 | 541 | 524 | 8.40 | 0.065 |
| ADG, kg/d | 1.75 | 1.49 | 0.14 | 0.073 |
| DMI, kg/d | 10.55 | 9.39 | 0.37 | 0.006 |
| Genus; OTU | Fold Change 1 | FDR p-Value 2 |
|---|---|---|
| F16; 277519 | −110.58 | 0.007 |
| Oscillospira; 290253 | −25.38 | 0.007 |
| Clostridiales; 207713 | −29.59 | 0.009 |
| Prevotella; 626329 | −20.11 | 0.01 |
| YRC22; 544154 | −18.98 | 0.03 |
| Ruminococcaceae; 196905 | −12.19 | 0.03 |
| Prevotella; 296753 | −11.17 | 0.03 |
| Prevotella; 169950 | −9.38 | 0.03 |
| Prevotella; 593357 | −6.93 | 0.03 |
| Bacteroidales; 152221 | −18.98 | 0.03 |
| Bacteroidales; 346529 | −12.76 | 0.03 |
| Bacteroidales; 325575 | −5.19 | 0.04 |
| BS11; 288543 | −19.66 | 0.04 |
| LD1-PB3; 329608 | −19.30 | 0.04 |
| BS11; 354615 | −9.93 | 0.05 |
| Clostridiales; 133719 | 17.82 | 0.007 |
| Prevotella; 109358 | 15.95 | 0.008 |
| S24-7; 4479793 | 15.48 | 0.009 |
| Ruminococcus; 823658 | 29.63 | 0.009 |
| Prevotella; 263905 | 41.04 | 0.009 |
| Bacteroidales; 254248 | 61.29 | 0.01 |
| Treponema; 699927 | 10.30 | 0.01 |
| Clostridiales; 529192 | 25.64 | 0.01 |
| Clostridiales; 157546 | 48.12 | 0.01 |
| Prevotella; 106839 | 30.20 | 0.01 |
| Clostridiales; 609589 | 5.92 | 0.02 |
| Lachnospiraceae; 289064 | 16.19 | 0.03 |
| Bacteroidales; 313083 | 9.81 | 0.03 |
| Ruminococcus; 590595 | 10.05 | 0.03 |
| Oscillospira; 270866 | 21.27 | 0.03 |
| [Paraprevotellaceae]; 2201 | 6.16 | 0.03 |
| Succinogenes; 302906 | 6.80 | 0.03 |
| Bacteroidales; 556424 | 71.54 | 0.04 |
| BS11; 240168 | 22.44 | 0.05 |
| Prevotella; 323625 | 11.35 | 0.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sidney, T.; Treon, E.; Taiwo, G.; Johnson, S.; Leal, Y.; Fan, P.; Ogunade, I.M. Differential Rumen Microbial Taxa in Charolais Bulls with Divergent Residual Feed Intake. Appl. Microbiol. 2025, 5, 56. https://doi.org/10.3390/applmicrobiol5030056
Sidney T, Treon E, Taiwo G, Johnson S, Leal Y, Fan P, Ogunade IM. Differential Rumen Microbial Taxa in Charolais Bulls with Divergent Residual Feed Intake. Applied Microbiology. 2025; 5(3):56. https://doi.org/10.3390/applmicrobiol5030056
Chicago/Turabian StyleSidney, Taylor, Emily Treon, Godstime Taiwo, Samanthia Johnson, Yarahy Leal, Peixin Fan, and Ibukun M. Ogunade. 2025. "Differential Rumen Microbial Taxa in Charolais Bulls with Divergent Residual Feed Intake" Applied Microbiology 5, no. 3: 56. https://doi.org/10.3390/applmicrobiol5030056
APA StyleSidney, T., Treon, E., Taiwo, G., Johnson, S., Leal, Y., Fan, P., & Ogunade, I. M. (2025). Differential Rumen Microbial Taxa in Charolais Bulls with Divergent Residual Feed Intake. Applied Microbiology, 5(3), 56. https://doi.org/10.3390/applmicrobiol5030056







