Wickerhamomyces pijperi: An Up-And-Coming Yeast with Pectinolytic Activity Suitable for Cocoa Bean Fermentation
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Strains and Maintenance of Strains
2.2. High-Throughput Screening of Yeasts for Pectinolytic Activity
2.3. Identification of Selected Strains
2.4. Verification of the Ability of Pectinolytic Yeasts to Reduce Viscosity in a Pectin-Rich Cocoa Simulation Medium
2.5. Investigation of Different Growth Conditions for W. pijperi Strains
2.6. Determination of Dynamic Viscosity (η)
2.7. Biomass Production in a 15 L Pre-Pilot-Scale Bioreactor
2.7.1. Inoculum Preparation, Reactor Preparation, Inoculation and Biomass Production
2.7.2. Measurements During and After Cultivation
2.7.3. Freeze-Drying of Produced Biomass
2.8. Growth of Pectinolytic Yeast Strains on Cocoa Beans in a Micro-Scale Fermentation (20 g)
2.9. Cocoa Bean Fermentation at the Lab-Scale (1 kg) with Pectinolytic Yeast Strains
2.10. Cocoa Bean Fermentation at a Small-Scale (20 kg) in Costa Rica with One Selected Pectinolytic Yeast
2.11. Analyses Carried out During Cocoa Bean Fermentations
2.11.1. Determination of Pulp Drainage (Runoff), Bean Weight, and Pulp Content During Fermentation
2.11.2. Determination of the pH of the Cocoa Pulp and Cotyledon
2.11.3. Microbiological Analysis
2.11.4. Cut Test
2.11.5. Quantification of Yeasts by qPCR
2.12. Statistical Analyses
3. Results
3.1. Selection of Yeasts with Pectinolytic Activity
3.1.1. In Vitro Determination of the Pectinolytic Activity of Yeasts on Nutrient Media Containing Pectic Substances
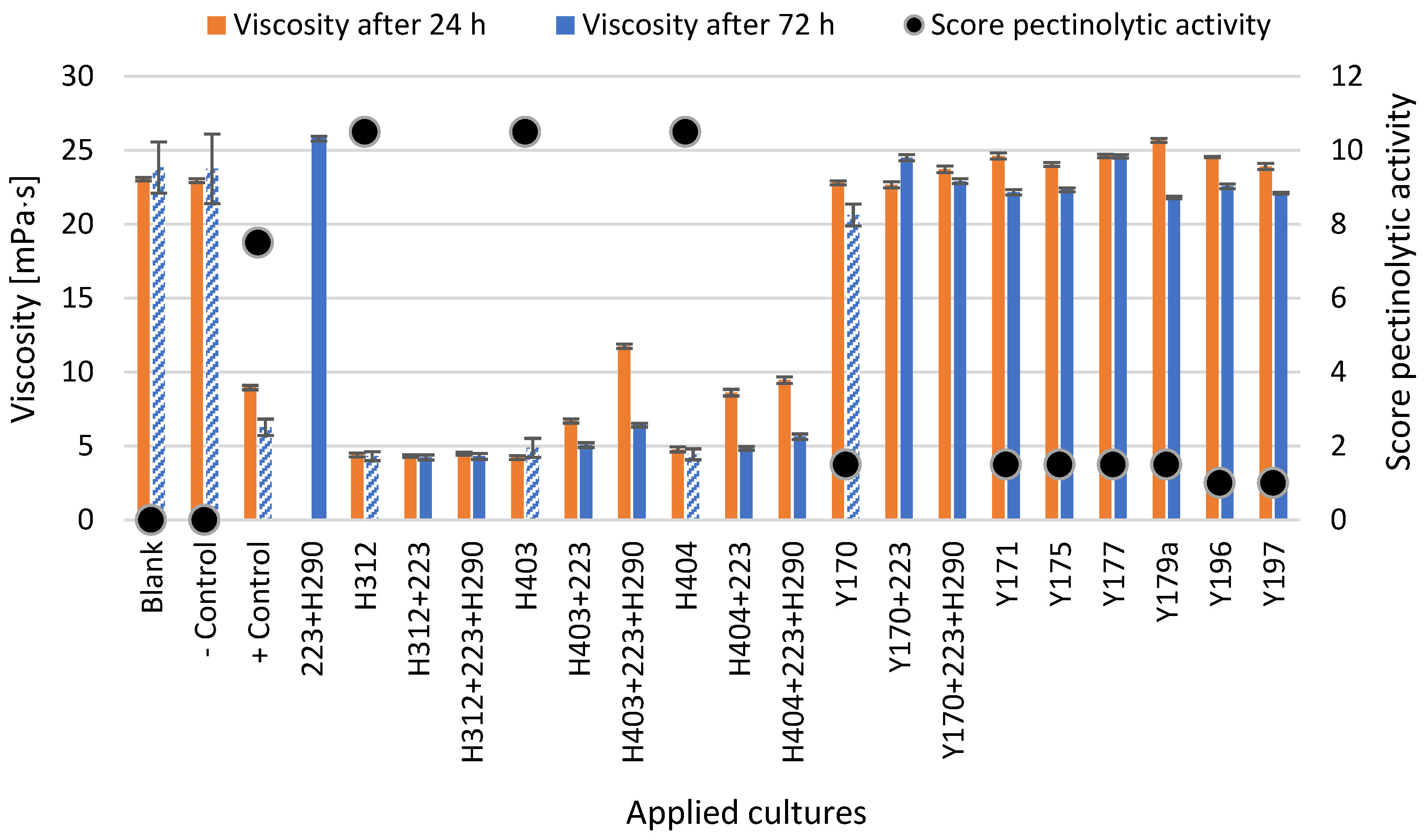
3.1.2. In Vitro Determination of the Pectinolytic Activity of Yeasts Based on Their Ability to Reduce the Viscosity of a Pectin-Rich Medium
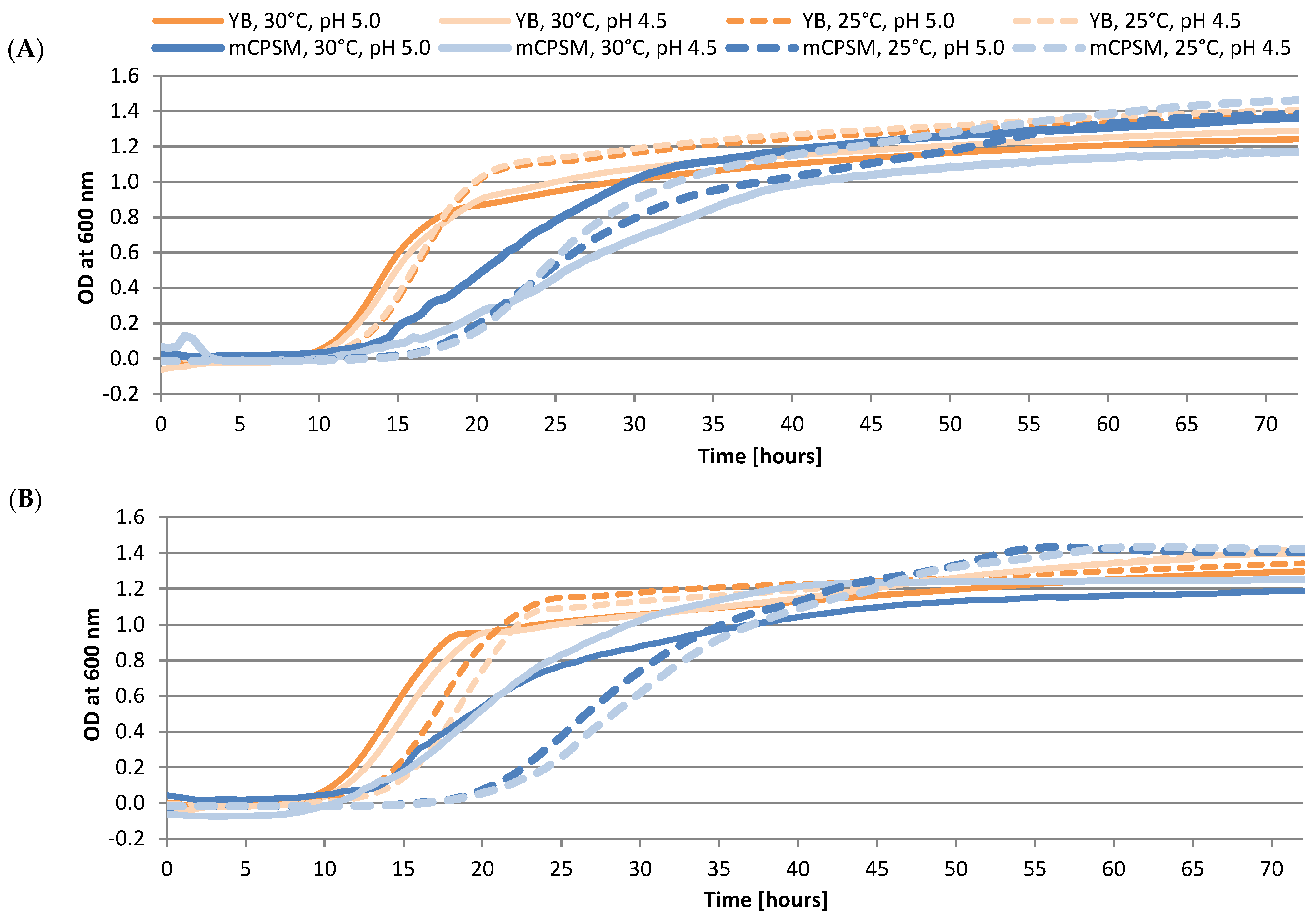
3.2. Growth of W. pijperi Under Different Conditions
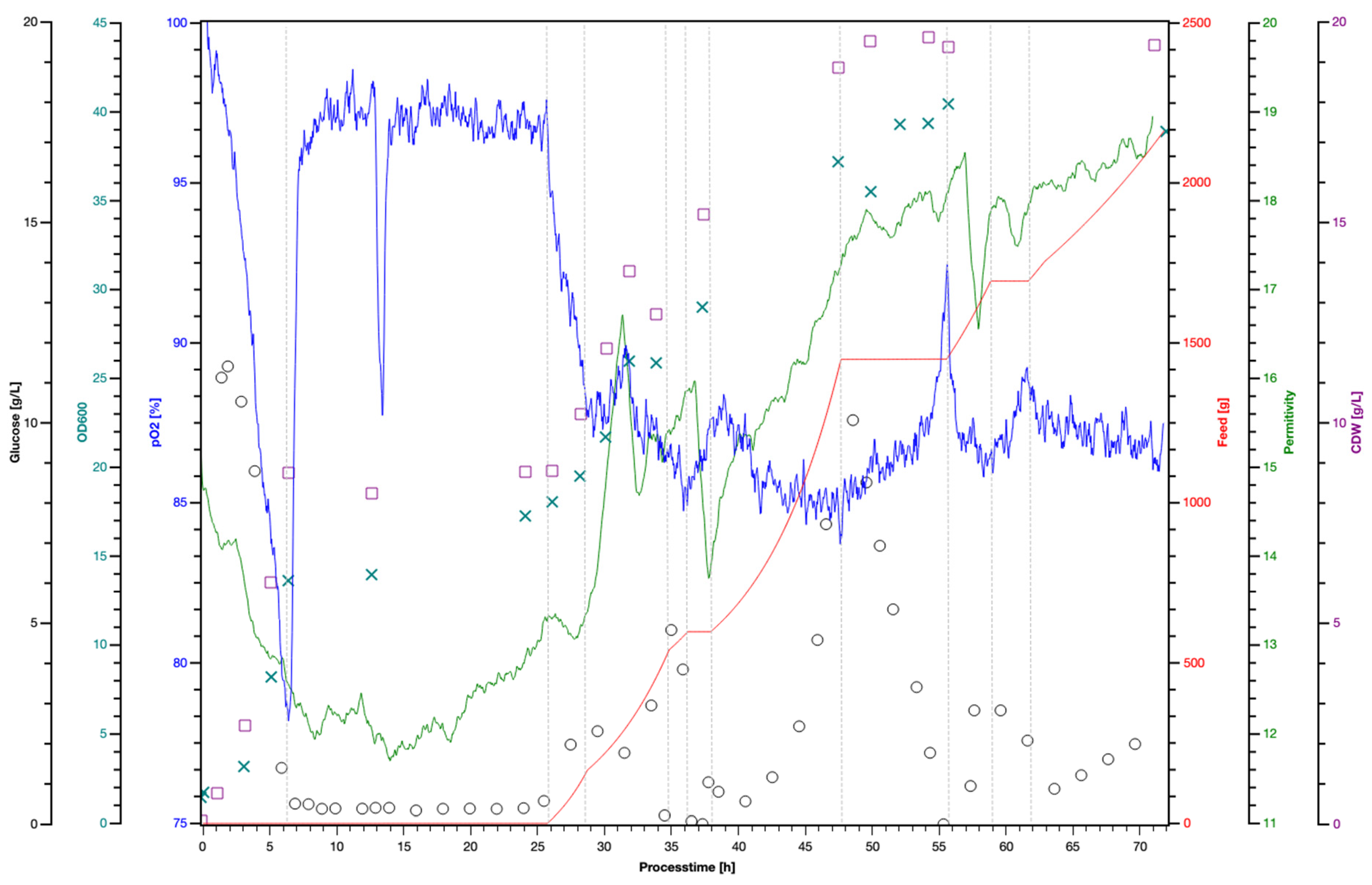
3.3. Production of a Freeze-Dried Ready-to-Use Culture of W. pijperi H403
3.4. In Vitro Growth of Pectinolytic Yeast on Cocoa Beans
3.5. Cocoa Bean Fermentation with Pectinolytic Yeasts in Lab-Scale (1 kg) and Small-Scale (20 kg) Fermentation
3.5.1. Impact of Pectinolytic Yeasts on Pulp Degradation During Cocoa Bean Fermentation
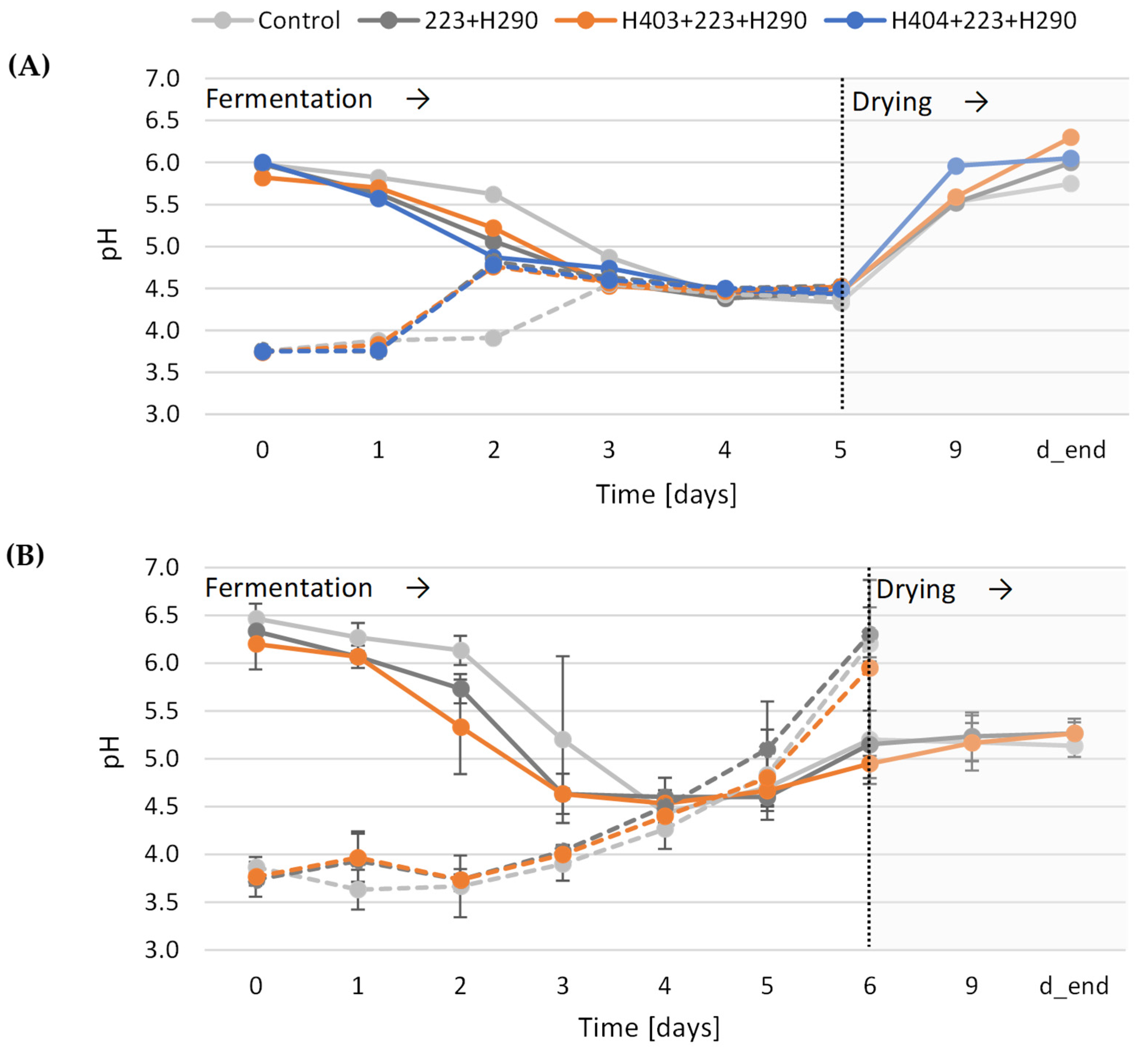
3.5.2. pH Development During the Fermentation of Cocoa Beans with Pectinolytic Yeast
3.5.3. Temperature Profile During Small-Scale Fermentation (20 kg) Inoculated with Pectinolytic Yeasts
3.5.4. Microbial Development of Inoculated Microorganisms During Cocoa Bean Fermentation
3.5.5. Culture-Independent Monitoring of Yeasts During Fermentation in Small-Scale Systems (20 kg)
3.5.6. Cut Test of Beans After Fermentation and Drying of Small-Scale Fermentations (20 kg)
4. Discussion
4.1. Relevance of Pectinolytic Yeasts in Pulp Degradation During Cocoa Bean Fermentation
4.2. Growth of W. pijperi Strains on Cocoa Beans and Overall Influence on Fermentation and Quality of Fermented and Dried Cocoa Beans
4.3. First Insights and Challenges in the Cultivation of W. pijperi
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Akinfala, T.O.; Houbraken, J.; Sulyok, M.; Adedeji, A.R.; Odebode, A.C.; Krska, R.; Ezekiel, C.N. Moulds and Their Secondary Metabolites Associated with the Fermentation and Storage of Two Cocoa Bean Hybrids in Nigeria. Int. J. Food Microbiol. 2020, 316, 108490. [Google Scholar] [CrossRef] [PubMed]
- Sarbu, I.; Csutak, O. The Microbiology of Cocoa Fermentation. In Caffeinated and Cocoa Based Beverages; Grumezescu, A.M., Holban, A.M., Eds.; Woodhead Publishing: Cambridge, UK, 2019; pp. 423–446. ISBN 978-0-12-815864-7. [Google Scholar]
- Schwan, R.F.; Wheals, A.E. The Microbiology of Cocoa Fermentation and Its Role in Chocolate Quality. Crit. Rev. Food Sci. Nutr. 2004, 44, 205–221. [Google Scholar] [CrossRef]
- Leal, G.A.; Gomes, L.H.; Efraim, P.; De Almeida Tavares, F.C.; Figueira, A. Fermentation of Cacao (Theobroma cacao L.) seeds with a Hybrid Kluyveromyces Marxianus Strain Improved Product Quality Attributes. FEMS Yeast Res. 2008, 8, 788–798. [Google Scholar] [CrossRef] [PubMed]
- Lopez, A.S.; Dimick, P.S. Cocoa Fermentation. In Biotechnology Set; Rehm, H.-J., Reed, G., Eds.; Wiley: Hoboken, NJ, USA, 2001; pp. 561–577. ISBN 978-3-527-25762-1. [Google Scholar]
- Meersman, E.; Struyf, N.; Kyomugasho, C.; Jamsazzadeh Kermani, Z.; Santiago, J.S.; Baert, E.; Hemdane, S.; Vrancken, G.; Verstrepen, K.J.; Courtin, C.M.; et al. Characterization and Degradation of Pectic Polysaccharides in Cocoa Pulp. J. Agric. Food Chem. 2017, 65, 9726–9734. [Google Scholar] [CrossRef] [PubMed]
- Romanens, E.; Näf, R.; Lobmaier, T.; Pedan, V.; Leischtfeld, S.F.; Meile, L.; Schwenninger, S.M. A Lab-Scale Model System for Cocoa Bean Fermentation. Appl. Microbiol. Biotechnol. 2018, 102, 3349–3362. [Google Scholar] [CrossRef]
- Pettipher, G.L. Analysis of Cocoa Pulp and the Formulation of a Standardised Artificial Cocoa Pulp Medium. J. Sci. Food Agric. 1986, 37, 297–309. [Google Scholar] [CrossRef]
- Schols, H.A.; Voragen, A.G.J. Pectic Polysaccharides. In Handbook of Food Enzymology; Whitaker, J.R., Voragen, A.G.J., Wong, D.W.S., Eds.; Marcel Dekker: New York, NY, USA; Basel, Switzerland, 2003; pp. 811–825. ISBN 978-0-8247-0686-9. [Google Scholar]
- Chaudhri, A. Microbially Derived Pectinases: A Review. IOSR J. Pharm Biol. Sci. 2012, 2, 1–5. [Google Scholar] [CrossRef]
- Mohnen, D. Pectin Structure and Biosynthesis. Curr. Opin. Plant Biol. 2008, 11, 266–277. [Google Scholar] [CrossRef]
- Chakraborty, S.; Fernandes, V.O.; Dias, F.M.V.; Prates, J.A.M.; Ferreira, L.M.A.; Fontes, C.M.G.A.; Goyal, A.; Centeno, M.S.J. Role of Pectinolytic Enzymes Identified in Clostridium Thermocellum Cellulosome. PLoS ONE 2015, 10, e0116787. [Google Scholar] [CrossRef]
- Pedrolli, D.B.; Carmona, E.C. Purification and Characterization of the Exopolygalacturonase Produced by Aspergillus Giganteus in Submerged Cultures. J. Ind. Microbiol. Biotechnol. 2010, 37, 567–573. [Google Scholar] [CrossRef]
- Swain, M.R.; Ray, R.C. Production, Characterization and Application of a Thermostable Exo-Polygalacturonase by Bacillus Subtilis CM5. Food Biotechnol. 2010, 24, 37–50. [Google Scholar] [CrossRef]
- Blanco, P.; Sieiro, C.; Villa, T.G. Production of Pectic Enzymes in Yeasts. FEMS Microbiol. Lett. 1999, 175, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Benen, J.A.E.; Visser, J. Polygalacturonases. In Handbook of Food Enzymology; Whitaker, J.R., Voragen, A.G.J., Wong, D.W.S., Eds.; Marcel Dekker: New York, NY, USA; Basel, Switzerland, 2003; pp. 838–847. ISBN 978-0-8247-0686-9. [Google Scholar]
- Dasilva, E.; Borges, M.; Medina, C.; Piccoli, R.; Schwan, R. Pectinolytic Enzymes Secreted by Yeasts from Tropical Fruits. FEMS Yeast Res. 2005, 5, 859–865. [Google Scholar] [CrossRef] [PubMed]
- Samagaci, L.; Ouattara, H.; Niamké, S.; Lemaire, M. Pichia Kudrazevii and Candida Nitrativorans Are the Most Well-Adapted and Relevant Yeast Species Fermenting Cocoa in Agneby-Tiassa, a Local Ivorian Cocoa Producing Region. Food Res. Int. 2016, 89, 773–780. [Google Scholar] [CrossRef]
- Schwan, R.F.; Cooper, R.M.; Wheals, A.E. Endopolygalacturonase Secretion by Kluyveromyces Marxianus and Other Cocoa Pulp-Degrading Yeasts. Enzym. Microb. Technol. 1997, 21, 234–244. [Google Scholar] [CrossRef]
- Lima, C.-C.; Vaz, A.-M.; De Castro, G.M.; Lobo, F.; Solar, R.; Rodrigues, C.; Martins Pinto, L.R.; Vandenberghe, L.; Pereira, G.; Miúra Da Costa, A.; et al. Integrating Microbial Metagenomics and Physicochemical Parameters and a New Perspective on Starter Culture for Fine Cocoa Fermentation. Food Microbiol. 2021, 93, 103608. [Google Scholar] [CrossRef]
- Romanens, E.; Pedan, V.; Meile, L.; Miescher Schwenninger, S. Influence of Two Anti-Fungal Lactobacillus Fermentum-Saccharomyces Cerevisiae Co-Cultures on Cocoa Bean Fermentation and Final Bean Quality. PLoS ONE 2020, 15, e0239365. [Google Scholar] [CrossRef]
- Figueroa-Hernández, C.; Mota-Gutierrez, J.; Ferrocino, I.; Hernández-Estrada, Z.J.; González-Ríos, O.; Cocolin, L.; Suárez-Quiroz, M.L. The Challenges and Perspectives of the Selection of Starter Cultures for Fermented Cocoa Beans. Int. J. Food Microbiol. 2019, 301, 41–50. [Google Scholar] [CrossRef]
- Díaz-Muñoz, C.; De Vuyst, L. Functional Yeast Starter Cultures for Cocoa Fermentation. J. Appl. Microbiol. 2022, 133, 39–66. [Google Scholar] [CrossRef]
- Taskila, S. Industrial Production of Starter Cultures. In Starter Cultures in Food Production; John Wiley & Sons: Hoboken, NJ, USA, 2017; pp. 79–100. ISBN 978-1-118-93379-4. [Google Scholar]
- Liu, C.-F.; Zhang, X.-F.; Yu, T.-L.; Lee, C.-L. Utilizing Deep Ocean Water in Yeast Fermentation for Enhanced Mineral-Rich Biomass Production and Fermentative Regulation by Proteomics Modulation. Heliyon 2024, 10, e31031. [Google Scholar] [CrossRef]
- Yüzgeç, U.; Türker, M.; Hocalar, A. On-Line Evolutionary Optimization of an Industrial Fed-Batch Yeast Fermentation Process. ISA Trans. 2009, 48, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Kasperski, A.; Miśkiewicz, T. Optimization of Pulsed Feeding in a Baker’s Yeast Process with Dissolved Oxygen Concentration as a Control Parameter. Biochem. Eng. J. 2008, 40, 321–327. [Google Scholar] [CrossRef]
- Guo, Y.-F.; Wang, M.-Q.; Wang, Y.-L.; Wang, H.-T.; Xu, J.-Z. Controlling the Formation of Foams in Broth to Promote the Co-Production of Microbial Oil and Exopolysaccharide in Fed-Batch Fermentation. Fermentation 2022, 8, 68. [Google Scholar] [CrossRef]
- Polo, L.; Mañes-Lázaro, R.; Olmeda, I.; Cruz-Pio, L.E.; Medina, Á.; Ferrer, S.; Pardo, I. Influence of Freezing Temperatures Prior to Freeze-Drying on Viability of Yeasts and Lactic Acid Bacteria Isolated from Wine. J. Appl. Microbiol. 2017, 122, 1603–1614. [Google Scholar] [CrossRef] [PubMed]
- Miescher Schwenninger, S.; Freimüller Leischtfeld, S.; Gantenbein-Demarchi, C. High-throughput Identification of the Microbial Biodiversity of Cocoa Bean Fermentation by MALDI-TOF MS. Lett. Appl. Microbiol. 2016, 63, 347–355. [Google Scholar] [CrossRef]
- Streule, S.; Freimüller Leischtfeld, S.; Galler, M.; Miescher Schwenninger, S. Monitoring of Cocoa Post-Harvest Process Practices on a Small-Farm Level at Five Locations in Ecuador. Heliyon 2022, 8, e09628. [Google Scholar] [CrossRef]
- Romanens, E.; Freimüller Leischtfeld, S.; Volland, A.; Stevens, M.J.A.; Krähenmann, U.; Isele, D.; Fischer, B.; Meile, L.; Miescher Schwenninger, S. Screening of Lactic Acid Bacteria and Yeast Strains to Select Adapted Anti-Fungal Co-Cultures for Cocoa Bean Fermentation. Int. J. Food Microbiol. 2019, 290, 262–272. [Google Scholar] [CrossRef]
- Silva, C.F.; Vilela, D.M.; De Souza Cordeiro, C.; Duarte, W.F.; Dias, D.R.; Schwan, R.F. Evaluation of a Potential Starter Culture for Enhance Quality of Coffee Fermentation. World J. Microbiol. Biotechnol. 2013, 29, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Martos, M.A.; Zubreski, E.R.; Combina, M.; Garro, O.A.; Hours, R.A. Isolation of a Yeast Strain Able to Produce a Polygalacturonase with Maceration Activity of Cassava Roots. Food Sci. Technol. 2013, 33, 332–338. [Google Scholar] [CrossRef]
- Glass, N.L.; Donaldson, G.C. Development of Primer Sets Designed for Use with the PCR to Amplify Conserved Genes from Filamentous Ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef]
- Schwendimann, L.; Kauf, P.; Fieseler, L.; Gantenbein-Demarchi, C.; Miescher Schwenninger, S. Development of a Quantitative PCR Assay for Rapid Detection of Lactobacillus Plantarum and Lactobacillus Fermentum in Cocoa Bean Fermentation. J. Microbiol. Methods 2015, 115, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Hierro, N.; Esteve-Zarzoso, B.; González, Á.; Mas, A.; Guillamón, J.M. Real-Time Quantitative PCR (QPCR) and Reverse Transcription-QPCR for Detection and Enumeration of Total Yeasts in Wine. Appl. Environ. Microbiol. 2006, 72, 7148–7155. [Google Scholar] [CrossRef] [PubMed]
- Díaz, C.; Molina, A.M.; Nähring, J.; Fischer, R. Characterization and Dynamic Behavior of Wild Yeast during Spontaneous Wine Fermentation in Steel Tanks and Amphorae. BioMed Res. Int. 2013, 2013, 540465. [Google Scholar] [CrossRef]
- Serra, J.L.; Moura, F.G.; Pereira, G.-M.; Soccol, C.R.; Rogez, H.; Darnet, S. Determination of the Microbial Community in Amazonian Cocoa Bean Fermentation by Illumina-Based Metagenomic Sequencing. LWT 2019, 106, 229–239. [Google Scholar] [CrossRef]
- Visintin, S.; Alessandria, V.; Valente, A.; Dolci, P.; Cocolin, L. Molecular Identification and Physiological Characterization of Yeasts, Lactic Acid Bacteria and Acetic Acid Bacteria Isolated from Heap and Box Cocoa Bean Fermentations in West Africa. Int. J. Food Microbiol. 2016, 216, 69–78. [Google Scholar] [CrossRef]
- Sanchez, J.; Guiraud, J.P.; Galzy, P. A Study of the Polygalacturonase Activity of Several Yeast Strains Isolated from Cocoa. Appl. Microbiol. Biotechnol. 1984, 20, 262–267. [Google Scholar] [CrossRef]
- Naumova, E.S.; Borovkova, A.N.; Shalamitskiy, M.Y.; Naumov, G.I. Natural Polymorphism of Pectinase PGU Genes in the Saccharomyces Yeasts. Microbiology 2021, 90, 349–360. [Google Scholar] [CrossRef]
- van Wyk, H.; Divol, B. Recovery of Endo-Polygalacturonase Activity in Wine Yeast and Its Effect on Wine Aroma. FEMS Yeast Res. 2009, 10, 58–71. [Google Scholar] [CrossRef][Green Version]
- Li, J.; Peng, C.; Mao, A.; Zhong, M.; Hu, Z. An Overview of Microbial Enzymatic Approaches for Pectin Degradation. Int. J. Biol. Macromol. 2024, 254, 127804. [Google Scholar] [CrossRef]
- Ouattara, H.G.; Elias, R.J.; Dudley, E.G. Microbial Synergy between Pichia Kudriazevii YS201 and Bacillus Subtilis BS38 Improves Pulp Degradation and Aroma Production in Cocoa Pulp Simulation Medium. Heliyon 2020, 6, e03269. [Google Scholar] [CrossRef]
- Haile, M.; Kang, W.H. Isolation, Identification, and Characterization of Pectinolytic Yeasts for Starter Culture in Coffee Fermentation. Microorganisms 2019, 7, 401. [Google Scholar] [CrossRef]
- De Vuyst, L.; Weckx, S. The Cocoa Bean Fermentation Process: From Ecosystem Analysis to Starter Culture Development. J. Appl. Microbiol. 2016, 121, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Streule, S.; Freimüller Leischtfeld, S.; Chatelain, K.; Miescher Schwenninger, S. Effect of Pod Storage and Drying Temperature on Fermentation Dynamics and Final Bean Quality of Cacao Nacional in Ecuador. Foods 2024, 13, 1536. [Google Scholar] [CrossRef] [PubMed]
- Streule, S.; André, A.; Freimüller Leischtfeld, S.; Chatelain, K.; Gillich, E.; Chetschik, I.; Miescher Schwenninger, S. Influences of Depulping, Pod Storage and Fermentation Time on Fermentation Dynamics and Quality of Ghanaian Cocoa. Foods 2024, 13, 2590. [Google Scholar] [CrossRef]
- Samagaci, L.; Ouattara, H.G.; Goualie, B.G.; Niamke, S.L. Polyphasic Analysis of Pectinolytic and Stress-Resistant Yeast Strains Isolated From Ivorian Cocoa Fermentation. J. Food Res. 2014, 4, 124. [Google Scholar] [CrossRef]
- Lefeber, T.; Papalexandratou, Z.; Gobert, W.; Camu, N.; De Vuyst, L. On-Farm Implementation of a Starter Culture for Improved Cocoa Bean Fermentation and Its Influence on the Flavour of Chocolates Produced Thereof. Food Microbiol. 2012, 30, 379–392. [Google Scholar] [CrossRef]
- Díaz-Muñoz, C.; Van de Voorde, D.; Comasio, A.; Verce, M.; Hernandez, C.E.; Weckx, S.; De Vuyst, L. Curing of Cocoa Beans: Fine-Scale Monitoring of the Starter Cultures Applied and Metabolomics of the Fermentation and Drying Steps. Front. Microbiol. 2020, 11, 616875. [Google Scholar] [CrossRef]
- Ouattara, H.G.; Koffi, B.L.; Karou, G.T.; Sangaré, A.; Niamke, S.L.; Diopoh, J.K. Implication of Bacillus Sp. in the Production of Pectinolytic Enzymes during Cocoa Fermentation. World J. Microbiol. Biotechnol. 2008, 24, 1753–1760. [Google Scholar] [CrossRef]
- Ardhana, M.M.; Fleet, G.H. The Microbial Ecology of Cocoa Bean Fermentations in Indonesia. Int. J. Food Microbiol. 2003, 86, 87–99. [Google Scholar] [CrossRef]
- Izawa, N.; Kudo, M.; Nakamura, Y.; Mizukoshi, H.; Kitada, T.; Sone, T. Production of Aroma Compounds from Whey Using Wickerhamomyces Pijperi. AMB Express 2015, 5, 23. [Google Scholar] [CrossRef]
- Falconí, C.E.; Yánez-Mendizábal, V.; Haro, R.J.; Claudio, D.R. Inoculum of a Native Microbial Starter Cocktail to Optimize Fine-Aroma Cocoa (Theobroma cacao) Bean Fermentation. Agronomy 2023, 13, 2572. [Google Scholar] [CrossRef]
- Sandhya, M.V.S.; Yallappa, B.S.; Varadaraj, M.C.; Puranaik, J.; Rao, L.J.; Janardhan, P.; Murthy, P.S. Inoculum of the Starter Consortia and Interactive Metabolic Process in Enhancing Quality of Cocoa Bean (Theobroma cacao) Fermentation. LWT 2016, 65, 731–738. [Google Scholar] [CrossRef]
- Chadha, Y.; Kukhtevich, I.; Padovani, F.; Schneider, R.; Schmoller, K. Single-Cell Imaging Reveals a Key Role of Bck2 in Budding Yeast Cell Size Adaptation to Nutrient Challenges. bioRxiv 2024. [Google Scholar] [CrossRef]
- Ramos, C.L.; Dias, D.R.; Miguel, M.G.D.C.P.; Schwan, R.F. Impact of Different Cocoa Hybrids (Theobroma cacao L.) and S. Cerevisiae UFLA CA11 Inoculation on Microbial Communities and Volatile Compounds of Cocoa Fermentation. Food Res. Int. 2014, 64, 908–918. [Google Scholar] [CrossRef] [PubMed]
- Zerva, I.; Remmas, N.; Melidis, P.; Tsiamis, G.; Ntougias, S. Microbial Succession and Identification of Effective Indigenous Pectinolytic Yeasts From Orange Juice Processing Wastewater. Waste Biomass Valorization 2021, 12, 4885–4899. [Google Scholar] [CrossRef]
- Gutierrez-Cano, A.R.; Jones, B.; Macario, J.; Martin, S.; Cardenas, D.; Simpson, H.; Boundy-Mills, K.; Edwards, M.C. Characterization of Pectinase-Producing Saccharomyces Cerevisiae UCDFST 09-448 and Its Effects on Cull Peach Fermentations. J. Ind. Microbiol. Biotechnol. 2024, 51, kuae037. [Google Scholar] [CrossRef]
- Vanags, J.; Hrynko, V.; Viesturs, U. Development and Application of a Flexible Controller in Yeast Fermentations Using pO2 Cascade Control. Eng. Life Sci. 2010, 10, 321–332. [Google Scholar] [CrossRef]
- Rosenfeld, E.; Beauvoit, B.; Blondin, B.; Salmon, J.-M. Oxygen Consumption by Anaerobic Saccharomyces Cerevisiae under Enological Conditions: Effect on Fermentation Kinetics. Appl. Environ. Microbiol. 2003, 69, 113–121. [Google Scholar] [CrossRef]
- Tiso, T.; Demling, P.; Karmainski, T.; Oraby, A.; Eiken, J.; Liu, L.; Bongartz, P.; Wessling, M.; Desmond, P.; Schmitz, S.; et al. Foam Control in Biotechnological Processes—Challenges and Opportunities. Discov. Chem. Eng. 2024, 4, 2. [Google Scholar] [CrossRef]
- De Deken, R.H. The Crabtree Effect: A Regulatory System in Yeast. J. Gen. Microbiol. 1966, 44, 149–156. [Google Scholar] [CrossRef]
- Elsemman, I.E.; Rodriguez Prado, A.; Grigaitis, P.; Garcia Albornoz, M.; Harman, V.; Holman, S.W.; Van Heerden, J.; Bruggeman, F.J.; Bisschops, M.M.M.; Sonnenschein, N.; et al. Whole-Cell Modeling in Yeast Predicts Compartment-Specific Proteome Constraints That Drive Metabolic Strategies. Nat. Commun. 2022, 13, 801. [Google Scholar] [CrossRef] [PubMed]
- Vahalová, P.; Červinková, K.; Cifra, M. Biological Autoluminescence for Assessing Oxidative Processes in Yeast Cell Cultures. Sci. Rep. 2021, 11, 10852. [Google Scholar] [CrossRef] [PubMed]
- Crabtree, H.G. The Carbohydrate Metabolism of Certain Pathological Overgrowths. Biochem. J. 1928, 22, 1289–1298. [Google Scholar] [CrossRef] [PubMed]
- Broach, J.R. Nutritional Control of Growth and Development in Yeast. Genetics 2012, 192, 73–105. [Google Scholar] [CrossRef]
- Duc, C.; Pradal, M.; Sanchez, I.; Noble, J.; Tesnière, C.; Blondin, B. A Set of Nutrient Limitations Trigger Yeast Cell Death in a Nitrogen-Dependent Manner during Wine Alcoholic Fermentation. PLoS ONE 2017, 12, e0184838. [Google Scholar] [CrossRef]



| Pectinolytic Activity | Description of the Extent of the Pectinolytic Activity | Radius of Clear Zone (mm) | Score Based on Extent of Pectinolytic Activity | |
|---|---|---|---|---|
| − | Absent | No visible degradation zone | 0 | 0.0 |
| +/− | Minimal | No clear degradation zone around the colony, but a clearing of medium | 0 | 0.5 |
| + | Weak | Small, but clear degradation zone around the colony | 1.0–4.9 | 1.5 |
| ++ | Middle | Medium-sized degradation zone around the colony | 5.0–9.9 | 2.5 |
| +++ | Strong | Large degradation zone around the colony | ≥10.0 | 3.5 |
| Target Organism/s | Target Gene | Name of Primers | Sequence 5′–3′ | Melting Temperature [°C] | Amplicon Length [bp] | Reference |
|---|---|---|---|---|---|---|
| Total yeast count | 26S rRNA | YeastF | GAGTCGAGTTGTTTGGGAATGC | 55 | 124 | Hierro et al. [37] |
| YeastR | TCTCTTTCCAAAGTTCTTTTCATCTTT | |||||
| S. cerevisiae | 26S rRNA | SC-5fw | AGGAGTGCGGTTCTTTCTAAAG | 55 * | 215 | Díaz et al. [38] |
| SC-3bw | TGAAATGCGAGATTCCCCCA | |||||
| W. pijperi | LSU rDNA | Wp2F | GGCGATATTCAGTCTCTCGTAGACTG | 60 | 154 | This study |
| Wp2R | GCAGAAGCCGCAGTCCTCGGTC |
| Parameter | Assays for the Quantification of | ||
|---|---|---|---|
| Total Yeast Count | S. cerevisiae | W. pijperi | |
| Slope | −3.51 [−3.61, −3.41] | −3.50 [−3.66, −3.34] | −3.56 [−3.62, −3.50] |
| Intercept * | 35.69 [35.24, 36.13] | 36.95 [36.23, 37.67] | 36.92 [36.67, 37.17] |
| R2 | 0.9892 | 0.9724 | 0.9976 |
| Efficiency ** | 92.71% | 93.07% | 90.94% |
| Species | Strain | Identification of Selected Yeast Species | Pectinolytic Activity | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MALDI-TOF MSScore | ITS Gene Sequence % Similarity (Accession Number) | MP5 | YNBpc | YNBpa | Total Score for Pectinolytic Activity | |||||
| Activity | Score | Activity | Score | Activity | Score | |||||
| fH. opuntiae | Y168a | na | na | − | 0.0 | − | 0.0 | − | 0.0 | 0.0 |
| K. marxianus | DSM 70292 | na | na | ++ | 2.5 | ++ | 2.5 | ++ | 2.5 | 7.5 |
| W. pijperi * | H312 | 2.24 | na | +++ | 3.5 | +++ | 3.5 | +++ | 3.5 | 10.5 |
| W. pijperi * | H403 | 2.15 | na | +++ | 3.5 | +++ | 3.5 | +++ | 3.5 | 10.5 |
| W. pijperi * | H404 | 2.17 | na | +++ | 3.5 | +++ | 3.5 | +++ | 3.5 | 10.5 |
| Not identified ** | Y166a | <1.7 | No reliable identification | ++ | 2.5 | − | − | + | 1.5 | 4.0 |
| Ustilago sp. *** | H405 | <1.7 | 99 (KY284846.1) | + | 1.5 | + | 1.5 | − | 0.0 | 3.0 |
| Ustilago sp. *** | H406 | <1.7 | 99 (KY284846.1) | + | 1.5 | + | 1.5 | − | 0.0 | 3.0 |
| S. cerevisiae | Y091 | <1.7 | 99 (KT64941.1) | +/− | 0.5 | +/− | 0.5 | +/− | 0.5 | 1.5 |
| S. cerevisiae | Y081 | <1.7 | 100 (KX434760.1) | +/− | 0.5 | +/− | 0.5 | +/− | 0.5 | 1.5 |
| S. cerevisiae * | Y175 | 2.15 | na | +/− | 0.5 | +/− | 0.5 | +/− | 0.5 | 1.5 |
| S. cerevisiae * | Y179a | 2.15 | na | +/− | 0.5 | +/− | 0.5 | +/− | 0.5 | 1.5 |
| S. cerevisiae | Y163 | 2.14 | na | +/− | 0.5 | +/− | 0.5 | +/− | 0.5 | 1.5 |
| S. cerevisiae | Y167a | 2.11 | na | +/− | 0.5 | +/− | 0.5 | +/− | 0.5 | 1.5 |
| S. cerevisiae * | Y171 | 2.10 | na | +/− | 0.5 | +/− | 0.5 | +/− | 0.5 | 1.5 |
| S. cerevisiae | Y076 | 2.09 | na | +/− | 0.5 | +/− | 0.5 | +/− | 0.5 | 1.5 |
| S. cerevisiae * | Y170 | 2.08 | na | +/− | 0.5 | +/− | 0.5 | +/− | 0.5 | 1.5 |
| S. cerevisiae | Y168b | 2.08 | na | +/− | 0.5 | +/− | 0.5 | +/− | 0.5 | 1.5 |
| S. cerevisiae * | Y177 | 2.05 | na | +/− | 0.5 | +/− | 0.5 | +/− | 0.5 | 1.5 |
| S. cerevisiae | Y135 | 2.02 | na | +/− | 0.5 | +/− | 0.5 | +/− | 0.5 | 1.5 |
| S. cerevisiae | Y082 | 2.01 | na | +/− | 0.5 | +/− | 0.5 | +/− | 0.5 | 1.5 |
| Saccharomyces sp. | Y180a | 1.95 | na | +/− | 0.5 | +/− | 0.5 | +/− | 0.5 | 1.5 |
| Saccharomyces sp. | Y093 | 1.94 | na | +/− | 0.5 | +/− | 0.5 | +/− | 0.5 | 1.5 |
| Saccharomyces sp. | Y089 | 1.93 | na | +/− | 0.5 | +/− | 0.5 | +/− | 0.5 | 1.5 |
| Saccharomyces sp. | Y169b | 1.91 | na | +/− | 0.5 | +/− | 0.5 | +/− | 0.5 | 1.5 |
| Saccharomyces sp. | Y174 | 1.9 | na | +/− | 0.5 | +/− | 0.5 | +/− | 0.5 | 1.5 |
| Saccharomyces sp. | Y239 | 1.88 | na | +/− | 0.5 | +/− | 0.5 | +/− | 0.5 | 1.5 |
| Saccharomyces sp. | Y158 | 1.87 | na | +/− | 0.5 | +/− | 0.5 | +/− | 0.5 | 1.5 |
| Saccharomyces sp. | Y172 | 1.85 | na | +/− | 0.5 | +/− | 0.5 | +/− | 0.5 | 1.5 |
| Saccharomyces sp. | Y095 | 1.73 | na | +/− | 0.5 | +/− | 0.5 | +/− | 0.5 | 1.5 |
| Saccharomyces sp. | Y094 | 1.72 | na | +/− | 0.5 | +/− | 0.5 | +/− | 0.5 | 1.5 |
| S. cerevisiae | Y212b | 2.00 | na | +/− | 0.5 | +/− | 0.5 | +/− | 0.5 | 1.5 |
| S. cerevisiae | Y156 | <1.7 | 99 (JX497730.1) | +/− | 0.5 | +/− | 0.5 | − | 0.0 | 1.0 |
| H. opuntiae | Y114a | 2.38 | na | +/− | 0.5 | − | 0.0 | +/− | 0.5 | 1.0 |
| S. cerevisiae | Y152 | 2.20 | na | +/− | 0.5 | +/− | 0.5 | − | 0.0 | 1.0 |
| S. cerevisiae * | Y197 | 2.12 | na | +/− | 0.5 | − | 0.0 | +/− | 0.5 | 1.0 |
| S. cerevisiae | Y234 | 2.07 | na | +/− | 0.5 | +/− | 0.5 | − | 0.0 | 1.0 |
| S. cerevisiae * | Y196 | 2.03 | na | +/− | 0.5 | − | 0.0 | +/− | 0.5 | 1.0 |
| Saccharomyces sp. | Y240 | 1.96 | na | +/− | 0.5 | +/− | 0.5 | − | 0.0 | 1.0 |
| Saccharomyces sp. | Y092 | 1.95 | na | +/− | 0.5 | − | 0.0 | +/− | 0.5 | 1.0 |
| Saccharomyces sp. | Y232 | 1.95 | na | +/− | 0.5 | +/− | 0.5 | − | 0.0 | 1.0 |
| Saccharomyces sp. | Y275b | 1.93 | na | +/− | 0.5 | − | 0.0 | +/− | 0.5 | 1.0 |
| Saccharomyces sp. | Y130 | 1.92 | na | +/− | 0.5 | +/− | 0.5 | − | 0.0 | 1.0 |
| Saccharomyces sp. | Y231 | 1.90 | na | +/− | 0.5 | +/− | 0.5 | − | 0.0 | 1.0 |
| Yeast Strain | Medium | Temperature | pH | Cell Count (log CFU/mL) | OD600 | pH | Viscosity η (mPa.s) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 h | 24 h | 48 h | 24 h | 48 h | 0 h | 24 h | 48 h | 0 h | 24 h | 48 h | ||||
| H312 | YB | 25 °C | 4.5 | 3.5 | 6.5 ± 0.3 b | 7.3 ± 0.2 ab | 1.59 ± 0.0 abc | 1.62 ± 0.0 acde | 4.45 ± 0.0 | 4.33 ± 0.0 c | 4.45 ± 0.0 ae | nd | nd | nd |
| 5.0 | 3.4 | 7.1 ± 0.3 ab | 7.2 ± 0.4 ab | 1.58 ± 0.0 abc | 1.74 ± 0.0 abcd | 5.03 ± 0.0 | 4.53 ± 0.0 a | 4.73 ± 0.1 a | nd | nd | nd | |||
| 30 °C | 4.5 | 3.8 | 7.4 ± 0.0 a | 6.9 ± 0.1 ab | 1.24 ± 0.0 abde | 1.50 ± 0.0 ae | 4.52 ± 0.1 | 4.11 ± 0.0 b | 3.66 ± 0.1 bc | nd | nd | nd | ||
| 5.0 | 3.9 | 7.4 ± 0.1 a | 6.6 ± 0.4 a | 1.31 ± 0.0 abcde | 1.55 ± 0.1 ae | 5.07 ± 0.0 | 4.35 ± 0.0 c | 3.81 ± 0.0 bc | nd | nd | nd | |||
| mCPSM | 25 °C | 4.5 | 3.6 | 7.5 ± 0.1 a | 7.0 ± 0.1 ab | 1.56 ± 0.1 abc | 1.99 ± 0.1 bc | 4.53 ± 0.0 | nd | nd | nd | nd | nd | |
| 5.0 | 3.7 | 7.2 ± 0.2 ab | 7.2 ± 0.1 ab | 1.34 ± 0.1 abcde | 1.96 ± 0.1 bcd | 5.03 ± 0.0 | nd | nd | nd | nd | nd | |||
| 30 °C | 4.5 | 4.1 | 7.5 ± 0.1 a | 7.1 ± 0.9 ab | 1.18 ± 0.3 bde | 1.53 ± 0.3 ae | 4.53 ± 0.1 | 4.09 ± 0.0 b | 3.66 ± 0.0 bc | 20.86 ± 5.6 b | 4.57 ± 0.5 bc | 2.87 ± 0.4 b | ||
| 5.0 | 3.9 | 7.4 ± 0.0 a | 6.8 ± 0.4 ab | 0.89 ± 0.1 d | 1.33 ± 0.1 e | 5.07 | 4.68 ± 0.0 d | 3.38 ± 0.6 b | 18.37 ± 1.3 ab | 5.71 ± 0.5 bc | 3.14 ± 0.3 b | |||
| H403 | YB | 25 °C | 4.5 | 3.4 | 7.0 ± 0.1 ab | 7.6 ± 0.4 ab | 1.68 ± 0.0 ac | 1.73 ± 0.0 abcd | 4.45 ± 0.0 | 4.21 ± 0.0 e | 4.35 ± 0.0 ade | nd | nd | nd |
| 5.0 | 3.4 | 7.1 ± 0.2 ab | 7.4 ± 0.2 ab | 1.71 ± 0.0 c | 1.76 ± 0.1 abcd | 5.03 ± 0.0 | 4.47 ± 0.0 af | 4.61 ± 0.0 a | nd | nd | nd | |||
| 30 °C | 4.5 | 4.4 | 7.5 ± 0.0 a | 7.3 ± 0.2 ab | 1.38 ± 0.2 abce | 1.59 ± 0.2 ade | 4.52 ± 0.1 | 4.17 ± 0.0 be | 3.70 ± 0.0 bc | nd | nd | nd | ||
| 5.0 | 4.4 | 7.6 ± 0.0 a | 6.6 ± 0.4 a | 1.45 ± 0.1 abce | 1.59 ± 0.1 ade | 5.07 ± 0.0 | 4.41 ± 0.0 cf | 3.89 ± 0.0 bcd | nd | nd | nd | |||
| mCPSM | 25 °C | 4.5 | 3.7 | 7.3 ± 0.0 a | 6.9 ± 0.6 ab | 0.93 ± 0.1 d | 2.04 ± 0.1 b | 4.53 ± 0.0 | nd | nd | nd | nd | nd | |
| 5.0 | 3.7 | 7.1 ± 0.1 ab | 6.8 ± 0.9 a | 1.02 ± 0.2 de | 2.04 ± 0.1 b | 5.03 ± 0.0 | nd | nd | nd | nd | nd | |||
| 30 °C | 4.5 | 4.5 | 7.0 ± 0.9 ab | 6.8 ± 0.2 ab | 1.04 ± 0.0 de | 1.43 ± 0.0 ae | 4.53 ± 0.1 | 4.12 ± 0.0 b | 3.62 ± 0.0 bc | 16.27 ± 2.9 ab | 3.00 ± 0.2 c | 2.59 ± 0.3 b | ||
| 5.0 | 4.3 | 7.6 ± 0.0 a | 8.0 ± 0.1 b | 1.02 ± 0.3 de | 1.25 ± 0.2 e | 5.07 | 4.66 ± 0.1 d | 3.99 ± 0.0 cde | 21.00 ± 5.7 b | 9.55 ± 0.9 ad | 3.25 ± 0.5 b | |||
| Feed Parameters | Feed Phase | |||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |
| μset (h−1) | 0.4 | 0.26 | 0.16 | 0.1 | 0.05 | 0.01 |
| F(0) (g/h) | 36.36 | 36.36 | 36.36 | 36.36 | 36.36 | 36.36 |
| Duration (h) | 24 | 6 | 1 | 10 | 3 | 10 |
| Experimental Condition to Evaluate Viscosity Reduction in mCPSMpc | Incubation Time | Viscosity η (mPa.s) | |
|---|---|---|---|
| Control | H403 | ||
| Using freeze-dried powder (5 log cells/mL) | 0 h | 11.2 ± 1.2 | - |
| 24 h | 10.6 ± 0.4 | 4.6 ± 1.2 | |
| 48 h | 12.8 ± 2.6 | 3.3 ± 0.1 | |
| Using CFS (1:3) after 0, 15, and 24 h incubation of freeze-dried powder in mCPSM | 0 h | 6.9 ± 0.9 | - |
| 15 h | 4.6 ± 0.3 | 3.2 ± 0.2 | |
| 24 h | 4.7 ± 0.2 | 2.4 ± 0.2 | |
| Applied Microorganisms | Cell Count after 96 h [log CFU/g] | |||
|---|---|---|---|---|
| Pectinolytic Yeast (H312, H403, H404, Y170) | S. cerevisiae H290 | Total Yeast * | L. fermentum 223 | |
| Non-inoculated beans | <2.7 | <2.7 | <2.7 | <2.7 |
| H290 | na | 7.8 | na | na |
| 223 + H290 | na | 7.9 | na | 9.4 |
| H312 | 6.9 | na | na | na |
| H312 + 223 | 6.6 | na | na | 9.2 |
| H312 + 223 + H290 | 6.4 | 8.1 | na | 9.3 |
| H403 | 6.6 | na | na | na |
| H403 + 223 | 6.5 | na | na | 9.4 |
| H403 + 223 + H290 | 7.5 | 7.9 | na | 10.0 |
| H404 | 5.6 | na | na | na |
| H404 + 223 | 4.3 | na | na | 9.4 |
| H404 + 223 + H290 | 6.4 | 7.7 | na | 9.2 |
| Y170 | 7.8 | na | na | na |
| Y170 + 223 | 7.7 | na | na | 9.2 |
| Y170 + 223 + H290 | na ** | na ** | 7.8 | 9.2 |
| Variation | Fermentation | Drying | ||||
|---|---|---|---|---|---|---|
| Tav (°C) | Tmax (°C) | Time until Tmax (h) | Tav (°C) | Tmax (°C) | Tmin (°C) | |
| Control | 37.0 ± 2.1 | 48.7 ± 1.2 | 83.1 ± 11.8 | 31.6 ± 1.8 | 49.9 ± 2.3 | 17.7 ± 1.2 |
| 223 + H290 | 38.7 ± 0.8 | 48.6 ± 0.8 | 59.6 ± 8.0 | 31.6 ± 1.9 | 49.7 ± 0.7 | 17.7 ± 1.1 |
| H403 + 223 + H290 | 38.6 ± 1.5 | 48.9 ± 1.3 | 59.4 ± 7.5 | 31.4 ± 1.8 | 50.2 ± 1.8 | 17.7 ± 1.2 |
| Microbial Group | Scale | Variation | Fermentation Duration (Day) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | |||
| LAB (log CFU/g) | 1 kg | Control | 2.7 | 5.2 | 7.9 | 9.0 | 8.3 | 8.3 | - |
| 223 + H290 | 6.0 | 7.7 | nd | 9.0 | 8.0 | 8.0 | - | ||
| H403 + 223 + H290 | 6.0 | 7.7 | 9.1 | 8.9 | 7.6 | 7.6 | - | ||
| H404 + 223 + H290 | 6.0 | 7.8 | 9.1 | 9.0 | 7.9 | 7.9 | - | ||
| 20 kg | Control | 2.0 ± 0.0 | 5.4 ± 1.4 | 7.7 ± 1.1 | 6.6 ± 0.6 | 7.4 ± 0.0 | 7.0 ± 0.9 | 8.4 ± 0.2 | |
| 223 + H290 | 5.3 ± 1.1 | 8.7 ± 0.3 | 8.9 ± 0.0 | 6.5 ± 0.5 | 6.5 ± 1.0 | 7.1 ± 1.3 | 7.7 ± 0.3 | ||
| H403 + 223 + H290 | 5.8 ± 0.6 | 8.7 ± 0.2 | 8.6 ± 0.2 | 6.5 ± 0.6 | 5.8 ± 0.7 | 6.6 ± 1.0 | 8.2 ± 0.5 | ||
| Yeasts (log CFU/g) | 1 kg | Control | 2.7 | 3.3 | 6.7 | 8.2 | 7.1 | 7.1 | - |
| 223 + H290 | 6.0 | 8.1 | 7.7 | 8.4 | 7.8 | 7.8 | - | ||
| H403 + 223 + H290 | 6.0 | 8.2 | 7.8 | 8.4 | 7.3 | 7.3 | - | ||
| H404 + 223 + H290 | 6.0 | 8.2 | 7.7 | 8.4 | 7.5 | 7.5 | - | ||
| 20 kg | Control | 2.9 ± 0.7 | 6.5 ± 0.1 | 7.7 ± 0.2 | 6.7 ± 0.9 | 4.6 ± 0.6 | 5.1 ± 0.5 | 5.4 ± 0.2 | |
| 223 + H290 | 5.4 ± 0.9 | 6.8 ± 0.3 | 7.1 ± 0.4 | 4.3 ± 0.5 | 4.4 ± 0.0 | 4.8 ± 0.9 | 5.3 ± 0.3 | ||
| H403 + 223 + H290 | 6.2 ± 0.6 | 6.6 ± 0.4 | 6.9 ± 0.4 | 3.9 ± 0.6 | 3.4 ± 0.6 | 4.1 ± 0.9 | 5.5 ± 0.6 | ||
| Step | Variation | Well Fermented (%) | Slightly Fermented (%) | Violet Beans (%) |
|---|---|---|---|---|
| Fermentation | Control | 2.0 ± 2.0 | 85.3 ± 8.1 | 12.7 ± 7.0 |
| 223 + H290 | 0.7 ± 1.2 | 92.7 ± 2.3 | 6.7 ± 3.1 | |
| H403 + 223 + H290 | 0.7 ± 1.2 | 90.0 ± 7.2 | 8.0 ± 6.9 | |
| Drying | Control | 9.0 ± 9.2 | 83.5 ± 10.3 | 7.6 ± 4.6 |
| 223 + H290 | 11.9 ± 7.2 | 83.6 ± 7.9 | 4.6 ± 2.5 | |
| H403 + 223 + H290 | 13.2 ± 11.4 | 83.1 ± 9.3 | 3.7 ± 2.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Freimüller Leischtfeld, S.; Hämmerli, A.; Lehmann, A.; Tönz, A.; Beck, B.M.; Wild, J.; Weis, S.; Neutsch, L.; Miescher Schwenninger, S. Wickerhamomyces pijperi: An Up-And-Coming Yeast with Pectinolytic Activity Suitable for Cocoa Bean Fermentation. Appl. Microbiol. 2025, 5, 43. https://doi.org/10.3390/applmicrobiol5020043
Freimüller Leischtfeld S, Hämmerli A, Lehmann A, Tönz A, Beck BM, Wild J, Weis S, Neutsch L, Miescher Schwenninger S. Wickerhamomyces pijperi: An Up-And-Coming Yeast with Pectinolytic Activity Suitable for Cocoa Bean Fermentation. Applied Microbiology. 2025; 5(2):43. https://doi.org/10.3390/applmicrobiol5020043
Chicago/Turabian StyleFreimüller Leischtfeld, Susette, Alexander Hämmerli, Armin Lehmann, Andrea Tönz, Barbara Maria Beck, Jessica Wild, Stefanie Weis, Lukas Neutsch, and Susanne Miescher Schwenninger. 2025. "Wickerhamomyces pijperi: An Up-And-Coming Yeast with Pectinolytic Activity Suitable for Cocoa Bean Fermentation" Applied Microbiology 5, no. 2: 43. https://doi.org/10.3390/applmicrobiol5020043
APA StyleFreimüller Leischtfeld, S., Hämmerli, A., Lehmann, A., Tönz, A., Beck, B. M., Wild, J., Weis, S., Neutsch, L., & Miescher Schwenninger, S. (2025). Wickerhamomyces pijperi: An Up-And-Coming Yeast with Pectinolytic Activity Suitable for Cocoa Bean Fermentation. Applied Microbiology, 5(2), 43. https://doi.org/10.3390/applmicrobiol5020043







