In Vitro Modelling of a Typical Dietary Intake in Restrictive Anorexia Nervosa Results in Changes to Gut Microbial Community and Metabolites
Abstract
1. Introduction
2. Materials and Methods
2.1. Modelling Dietary Intake R-AN and HCs
2.2. Three-Stage Continuous Culture System
2.3. Total Bacteria and SCFA Analysis
2.4. DNA Extraction, Quantification, and Qualification
2.5. 16S rRNA Gene-Based Next-Generation Sequencing (NGS) and Bioinformatics
2.6. Data Processing and Bioinformatic Analysis
2.7. Statistical Analysis
2.7.1. 16S Metagenomic Sequencing and Analysis
2.7.2. 16S rRNA Gene-Based Next-Generation Sequencing (NGS) and Bioinformatics
3. Results
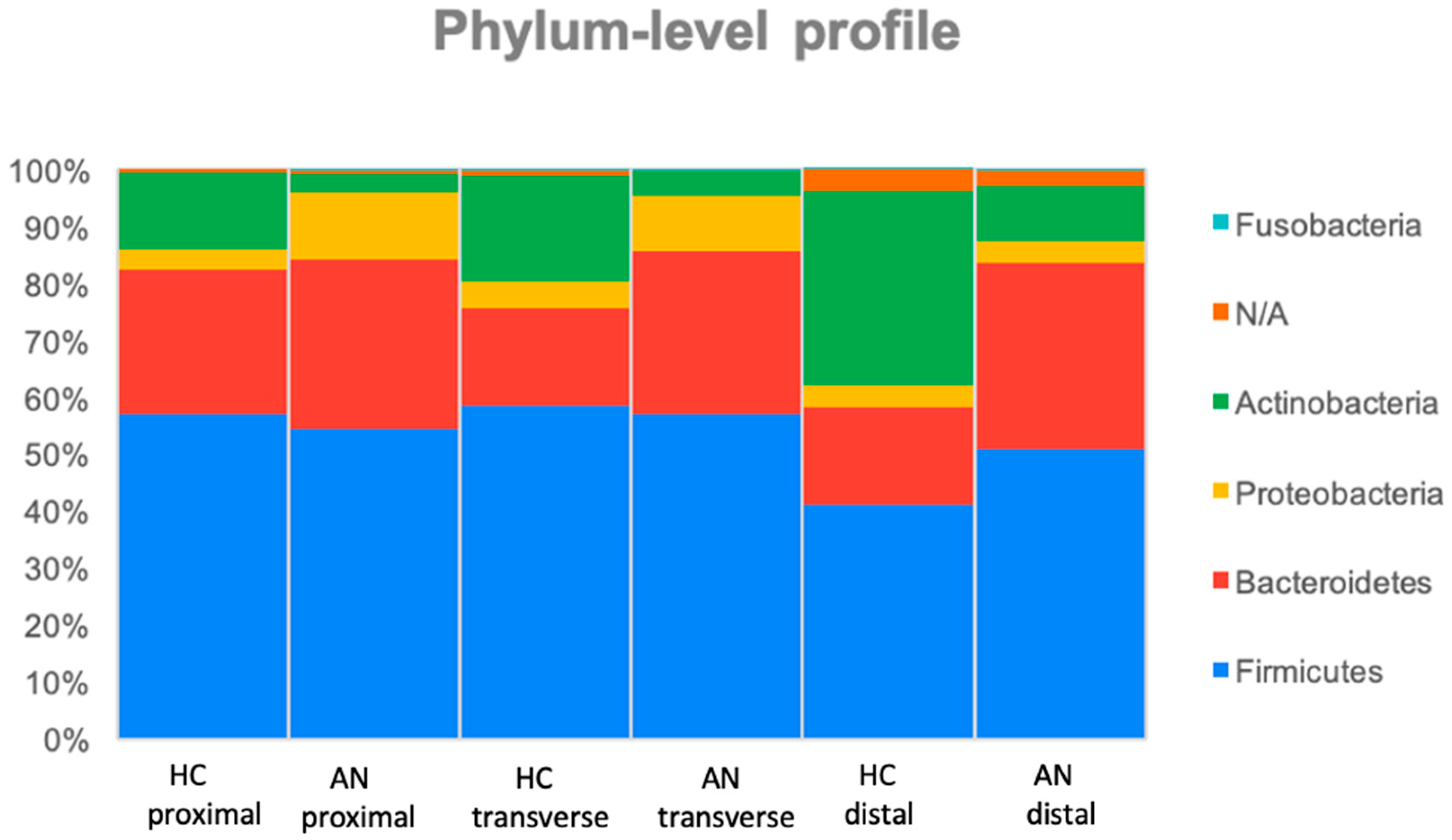
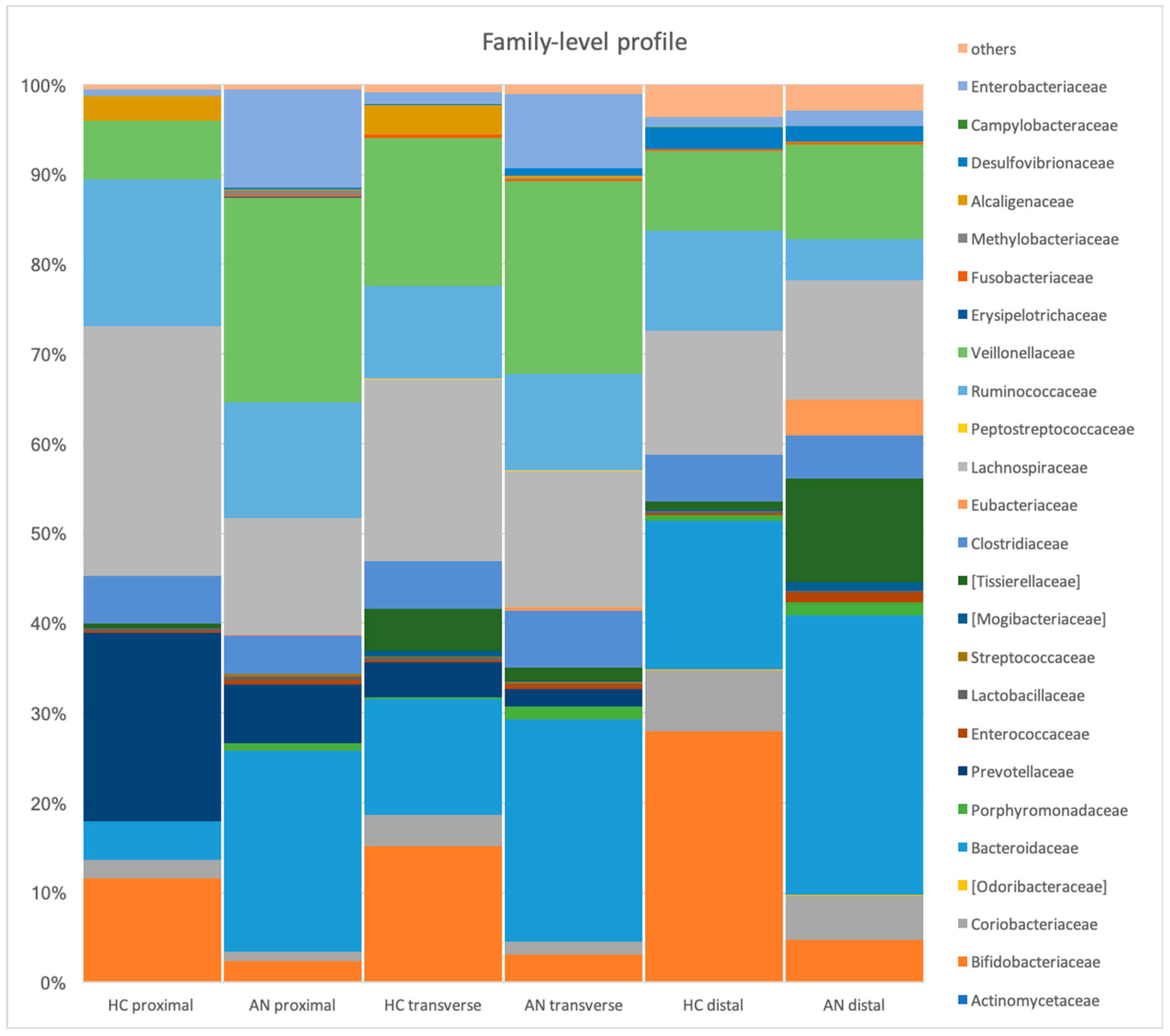
3.1. Microbial Profiles in the Stimulation of Proximal, Transverse, and Distal Colon (HC Versus R-AN)
3.2. Total Bacteria, SCFA, and BCFA Production
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Eddy, K.T.; Dorer, D.J.; Franko, D.L.; Tahilani, K.; Thompson-Brenner, H.; Herzog, D.B. Diagnostic crossover in anorexia nervosa and bulimia nervosa: Implications for DSM-V. Am. J. Psychiatry 2008, 165, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Mehler, P.S.; Brown, C. Anorexia nervosa—Medical complications. J. Eat. Disord. 2015, 3, 11. [Google Scholar] [CrossRef] [PubMed]
- ICD-11 for Mortality and Morbidity Statistics. Available online: https://icd.who.int/browse11/l-m/en (accessed on 19 November 2023).
- Borgo, F.; Riva, A.; Benetti, A.; Casiraghi, M.C.; Bertelli, S.; Garbossa, S.; Anselmetti, S.; Scarone, S.; Pontiroli, A.E.; Morace, G.; et al. Microbiota in anorexia nervosa: The triangle between bacterial species, metabolites, and psychological tests. PLoS ONE 2017, 12, e0179014. [Google Scholar] [CrossRef] [PubMed]
- Morita, C.; Tsuji, H.; Hata, T.; Gondo, M.; Takakura, S.; Kawai, K.; Yoshihara, K.; Ogata, K.; Nomoto, K.; Miyazaki, K.; et al. Gut Dysbiosis in Patients with Anorexia Nervosa. PLoS ONE 2015, 10, e0145274. [Google Scholar] [CrossRef]
- Monteleone, A.M.; Troisi, J.; Fasano, A.; Dalle Grave, R.; Marciello, F.; Serena, G.; Calugi, S.; Scala, G.; Corrivetti, G.; Cascino, G.; et al. Multi-Omics Data Integration in Anorexia Nervosa Patients Before and After Weight Regain: A Microbiome-Metabolomics Investigation. Clin. Nutr. 2021, 40, 1137–1146. [Google Scholar] [CrossRef]
- Prochazkova, P.; Roubalova, R.; Dvorak, J.; Kreisinger, J.; Hill, M.; Tlaskalova-Hogenova, H.; Tomasova, P.; Pelantova, H.; Cermakova, M.; Kuzma, M.; et al. The Intestinal Microbiota and Metabolites in Patients with Anorexia Nervosa. Gut Microbes 2021, 13, e1902771. [Google Scholar] [CrossRef]
- Fernandez-Aranda, F.; Pinheiro, A.P.; Tozzi, F.; Thornton, L.M.; Fichter, M.M.; Halmi, K.A.; Kaplan, A.S.; Klump, K.L.; Strober, M.; Woodside, D.B.; et al. Symptom profile of major depressive disorder in women with eating disorders. Aust. N. Z. J. Psychiatry 2007, 41, 24–31. [Google Scholar] [CrossRef]
- Garcia, N.; Gutierrez, E. Anorexia Nervosa and Microbiota: Systematic Review and Critical Appraisal. Eat. Weight Disord. 2023, 28, 1. [Google Scholar] [CrossRef]
- Godart, N.T.; Flament, M.; Perdereau, F.; Jeammet, P. Comorbidity between eating disorders and anxiety disorders: A review. Int. J. Eat. Disord. 2002, 32, 253–270. [Google Scholar] [CrossRef] [PubMed]
- Kaye, W.H.; Bulik, C.M.; Thornton, L.; Barbarich, N.; Masters, K.; PFC Group. Comorbidity of anxiety disorders with anorexia and bulimia nervosa. Am. J. Psychiatry 2004, 161, 2215–2221. [Google Scholar] [CrossRef]
- Diagnostic and Statistical Manual of Mental Disorders, 5th ed.; text rev.; DSM-5-TR; American Psychiatric Association: Washington, DC, USA, 2022.
- Baxter, N.T.; Schmidt, A.W.; Venkataraman, A.; Kim, K.S.; Waldron, C.; Schmidt, T.M. Dynamics of human gut microbiota and short-chain fatty acids in response to dietary interventions with three fermentable fibers. mBio 2019, 10, e02566-18. [Google Scholar] [CrossRef] [PubMed]
- Landini, L.; Dadson, P.; Gallo, F.; Honka, M.-J.; Cena, H. Microbiota in anorexia nervosa: Potential for treatment. Nutr. Res. Rev. 2023, 36, 372–391. [Google Scholar] [CrossRef] [PubMed]
- Kaelin, W.G., Jr.; McKnight, S.L. Influence of metabolism on epigenetics and disease. Cell 2013, 153, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the human intestinal microbial flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef]
- Cristofori, F.; Dargenio, V.N.; Dargenio, C.; Miniello, V.L.; Barone, M.; Francavilla, R. Anti-Inflammatory and Immunomodulatory Effects of Probiotics in Gut Inflammation: A Door to the Body. Front. Immunol. 2021, 12, 578386. [Google Scholar] [CrossRef]
- Grau-Del Valle, C.; Fernández, J.; Solá, E.; Montoya-Castilla, I.; Morillas, C.; Bañuls, C. Association between gut microbiota and psychiatric disorders: A systematic review. Front. Psychol. 2023, 14, 1215674. [Google Scholar] [CrossRef]
- Roubalova, R.; Prochazkova, P.; Papezova, H.; Smitka, K.; Bilej, M.; Tlaskalova-Hogenova, H. Anorexia nervosa: Gut microbiota-immune-brain interactions. Clin. Nutr. 2020, 39, 676–684. [Google Scholar] [CrossRef]
- Sweeney, T.E.; Morton, J.M. The human gut microbiome: A review of the effect of obesity and surgically induced weight loss. JAMA Surg. 2013, 148, 563–569. [Google Scholar] [CrossRef]
- Koliada, A.; Syzenko, G.; Moseiko, V.; Budovska, L.; Puchkov, K.; Perederiy, V.; Gavalko, Y.; Dorofeyev, A.; Romanenko, M.; Tkach, S.; et al. Association between body mass index and Firmicutes/Bacteroidetes ratio in an adult Ukrainian population. BMC Microbiol. 2017, 17, 120. [Google Scholar] [CrossRef]
- Mack, I.; Cuntz, U.; Grämer, C.; Niedermaier, S.; Pohl, C.; Schwiertz, A.; Zimmermann, K.; Zipfel, S.; Enck, P.; Penders, J. Weight gain in anorexia nervosa does not ameliorate the fecal microbiota, branched-chain fatty acid profiles, and gastroin-testinal complaints. Sci. Rep. 2016, 6, 26752. [Google Scholar] [CrossRef] [PubMed]
- Fetissov, S.O.; Hökfelt, T. On the origin of eating disorders: Altered signaling between gut microbiota, adaptive immunity, and the brain melanocortin system regulating feeding behavior. Curr. Opin. Pharmacol. 2019, 48, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Tennoune, N.; Chan, P.; Breton, J.; Legrand, R.; Chabane, Y.N.; Akkermann, K.; Jarv, A.; Ouelaa, W.; Takagi, K.; Ghouzali, I.; et al. Bacterial ClpB heat-shock protein, an antigen-mimetic of the anorexigenic peptide alpha-MSH, at the origin of eating disorders. Transl. Psychiatry 2014, 4, e458. [Google Scholar] [CrossRef] [PubMed]
- Flint, H.J.; Scott, K.P.; Duncan, S.H.; Louis, P.; Forano, E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 2012, 3, 289–306. [Google Scholar] [CrossRef]
- Cremer, J.; Segota, I.; Yang, C.Y.; Arnoldini, M.; Sauls, J.T.; Zhang, Z.; Gutierrez, E.; Groisman, A.; Hwa, T. Effect of flow and peristaltic mixing on bacterial growth in a gut-like channel. Proc. Natl. Acad. Sci. USA 2016, 113, 11414–11419. [Google Scholar] [CrossRef]
- Fine, K.D.; Santa Ana, C.A.; Porter, J.L.; Fordtran, J.S. Effect of changing intestinal flow rate on a measurement of intestinal permeability. Gastroenterology 1995, 108, 983–989. [Google Scholar] [CrossRef]
- Macfarlane, G.T.; Gibson, G.R. Validation of a three-stage compound continuous culture system for investigating the effect of retention time on the ecology and metabolism of bacteria in the human colon. Microb. Ecol. 1998, 35, 180–187. [Google Scholar] [CrossRef]
- Macfarlane, G.T.; Gibson, G.R. Metabolic activities of the normal colonic flora. In Human Health—The Contribution of Microorganisms; Gibson, S.A.W., Ed.; Springer: London, UK, 1994. [Google Scholar]
- Macfarlane, G.T.; Gibson, G. Microbiological Aspects of Short Chain Fatty Acid Production in the Large Bowel; Cambridge University Press: Cambridge, UK, 1995. [Google Scholar]
- Murphy, K.G.; Bloom, S.R. Gut hormones in the control of appetite. Exp. Physiol. 2004, 89, 507–516. [Google Scholar] [CrossRef]
- Raatz, S.K.; Jahns, L.; Johnson, L.K.; Crosby, R.; Mitchell, J.E.; Crow, S.; Peterson, C.; Le Grange, D.; Wonderlich, S.A. Nutritional adequacy of dietary intake in women with anorexia nervosa. Nutrients 2015, 7, 3652–3665. [Google Scholar] [CrossRef]
- Jauregui Lobera, I.; Bolanos Rios, P. Choice of diet in patients with anorexia nervosa. Nutr. Hosp. 2009, 24, 682–687. [Google Scholar]
- British Nutrition Foundation. Nutrition Requirements. Available online: https://www.nutrition.org.uk/nutritional-information/nutrient-requirements/ (accessed on 26 October 2019).
- Macfarlane, G.T.; Gibson, G.R. Carbohydrate Fermentation, Energy Transduction and Gas Metabolism in the Human Large Intestine. In Gastrointestinal Microbiology; Chapman & Hall Microbiology Series; Mackie, R.I., White, B.A., Eds.; Springer: Boston, MA, USA, 1997. [Google Scholar] [CrossRef]
- Liu, L.; Poveda, C.; Jenkins, P.E.; Walton, G.E. An In Vitro Approach to Studying the Microbial Community and Impact of Pre and Probiotics under Anorexia Nervosa Related Dietary Restrictions. Nutrients 2021, 13, 4447. [Google Scholar] [CrossRef] [PubMed]
- Harrison, M.; Miller, L.; AlEnazi, N.; Herron, D. Pregnancy and Weight Loss Surgery. In Handbook of Nutrition and Pregnancy, 2nd ed.; Lammi-Keefe, C., Couch, S., Kirwan, J., Eds.; Humana Press: Cham, Switzerland, 2018; pp. 421–428. [Google Scholar] [CrossRef]
- Kamal, N.; Chami, T.; Andersen, A.; Rosell, F.A.; Schuster, M.M.; Whitehead, W.E. Delayed gastrointestinal transit times in anorexia nervosa and bulimia nervosa. Gastroenterology 1991, 101, 1320–1324. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2012, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Huse, S.M.; Welch, D.M.; Morrison, H.G.; Sogin, M.L. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol. 2010, 12, 1889–1898. [Google Scholar] [CrossRef]
- Nissen, L.; Casciano, F.; Gianotti, A. Intestinal fermentation in vitro models to study food-induced gut microbiota shift: An updated review. FEMS Microbiol. Lett. 2020, 367, fnaa097. [Google Scholar] [CrossRef]
- Williams, C.F.; Walton, G.E.; Jiang, L.; Plummer, S.; Garaiova, I.; Gibson, G.R. Comparative analysis of intestinal tract models. Annu. Rev. Food Sci. Technol. 2015, 6, 329–350. [Google Scholar] [CrossRef]
- Biagi, E.; Nylund, L.; Candela, M.; Ostan, R.; Bucci, L.; Pini, E.; Nikkïla, J.; Monti, D.; Satokari, R.; Franceschi, C.; et al. Through ageing, and beyond: Gut microbiota and inflammatory status in seniors and centenarians. PLoS ONE 2010, 5, e10667. [Google Scholar] [CrossRef]
- Moon, C.D.; Young, W.; Maclean, P.H.; Cookson, A.L.; Bermingham, E.N. Metagenomic insights into the roles of Proteobacteria in the gastrointestinal microbiomes of healthy dogs and cats. Microbiologyopen 2018, 7, e00677. [Google Scholar] [CrossRef]
- Fouladi, F.; Bulik-Sullivan, E.C.; Glenny, E.M.; Thornton, L.M.; Reed, K.K.; Thomas, S.; Kleiman, S.; Watters, A.; Oakes, J.; Huh, E.-Y.; et al. Reproducible changes in the anorexia nervosa gut microbiota following inpatient therapy remain distinct from non-eating disorder controls. Gut Microbes 2022, 14, 2143217. [Google Scholar] [CrossRef]
- Schulz, N.; Belheouane, M.; Dahmen, B.; Ruan, V.A.; Specht, H.E.; Dempfle, A.; Herpertz-Dahlmann, B.; Baines, J.F.; Seitz, J. Gut microbiota alteration in adolescent anorexia nervosa does not normalize with short-term weight restoration. Int. J. Eat. Disord. 2021, 54, 969–980. [Google Scholar] [CrossRef]
- Fan, Y.; Stoving, R.K.; Berreira Ibraim, S.; Hyotylainen, T.; Thirion, F.; Arora, T.; Lyu, L.; Stankevic, E.; Hansen, T.H.; Dechelotte, P.; et al. The gut microbiota contributes to the pathogenesis of anorexia nervosa in humans and mice. Nat. Micro-Biol. 2023, 8, 787–802. [Google Scholar] [CrossRef] [PubMed]
- Binda, C.; Lopetuso, L.R.; Rizzatti, G.; Gibiino, G.; Cennamo, V.; Gasbarrini, A. Actinobacteria: A relevant minority for the maintenance of gut homeostasis. Dig. Liver Dis. 2018, 50, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Grundy, M.M.L.; Edwards, C.H.; Mackie, A.R.; Gidley, M.J.; Butterworth, P.J.; Ellis, P.R. Re-evaluation of the mechanisms of dietary fibre and implications for macronutrient bioaccessibility, digestion and postprandial metabolism. Br. J. Nutr. 2016, 116, 816–833. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Gordon, J.I. The core gut microbiome, energy balance and obesity. J. Physiol. 2009, 587 Pt 17, 4153–4158. [Google Scholar] [CrossRef]
- Macfarlane, G.T.; Macfarlane, S. Fermentation in the human large intestine: Its physiologic consequences and the potential contribution of prebiotics. J. Clin. Gastroenterol. 2011, 45, S120–S127. [Google Scholar] [CrossRef]
- Roberfroid, M.; Gibson, G.R.; Hoyles, L.; McCartney, A.L.; Rastall, R.; Rowland, I.; Wolvers, D.; Watzl, B.; Szajewska, H.; Stahl, B.; et al. Prebiotic effects: Metabolic and health benefits. Br. J. Nutr. 2010, 104 (Suppl. S2), S1–S63. [Google Scholar] [CrossRef]
- Topping, D.L.; Clifton, P.M. Short-chain fatty acids and human colonic function: Roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 2001, 81, 1031–1064. [Google Scholar] [CrossRef] [PubMed]
- Winter, S.E.; Baumler, A.J. Dysbiosis in the inflamed intestine: Chance favors the prepared microbe. Gut Microbes 2014, 5, 71–73. [Google Scholar] [CrossRef]
- Breton, J.; Legrand, R.; Akkermann, K.; Jarv, A.; Harro, J.; Dechelotte, P.; Fetissov, S.O. Elevated Plasma Concentrations of Bacterial ClpB Protein in Patients with Eating Disorders. Int. J. Eat. Disord. 2016, 49, 805–808. [Google Scholar] [CrossRef]
- Adan, R.A.H.; Vink, T. Drug target discovery by pharmacogenetics: Mutations in the melanocortin system and eating disor-ders. Eur. Neuropsychopharmacol. 2001, 11, 483–490. [Google Scholar] [CrossRef]
- Markowiak-Kopeć, P.; Śliżewska, K. The Effect of Probiotics on the Production of Short-Chain Fatty Acids by Human Intesti-nal Microbiome. Nutrients 2020, 12, 1107. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.-P.-T.; Tidjani Alou, M.; Bachar, D.; Levasseur, A.; Brah, S.; Alhousseini, D.; Sokhna, C.; Diallo, A.; Wieringa, F.; Mil-lion, M.; et al. Gut Microbiota Alteration is Characterized by a Proteobacteria and Fusobacteria Bloom in Kwashiorkor and a Bacteroidetes Paucity in Marasmus. Sci. Rep. 2019, 9, 9084. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.I.; Yatsunenko, T.; Manary, M.J.; Trehan, I.; Mkakosya, R.; Cheng, J.; Kau, A.L.; Rich, S.S.; Concannon, P.; Mychaleckyj, J.C.; et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science 2013, 339, 548–554. [Google Scholar] [CrossRef] [PubMed]
- van de Wouw, M.; Schellekens, H.; Dinan, T.G.; Cryan, J.F. Microbiota-Gut-Brain Axis: Modulator of Host Metabolism and Appetite. J. Nutr. 2017, 147, 727–745. [Google Scholar] [CrossRef]
- Sanmiguel, C.P.; Jacobs, J.; Gupta, A.; Ju, T.; Stains, J.; Coveleskie, K.; Lagishetty, V.; Balioukova, A.; Chen, Y.; Dutson, E.; et al. Surgically Induced Changes in Gut Microbiome and Hedonic Eating as Related to Weight Loss: Prelimi-nary Findings in Obese Women Undergoing Bariatric Surgery. Psychosom. Med. 2017, 79, 880–887. [Google Scholar] [CrossRef]
- Armougom, F.; Henry, M.; Vialettes, B.; Raccah, D.; Raoult, D. Monitoring bacterial community of human gut microbiota reveals an increase in Lactobacillus in obese patients and Methanogens in anorexic patients. PLoS ONE 2009, 4, e7125. [Google Scholar] [CrossRef]
- Aizawa, E.; Tsuji, H.; Asahara, T.; Takahashi, T.; Teraishi, T.; Yoshida, S.; Ota, M.; Koga, N.; Hattori, K.; Kunugi, H. Possible association of Bifidobacterium and Lactobacillus in the gut microbiota of patients with major depressive disorder. J. Affect. Disord. 2016, 202, 254–257. [Google Scholar] [CrossRef]
- Cryan, J.F.; O’Riordan, K.J.; Cowan, C.S.M.; Sandhu, K.V.; Bastiaanssen, T.F.S.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V.; et al. The microbiota-gut-brain axis. Physiol. Rev. 2019, 99, 1877–2013. [Google Scholar] [CrossRef]
- Foster, J.A.; McVey Neufeld, K.A. Gut-brain axis: How the microbiome influences anxiety and depression. Trends Neurosci. 2013, 36, 305–312. [Google Scholar] [CrossRef]
- Yang, Z.; Li, J.; Gui, X.; Shi, X.; Bao, Z.; Han, H.; Li, M.D. Updated review of research on the gut microbiota and their relation to depression in animals and human beings. Mol. Psychiatry 2020, 25, 2759–2772. [Google Scholar] [CrossRef]
- Strandwitz, P.; Kim, K.H.; Terekhova, D.; Liu, J.K.; Sharma, A.; Levering, J.; McDonald, D.; Dietrich, D.; Ramadhar, T.R.; Lekbua, A.; et al. GABA-modulating bacteria of the human gut microbiota. Nat. Microbiol. 2019, 4, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Queipo-Ortuño, M.I.; Seoane, L.M.; Murri, M.; Pardo, M.; Gomez-Zumaquero, J.M.; Cardona, F.; Casanueva, F.; Tinahones, F.J. Gut microbiota composition in male rat models under different nutritional status and physical activity and its association with serum leptin and ghrelin levels. PLoS ONE 2013, 8, e65465. [Google Scholar] [CrossRef] [PubMed]
- Kleiman, S.C.; Carroll, I.M.; Tarantino, L.M.; Bulik, C.M. Gut feelings: A role for the intestinal microbiota in anorexia nervosa? Int. J. Eat. Disord. 2015, 48, 449–451. [Google Scholar] [CrossRef] [PubMed]
- Morkl, S.; Lackner, S.; Muller, W.; Gorkiewicz, G.; Kashofer, K.; Oberascher, A.; Painold, A.; Holl, A.; Holzer, P.; Meinitzer, A.; et al. Gut microbiota and body composition in anorexia nervosa inpatients in comparison to athletes, overweight, obese, and normal weight controls. Int. J. Eat. Disord. 2017, 50, 1421–1431. [Google Scholar] [CrossRef]
- Clavel, T.; Charrier, C.; Braune, A.; Wenning, M.; Blaut, M.; Haller, D. Isolation of bacteria from the ileal mucosa of TNFdeltaARE mice and description of Enterorhabdus mucosicola gen. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 2009, 59 Pt 7, 1805–1812. [Google Scholar] [CrossRef]
- Hanachi, M.; Manichanh, C.; Schoenenberger, A.; Pascal, V.; Levenez, F.; Cournede, N.; Dore, J.; Melchior, J.-C. Altered host-gut microbes symbiosis in severely malnourished anorexia nervosa (AN) patients undergoing enteral nutrition: An explicative factor of functional intestinal disorders? Clin. Nutr. 2019, 38, 2304–2310. [Google Scholar] [CrossRef]
- Louis, P.; Young, P.; Holtrop, G.; Flint, H.J. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environ. Microbiol. 2010, 12, 304–314. [Google Scholar] [CrossRef]
- Imhann, F.; Vich Vila, A.; Bonder, M.J.; Fu, J.; Gevers, D.; Visschedijk, M.C.; Spekhorst, L.M.; Alberts, R.; Franke, L.; van Dul-lemen, H.M.; et al. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut 2018, 67, 108–119. [Google Scholar] [CrossRef]
- Jesus, P.; Ouelaa, W.; Francois, M.; Riachy, L.; Guerin, C.; Aziz, M.; Do Rego, J.C.; Dechelotte, P.; Fetissov, S.O.; Coeffier, M. Al-teration of intestinal barrier function during activity-based anorexia in mice. Clin. Nutr. 2014, 33, 1046–1053. [Google Scholar] [CrossRef]
- Belmonte, L.; Achamrah, N.; Nobis, S.; Guerin, C.; Riou, G.; Bole-Feysot, C.; Boyer, O.; Richard, V.; Do Rego, J.C.; Dechelotte, P.; et al. A role for intestinal TLR4-driven inflammatory response during activity-based anorexia. Sci. Rep. 2016, 6, 35813. [Google Scholar] [CrossRef]
- Abbate-Daga, G.; Buzzichelli, S.; Amianto, F.; Rocca, G.; Marzola, E.; McClintock, S.M.; Fassino, S. Cognitive flexibility in verbal and nonverbal domains and decision-making in anorexia nervosa patients: A pilot study. BMC Psychiatry 2011, 11, 162. [Google Scholar] [CrossRef] [PubMed]
- Calugi, S.; El Ghoch, M.; Conti, M.; Dalle Grave, R. Depression and treatment outcome in anorexia nervosa. Psychiatry Res. 2014, 218, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Sanders, J.W.; Martin, J.W.; Hooke, M.; Hooke, J. Methylobacterium mesophilicum infection: Case report and literature review of an unusual opportunistic pathogen. Clin. Infect. Dis. 2000, 30, 936–938. [Google Scholar] [CrossRef] [PubMed]
- Thornton, L.M.; Welch, E.; Munn-Chernoff, M.A.; Lichtenstein, P.; Bulik, C.M. Anorexia Nervosa, Major Depression, and Suicide Attempts: Shared Genetic Factors. Suicide Life Threat. Behav. 2016, 46, 525–534. [Google Scholar] [CrossRef]
- Li, Z.; Bi, T. Causal effects of gut microbiota, metabolites, immune cells, liposomes, and inflammatory proteins on anorexia nervosa: A mediation joint multi-omics Mendelian randomization analysis. J. Affect. Disord. 2025, 368, 343–358. [Google Scholar] [CrossRef]
- Agus, A.; Clément, K.; Sokol, H. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut 2021, 70, 1174–1182. [Google Scholar] [CrossRef]
- Lai, C.-C.; Cheng, A.; Liu, W.-L.; Tan, C.-K.; Huang, Y.-T.; Chung, K.-P.; Lee, M.-R.; Hsueh, P.-R. Infections Caused by Unusual Methylobacterium Species. J. Clin. Microbiol. 2011, 49, 3329–3331. [Google Scholar] [CrossRef]
- Martens, E.C.; Kelly, A.G.; Tauzin, A.S.; Brumer, H. The Devil Lies in the Details: How Variations in Polysaccharide Fi-ne-Structure Impact the Physiology and Evolution of Gut Microbes. J. Mol. Biol. 2014, 426, 3851–3865. [Google Scholar] [CrossRef]
- Tailford, L.E.; Crost, E.H.; Kavanaugh, D.; Juge, N. Mucin Glycan Foraging in the Human Gut Microbiome. Front. Genet. 2015, 6, 81. [Google Scholar] [CrossRef]
- Speranza, E.; Cioffi, I.; Santarpia, L.; Del Piano, C.; De Caprio, C.; Naccarato, M.; Marra, M.; De Filippo, E.; Contaldo, F.; Pasanisi, F. Fecal Short Chain Fatty Acids and Dietary Intake in Italian Women With Restrictive Anorexia Nervosa: A Pilot Study. Front. Nutr. 2018, 5, 119. [Google Scholar] [CrossRef]
- Bailey, M.T.; Dowd, S.E.; Galley, J.D.; Hufnagle, A.R.; Allen, R.G.; Lyte, M. Exposure to a social stressor alters the structure of the intestinal microbiota: Implications for stressor-induced immunomodulation. Brain Behav. Immun. 2011, 25, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, P.E.; Proctor, K.; Snuggs, S. Dietary intake of adults with eating disorders: A systematic review and me-ta-analysis. J. Psychiatr. Res. 2024, 175, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Million, M.; Angelakis, E.; Maraninchi, M.; Henry, M.; Giorgi, R.; Valero, R.; Vialettes, B.; Raoult, D. Correlation between body mass index and gut concentrations of Lactobacillus reuteri, Bifidobacterium animalis, Methanobrevibacter smithii, and Escherichia coli. Int. J. Obes. 2013, 37, 1460–1466. [Google Scholar] [CrossRef] [PubMed]

| Daily Dietary Composition | Estimated Average Requirement for Adults (Normal Value) | R-AN Diet (n = 12) [33] | Sites of Absorption [38] | |
|---|---|---|---|---|
| Energy intake | Kcal | 2175 | 736 | - |
| Protein intake | g | 50 | 33 | Ileum and colon |
| Carbohydrate | g | 260 | 110 | - |
| Dietary fibre | g | 30 | 12 | Colon |
| Fat | g | 70 | 17 | Ileum |
| Potassium | mg | 3500 | 2600 | Ileum and colon |
| Chloride | mg | 2500 | 1825 | Duodenum and colon |
| Sodium | mg | 1600 | 1168 | Colon |
| Magnesium | mg | 270 | 227 | Duodenum |
| Phosphorus | mg | 550 | 607 | Jejunum |
| Calcium | mg | 700 | 545 | Jejunum |
| Iron | mg | 14.8 | 7.9 | Duodenum |
| Vitamin K | μg | - | - | Ileum and colon |
| Vitamin B12 | μg | 1.5 | 2.2 | Ileum |
| Thiamin | mg | 0.8 | 0.704 | Jejunum |
| Riboflavin | mg | 1.1 | 1.1 | Jejunum |
| Niacin | mg | 13 | 8.6 | Duodenum |
| Folate | μg | 200 | 234 | Duodenum |
| Vitamin B6 | mg | 1.2 | 0.9 | Jejunum |
| Vitamin A | mg | 600 | 503 | Ileum |
| Vitamin C | mg | 40 | 70 | Ileum |
| Vitamin E | mg | 3 | 2.7 | Ileum |
| Vitamin D | μg | 10 | 1.7 | Ileum |
| Selenium | μg | 60 | 39 | Duodenum |
| Zinc | mg | 7 | 5.4 | Jejunum |
| Medium Ingredient | HC Gut Model Medium (g/L) SS1 [29] | R-AN Medium (g/L) SS2 [37] |
|---|---|---|
| Starch | 5 | 2.1 |
| Peptone water | 5 | 3.3 |
| Tryptone | 5 | 3.3 |
| Yeast extract | 4.5 | 2.97 |
| Casein | 3 | 1.98 |
| Guar gum | 1 | 0.4 |
| Inulin | 1 | 0.4 |
| Pectin | 2 | 0.8 |
| Arabinogalactan | 2 | 0.8 |
| Xylan | 2 | 0.8 |
| KCI | 4.5 | 3.28 |
| NaCI | 4.5 | 3.28 |
| NaHCO3 | 1.5 | 1.095 |
| MgSO4·7H2O | 1.25 | 1.05 |
| KH2PO4 | 0.5 | 0.55 |
| K2HPO4 | 0.5 | 0.55 |
| CaCI2·6H2O | 0.15 | 0.117 |
| Hemin | 0.5 | 0.0265 |
| FeSO4·7H2O | 0.0005 | 0.000795 |
| Vitamin K | 10 μL | 6.68 μL |
| L-cystiene HCl | 0.8 | 0.8 |
| Tween 80 | 1 mL | 1 mL |
| Resazurin (0.25 g/L) | 4 mL | 4 mL |
| Mucin (porcine gastric type III) | 4 | 4 |
| Bile salts | 0.4 | 0.4 |
| Taxonomic Level | Classification | Colon Site | HC Relative Abundance | AN Relative Abundance | AN | HC Versus AN p-Value |
|---|---|---|---|---|---|---|
| Phylum | Proteobacteria | Proximal | 3.50 ± 3.80 | 11.70 ± 18.80 | + | 0.04 |
| Actinobacteria | Proximal Transverse Distal | 13.60 ± 7.80 18.60 ± 9.00 34.30 ± 25.90 | 3.40 ± 3.10 4.60 ± 4.80 9.60 ± 10.90 | - - - | <0.001 0.01 0.01 | |
| Family | Eubacteriaceae | Proximal Transverse Distal | 0.00 ± 0.00 0.02 ± 0.04 0.00 ± 0.00 | 0.14 ± 0.23 0.42 ± 0.73 3.97 ± 4.93 | + + + | 0.01 0.03 0.001 |
| Tissierellaceae | Transverse Distal | 4.70 ± 11.50 0.91 ± 0.31 | 1.40 ± 1.70 11.60 ± 7.10 | - + | 0.05 <0.001 | |
| Mogibacteriaceae | Transverse Distal | 0.70 ± 1.50 0.13 ± 0.11 | 0.16 ± 0.30 0.94 ± 1.00 | - + | 0.05 0.01 | |
| Actinomycetaceae | Distal | 0.01 ± 0.02 | 0.00 | - | 0.04 | |
| Porphyromonadaceae | Proximal Transverse | 0.09 ± 0.009 0.24 ± 0.19 | 0.76 ± 0.76 1.40 ± 1.59 | + + | <0.001 0.01 | |
| Lactobacillaceae | Transverse Distal | 0.30 ± 0.30 0.28 ± 0.09 | 0.13 ± 0.15 0.13 ± 0.03 | - - | 0.02 0.01 | |
| Desulfovibrionaceae | Proximal Transverse | 0.00 0.11 ± 0.15 | 0.16 ± 0.20 0.81 ± 0.83 | + + | <0.001 0.03 | |
| Campylobacteraceae | Transverse Distal | 0.03 ± 0.05 0.13 ± 0.19 | 0.00 0.00 | - - | 0.02 0.003 | |
| Enterobacteriaceae | Proximal Transverse | 0.70 ± 0.87 13.19 ± 18.88 | 10.97 ± 17.91 8.37 ± 12.74 | + - | 0.05 0.03 | |
| Bifidobacteriaceae | Transverse | 15.10 ± 9.70 | 3.10 ± 4.10 | - | 0.001 | |
| Coriobacteriaceae | Transverse | 3.50 ± 2.40 | 1.50 ± 0.80 | - | 0.006 | |
| Veillonellaceae | Proximal | 6.62 ± 5.87 | 22.90 ± 18.75 | + | 0.002 | |
| Methylobacteriaceae | Proximal Transverse Distal | 0.00 0.00 0.00 | 0.46 ± 0.82 0.13 ± 0.25 0.00 ± 0.00 | + + + | 0.004 0.003 0.003 | |
| Fusobacteriaceae | Proximal | 0.00 | 0.26 ± 0.74 | + | <0.001 | |
| Alcaligenaceae | Proximal | 2.74 ± 3.80 | 0.16 ± 0.16 | - | 0.00 | |
| Dethiosulfovibrionaceae | Transverse Distal | 0.00 0.00 | 0.11 ± 0.14 0.09 ± 0.05 | + + | 0.03 0.04 |
| Genus | Colon Site | HC Relative Abundance | R-AN Relative Abundance | R-AN | p Value |
|---|---|---|---|---|---|
| Lachnospira | Proximal Transverse | 2.39 ± 1.95 1.86 ± 2.05 | 0.12 ± 0.11 0.08 ± 0.08 | - - | <0.001 <0.001 |
| Methylobacteriaceae | Proximal Transverse Distal | 0.00 0.00 0.00 | 0.41 ± 0.82 0.12 ± 0.24 0.00 ± 0.00 | + + + | <0.001 <0.001 0.03 |
| Sutterella | Proximal | 2.45 ± 3.80 | 0.14 ± 0.16 | - | 0.001 |
| Fusobacterium | Proximal | 0.00 | 0.30 ± 0.74 | + | 0.002 |
| Parabacteroides | Proximal | 0.01 ± 0.01 | 0.64 ± 0.76 | + | 0.002 |
| Desulfovibrio | Proximal | 0.00 | 0.12 ± 0.20 | + | 0.002 |
| Atopobium | Proximal Transverse Distal | 0.00 ± 0.01 0.00 0.00 | 0.21 ± 0.04 0.36 ± 0.71 0.91 ± 1.22 | + + + | 0.01 <0.001 0.007 |
| Ruminococcus | Proximal | 13.09 ± 7.87 | 6.17 ± 4.71 | - | 0.02 |
| Peptoniphilus | Proximal Distal | 0.53 ± 0.81 0.61 ± 0.56 | 0.02 ± 0.03 5.03 ± 6.51 | - + | 0.03 0.005 |
| Clostridium | Proximal Transverse Distal | 0.12 ± 0.18 4.92 ± 1.85 0.00 | 0.94 ± 1.23 6.49 ± 6.18 0.01 ± 0.01 | + + + | 0.04 0.01 0.02 |
| Bifidobacterium | Proximal | 12.18 ± 7.56 | 2.69 ± 3.09 | - | 0.04 |
| Dialister | Proximal | 5.16 ± 4.90 | 24.65 ± 18.75 | + | 0.04 |
| Anaerofustis | Proximal | 0.00 | 0.01 ± 0.00 | + | 0.04 |
| Lachnobacterium | Proximal Distal | 0.45 ± 0.88 0.11 ± 0.12 | 0.01 ± 0.02 0.00 ± 0.01 | - - | 0.05 0.01 |
| Lactobacillales | Proximal Transverse Distal | 0.01 ± 0.00 0.02 ± 0.04 0.09 ± 0.13 | 0.00 0.00 0.00 | - - - | 0.05 0.01 0.006 |
| Porphyromonas | Transverse | 0.00 ± 0.01 | 0.81 ± 1.61 | + | 0.01 |
| Campylobacter | Transverse Distal | 0.02 ± 0.05 0.13 ± 0.18 | 0.00 0.00 | - - | 0.02 0.01 |
| Enterobacteriaceae | Transverse | 0.25 ± 0.35 | 1.83 ± 2.95 | + | 0.03 |
| Faecalibacterium | Transverse | 3.56 ± 6.43 | 5.45 ± 9.07 | + | 0.03 |
| Pyramidobacter | Transverse | 0.00 | 0.10 ± 0.10 | + | 0.03 |
| Tepidimicrobium | Distal | 0.01 ± 0.01 | 2.67 ± 3.77 | + | 0.003 |
| Pseudoramibacter_ Eubacterium | Distal | 0.00 | 3.46 ± 4.90 | + | 0.003 |
| Mitsuokella | Distal | 1.12 ± 1.57 | 0.02 ± 0.03 | - | 0.01 |
| Alkaliphilus | Distal | 0.00 | 0.18 ± 0.26 | + | 0.02 |
| Coriobacteriaceae | Distal | 0.04 ± 0.05 | 0.08 ± 0.08 | + | 0.02 |
| Pyramidobacter | Distal | 0.00 | 0.10 ± 0.14 | + | 0.03 |
| Mogibacteriaceae | Distal | 0.10 ± 0.07 | 0.83 ± 1.03 | + | 0.03 |
| Oxalobacter | Distal | 0.00 | 0.07 ± 0.09 | + | 0.03 |
| Roseburia | Distal | 0.11 ± 0.05 | 0.01 ± 0.02 | - | 0.04 |
| Colon Site | HC | R-AN | R-AN | p Value | |
|---|---|---|---|---|---|
| Total bacteria (log10 cells/mL) | Proximal | 8.50 ± 0.12 | 8.16 ± 0.05 | - | 0.0018 |
| Transverse | 8.23 ± 0.23 | 7.73 ± 0.32 | - | 0.05 | |
| Distal | 8.00 ± 0.09 | 7.28 ± 0.40 | - | 0.01 | |
| Acetate (mM) | Proximal | 30.02 ± 8.59 | 16.79 ± 1.96 | - | 0.023 |
| Transverse | 42.64 ± 17.17 | 21.31 ± 9.98 | - | n.s. | |
| Distal | 46.83 ± 28.44 | 31.94 ± 13.40 | - | n.s. | |
| Butyrate (mM) | Proximal | 28.80 ± 3.26 | 16.82 ± 5.98 | - | 0.012 |
| Transverse | 37.37 ± 6.08 | 17.16 ± 9.57 | - | 0.011 | |
| Distal | 36.61 ± 12.20 | 20.29 ± 7.76 | - | n.s. | |
| Propionate (mM) | Proximal | 17.54 ± 9.40 | 14.27 ± 13.78 | - | n.s. |
| Transverse | 28.82 ± 8.11 | 15.03 ± 6.66 | - | 0.039 | |
| Distal | 27.44 ± 10.21 | 19.54 ± 9.97 | - | n.s. | |
| BCFA (Iso-butyrate + Isovalerate) (mM) | Proximal | 2.91 ± 3.38 | 2.67 ± 3.61 | - | n.s. |
| Transverse | 6.11 ± 1.64 | 3.08 ± 1.86 | - | 0.05 | |
| Distal | 5.70 ± 1.84 | 7.06 ± 4.16 | + | n.s. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Poveda, C.; Jenkins, P.E.; Iddrisu, I.; Walton, G.E. In Vitro Modelling of a Typical Dietary Intake in Restrictive Anorexia Nervosa Results in Changes to Gut Microbial Community and Metabolites. Appl. Microbiol. 2024, 4, 1642-1660. https://doi.org/10.3390/applmicrobiol4040112
Liu L, Poveda C, Jenkins PE, Iddrisu I, Walton GE. In Vitro Modelling of a Typical Dietary Intake in Restrictive Anorexia Nervosa Results in Changes to Gut Microbial Community and Metabolites. Applied Microbiology. 2024; 4(4):1642-1660. https://doi.org/10.3390/applmicrobiol4040112
Chicago/Turabian StyleLiu, Litai, Carlos Poveda, Paul E. Jenkins, Ishawu Iddrisu, and Gemma E. Walton. 2024. "In Vitro Modelling of a Typical Dietary Intake in Restrictive Anorexia Nervosa Results in Changes to Gut Microbial Community and Metabolites" Applied Microbiology 4, no. 4: 1642-1660. https://doi.org/10.3390/applmicrobiol4040112
APA StyleLiu, L., Poveda, C., Jenkins, P. E., Iddrisu, I., & Walton, G. E. (2024). In Vitro Modelling of a Typical Dietary Intake in Restrictive Anorexia Nervosa Results in Changes to Gut Microbial Community and Metabolites. Applied Microbiology, 4(4), 1642-1660. https://doi.org/10.3390/applmicrobiol4040112






