Abstract
Filamentous fungi exhibit unparalleled potential as cell factories for protein production, owing to their adeptness in protein secretion and remarkable proficiency in post-translational modifications. This review delineates the role of filamentous fungi in bio-input technology across different generations and explores their capacity to generate secondary metabolites. Our investigation highlights filamentous fungi as frontrunners in the production of bioactive compounds, emphasizing the imperative nature of elucidating their metabolic repertoire. Furthermore, we delve into common strategies for genetic transformation in filamentous fungi, elucidating the underlying principles, advantages, and drawbacks of each technique. Taking a forward-looking approach, we explore the prospects of genome engineering, particularly the CRISPR-Cas9 technique, as a means to propel protein secretion in filamentous fungi. Detailed examination of the protein secretion pathways in these fungi provides insights into their industrial applications. Notably, extensive research within the scientific community has focused on Aspergillus and Trichoderma species for the industrial production of proteins and enzymes. This review also presents practical examples of genetic engineering strategies aimed at augmenting enzyme secretion in filamentous fungi for various industrial applications. These findings underscore the potential of filamentous fungi as versatile platforms for protein production and highlight avenues for future research and technological advancement in this field.
1. Introduction
The metabolic machinery within filamentous fungi cells serves as a potent and efficient apparatus for the large-scale production of proteins and secondary metabolites in industrial contexts. Presently, filamentous fungi contribute to the synthesis of more than half of the proteins available commercially [1]. Several factors designate them as prime candidates for industrial protein and secondary metabolite production. Notably, filamentous fungi are regarded as safe (GRAS—Generally Regarded as Safe), bolstering their suitability for industrial processes. Moreover, they exhibit a superior capacity for protein secretion compared to other microorganisms such as bacteria and yeast. Additionally, filamentous fungi possess well-characterized systems for post-translational processing, pivotal for modulating the activity and functionality of synthesized proteins. Furthermore, their metabolic versatility enables efficient utilization of various monosaccharides including xylose, arabinose, and galactose [2,3,4].
In Brazil, numerous species of filamentous fungi are enlisted with the Ministry of Agriculture, Livestock, and Supply (MAPA) as bio-inputs for pest and disease management in agricultural fields. These bio-inputs are strategically developed for commercial purposes, typically comprising the vegetative propagation structure of fungi, such as the fungus Beauveria bassiana IBCB 66. Notably, this product, comprising B. bassiana, demonstrates efficacy in controlling various pests including whitefly, rhizome borer, striped mite, sugarcane weevil, and corn leafhopper across diverse crops such as soybeans, beans, bananas, sugar cane, and corn. Another noteworthy example is a commercial product containing Metarhizium anisopliae, strain IBCB 425, which effectively targets root and grasshopper leafhoppers [5]. Moreover, products based on Cordyceps javanica, strain BRM 27666 [6], and C. fumosorosea exhibit notable effectiveness against insect pests [7] (Supplementary Table S1).
Within filamentous fungi, Trichoderma sp. emerges prominently as a biocontrol agent, its presence solidified in the global market with 144 registrations spanning 40 countries as of 2022 [8]. An extensive survey reveals Latin America as the most active market, with Brazil positioned at the forefront. Among the plethora of registered products in Brazil containing filamentous fungi as active ingredients, Trichoderma sp. accounts for 38% of the products, followed by Beauveria sp. (20%) and Metarhizium sp. (11%). The Trichoderma species authorized for disease management in Brazil predominantly include T. afroharzianum, T. asperellum, T. atroviride, T. endophyticum, T. hamatum, T. harzianum, T. koningiopsis, T. reesei, and T. stromaticum [9].
In the contemporary biological products market, all enlisted products adhere to either the first- or second-generation classification, characterized by the presence of fungal structures, namely spores (conidia) of the active ingredient (1st generation), or a blend of diverse microorganism species (2nd generation). Nevertheless, the burgeoning fields of genomics, proteomics, and the substantial progress in molecular biology and genetic techniques have facilitated the identification of diverse proteins or regulatory factors implicated in stress responses and virulence against insect pests within certain fungi [10]. This progress paves the way for the development of formulated biological products for subsequent generations. For instance, the toxins synthesized by B. bassiana primarily consist of secondary metabolites and low molecular weight compounds, including beauvericin, bassianin, bassianolide, beauverolides, tenelin, oosporein, oxalic acid, calcium oxalate crystals, and various analogues of beauvericin. Among these compounds, beauvericin secreted by mycelia emerges as one of the most significant toxins [11,12,13]. In a recent investigation, researchers undertook a chemical analysis of Cordyceps sp. BCC 1788, leading to the isolation of a cyclopeptide designated cordyheptapeptide A. This isolation was achieved from the desiccated extract of the mycelia, revealing the intracellular localization of the peptide in question [14]. Apart from serving as a metabolite of Cordyceps sp. BCC 1788, recognized for its antimalarial properties, cordyheptapeptide A demonstrates efficacy against malaria [15].
Conventional molecular methodologies encompass strategies such as optimizing the fermentation process and inducing random mutagenesis to further enhance desired traits, such as augmenting protein production via novel mutants derived from filamentous fungi. However, certain genetic engineering strategies hold promise in elevating protein expression and secretion in a targeted manner [4,16,17]. The methodologies involving knock-out, knock-in, gene replacement, and conditional gene expression have become standard procedures, facilitating the swift generation of genetically modified filamentous fungi harboring targeted genetic modifications. Moreover, the simultaneous expression of all genes constituting a complete biosynthetic pathway via a polycistronic expression cassette has demonstrated feasibility in filamentous fungi. This approach is employed for the production of bioactive secondary metabolites, including antibiotics such as penicillin and eniatin, as well as austinoid insecticides, across various species of Aspergillus sp. [18,19,20,21].
The demand for bio-insecticides (for insect control) and bio-fungicides (for fungal control) has exhibited notable growth, indicating an escalating prevalence of biological products in agricultural settings. These products are instrumental in the prevention, surveillance, and management of pests and diseases. Moreover, they align with the principles of the bioeconomy and underscore the value of biodiversity, reflecting a global perspective aimed at fostering increasingly sustainable production systems. This approach integrates environmental, economic, social, and productivity considerations [22]. For biofuels, which are pivotal for sustainable energy production, fourth-generation biofuels seek to integrate biofuel synthesis with carbon dioxide (CO2) capture and storage methodologies, employing techniques such as oxy-fuel combustion or leveraging genetic engineering and nanotechnology [23]. Analogous to the evolution observed in biofuel technology, the utilization of genetic engineering to procure agriculturally significant biological products can be likened to fourth-generation bio-inputs. In this context, the objective of this review is to elucidate potential genetic engineering frameworks applicable to filamentous fungi, with a specific emphasis on realizing fourth-generation bio-inputs in the near future.
2. The Generations of Bio-Input Technology
Bio-input can be defined as a product, process or technology of plant, animal or microbial origin, intended for the production, storage and processing of agricultural products, in aquatic production systems or planted forests, which positively interfere in the growth, development and response mechanism of animals, plants, microorganisms and derived substances that interact with physical-chemical and biological products and processes [24,25].
Microbiological products or products of microbial origin are live or inactivated microorganisms, including viruses, as well as those resulting from techniques involving changing hereditary material, which should prevent, destroy, repel, or mitigate any pest or to be used as a plant regulator, stimulator, defoliant, or desiccant [26].
Similar to bio-inputs, biofuels refer to fuels that are produced from living plant matter or by-products of agricultural production. We can divide biofuel technology into conventional and advanced. Conventional biofuels (also known as first-generation biofuels) are ethanol and biodiesel produced from food crops. The need for advanced biofuels arose from a concern about competition for natural resources (e.g. water, energy, land) between fuel and food production. Thus, advanced biofuels cannot compete with food production. In addition, they need to meet higher sustainability requirements, i.e. contribute to reducing greenhouse gas (GHG) emissions by a greater percentage than conventional biofuels [27,28].
The designation of generations of biofuels depends on the specific technology and raw material used to produce biofuels. It also links to temporal development trends over the years and the complexity of the biofuel market, which includes an increasing number of potential raw materials to be used in biofuel production [23]. Similarly, researchers are constantly searching for raw materials and technologies to produce them at lower costs while maintaining the quality and efficiency of the biological agent in the production areas. Thus, we can categorize bio-input technology into generations. Figure 1 shows bio-input technology and its stages of development in generations.
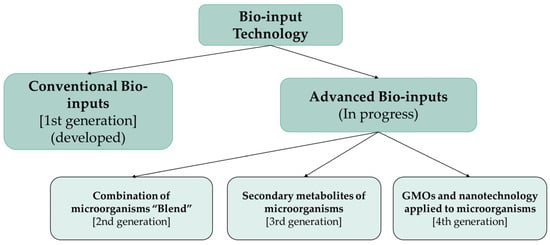
Figure 1.
The different generations of bio-input technology.
Table 1 shows some definitions and similarities between biofuel and bio-input technology and how important it is to understand and separate the generations of these technologies to have a holistic view based on science.

Table 1.
The generations of conventional and advanced technologies for biofuels and Bio-inputs.
Currently, products registered in Brazil whose active ingredient is filamentous fungi are the first and second generation of bio-inputs. Most biological assets used in agriculture are the fungal species Trichoderma sp., Beauveria bassiana and Metarhizium anisopliae (Figure 2) [9].
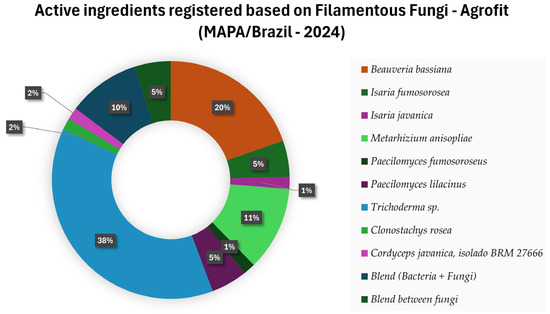
Figure 2.
Active ingredients registered in Brazil based on Filamentous Fungi for pest and disease control [9].
3. Secondary Metabolites from Filamentous fungi
In the early 1940s, antibiotics of fungal (penicillin, griseofulvin) and bacterial (gramicidin) were at the forefront of interest, but after the discovery of streptomycin and later chloramphenicol, tetracyclines and macrolides, attention turned to the actinobacteria of the genus Streptomyces spp. These species yielded the majority (70%) of antibiotics discovered in the 1950s and 1960s. Over the next two decades, the importance of actinobacteria increased to between 25 and 30% of all antibiotics discovered. And since the early 1990s, the number of bioactive compounds isolated from various filamentous fungi and other species of microscopic and higher fungi has increased by over 50% at the turn of the millennium [34].
Regarding the number of bioactive metabolites produced by microorganisms, it is possible to verify that fungi are in second place in obtaining these metabolites, behind only actinobacteria. And within the group of fungi, filamentous fungi are in first place in the ranking, thus showing the importance of exploring the metabolites produced by them (Figure 3) [35].
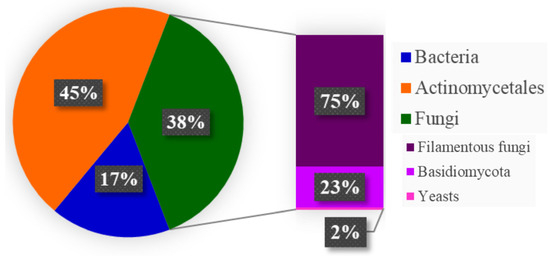
Figure 3.
Estimate of bioactive metabolites got from microorganisms, adapted from [35].
Filamentous fungi secrete a variety and quantity of enzymes. In addition, they can secrete primary and secondary metabolites, such as organic acids and antimicrobial compounds, also known as idiolites. Organic acids also work to release nutrients for fungal growth. For example, they solubilize minerals in the soil and may be involved in the degradation of cellulose [36]. In addition, fungi secrete organic acids to lower the environmental pH, promoting fungal growth and inhibiting bacterial growth [37].
Secondary metabolism and primary metabolism have a strong link in the sense that the precursors and cofactors for secondary metabolites are derived from processes in carbon metabolism. Figure 4 below illustrates the production of secondary metabolites during the growth of filamentous fungi in a representative and generalized manner. For example, the cell only produces penicillin after the logarithmic growth phase (trophophase). The production of the secondary metabolite occurs during the stationary phase of the cell (idiophase), in which the cells do not divide but are metabolically active [38].
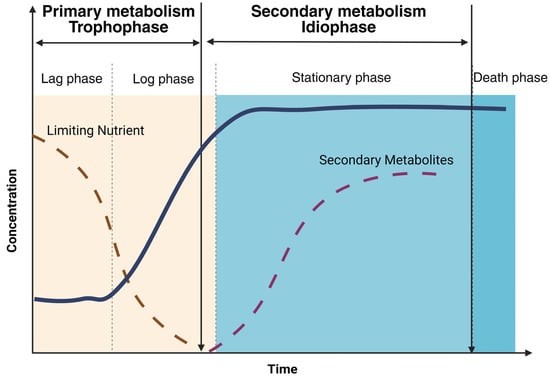
Figure 4.
Diagram of the primary and secondary metabolism of fungi showing when the production of secondary metabolites occurs, adapted from [38]. The solid blue line represents the growth curve of a hypothetical microorganism. The dashed brown line represents the nutrient concentration. And the dashed purple line represents the concentration of secondary metabolites, which occurs during the stationary growth phase of the microorganism.
The genes that are necessary for synthesizing a primary metabolite disperse throughout the fungal genome. About that, biosynthetic gene clusters (BGCs), such as aflatoxin, arrange the genes coding for the enzymatic activities necessary to produce any secondary metabolite [39]. Secondary metabolites are crucial in the development of fungi and actively shape their interactions with other organisms [40,41,42].
Researchers focus on the secondary metabolites that filamentous fungi can produce because they can use some of them as anti-cancer compounds, antibacterials, and mycotoxins. These metabolites play an extremely important role in the fields of health and agriculture [43]. Fungi produce two main classes of secondary metabolites: polyketides (PKs), which derive from short-chain acyl-coenzyme A (acyl-CoA) units and are synthesized by polyketide synthases (PKSs), and non-ribosomal peptides (NRPs), which derive from amino acids and are synthesized by non-ribosomal peptide synthases (NRPSs). In addition, the secondary metabolite pathways use cofactors such as ATP and NADPH derived from energy metabolism. Besides these dominant classes, there are ribosomal peptides (RiPPs) and terpenoids [44,45,46].
To illustrate, seven linear chain NRPs were reported in some species of the genera Cordyceps, Paecilomyces, Metarhizium and Hirsutella. Cicadapeptins obtained from Cordyceps heteropoda, leucinostatins can be found in Purpureocillium lilacinum (Paecilomyces lilacinus), Metarhizium marquandii (Paecilomyces marquandii) and Acremonium sp. Efrapeptins, which can be found in Acremonium sp. and Metarhizium anisopliae, peptaibols, culicinins, metanicins and LP237 analogues [47].
In fungi, polyketides (PKs) comprise CH2 (C=O) units and are the most abundant secondary metabolites [44,48]. Different routes synthesize fungal polyketides, each involving a specific class of polyketide synthase (PKS). Type I iterative PKSs are multidomain enzymes responsible for producing most fungal polyketide compounds [49]. A second route, dependent on type III iterative PKSs, are enzymes comprising a single keto-synthase (KS) domain [50]. While type I iterative PKSs have been well characterized and found in fungal genomes, some type III fungal iterative PKSs have already been characterized [51,52,53,54,55,56].
Fungi are a prolific source of polyketides (PKs), which exhibit remarkable structural diversity and biological activity. Representative examples include the cholesterol-lowering drug lovastatin, the antifungal drug griseofulvin, the immunosuppressive drug mycophenolic acid, and the phytopathogenic fungal virulence factor T toxin [57].
Genetic engineering tools with bioinformatics and molecular biology are used to change the metabolic pathways of microorganisms. This may involve the introduction or overexpression of genes related to the metabolite biosynthesis pathway, removal of competing pathways, or increasing the supply of precursors. These modifications aim to increase the yield and productivity of the desired metabolite [58].
4. Tools for the Genetic Transformation of Filamentous fungi
For a genetic transformation tool of a microorganism to achieve industrial viability, the expression system necessitates fundamental components: (a) A host strain with a history of safe utilization (Generally Regarded as Safe—GRAS), characterized by robust growth and proficient protein production in cost-effective media and at industrial scales. The implementation of a selection system ensures the successful integration of the gene into the host. Various nutritional marker genes (e.g., pyrG, amdS, and niaD) and antibiotic resistance markers (e.g., hygB) can serve this purpose. In the industrial production of food enzymes, avoidance of antibiotic resistance markers is advisable due to regulatory requirements for the final strain. (b) Utilization of a transformation tool to introduce one or more copies of the expression cassette containing the gene of interest into the strain’s genome, preferably at specific loci. (c) Incorporation of a robust promoter facilitating gene expression under fermentation conditions. Table 2 presents a comprehensive overview of the principles, advantages, and disadvantages of the most prevalent and established tools employed for the genetic transformation of filamentous fungi [59].
Post-genome sequencing, gene editing technologies have emerged as efficient tools for elucidating gene function. Among nucleases-based gene manipulation methods, the CRISPR/Cas9 system stands out as the most prominent. Originating from the adaptive immune system of bacteria, the CRISPR/Cas9 system serves as a sophisticated genomic engineering tool, evolved as a defense mechanism against viral and plasmid invasion [55,56]. Comprising two principal components, the Cas9 nuclease and a guide RNA (gRNA) molecule, this gene editing system directs the nuclease to a specific genomic location, termed the target site. The chimeric guide RNA (sgRNA) is formed through the fusion of a CRISPR RNA (crRNA) with a trans-activating RNA (tracrRNA), processed by endogenous bacterial machinery to generate the functional gRNA [60,61].
The guide RNA (gRNA) directs the Cas9 endonuclease to a precise genomic locus, facilitating the formation of Watson-Crick base pairs with the target DNA sequence. This interaction allows Cas9 to induce double-stranded DNA breaks at specific genomic sites. Binding of the Cas9 nuclease to the crRNA/tracrRNA complex induces a structural alteration in the protein, activating the Protospacer Adjacent Motif (PAM) recognition site. PAMs, consisting of 2 to 5 nucleotides (5’NGG3’ and 5’NNGRRT3’), are essential for anchoring the Cas9 nuclease to the cleavage site [61,62].
Following the induction of DNA double-strand breaks (DSBs), two distinct repair mechanisms ensue: non-homologous end joining (NHEJ) and homology-directed repair (HDR). NHEJ represents an error-prone and predominant pathway for DSB repair, wherein the broken ends are directly ligated without reliance on a homologous template. Consequently, this pathway may lead to targeted mutations, such as random deletions, insertions, base substitutions, targeted chromosomal rearrangements, or mutations at DNA breakpoints, resulting in premature stop codons within the Open Reading Frame (ORF) of the target gene. In contrast to the NHEJ pathway, which predominates as the primary DSB repair mechanism in microorganisms, HDR is a less efficient but notably reliable pathway [63,64,65]. Figure 5 illustrates the anchoring of the Cas9 nuclease to the cleavage site and subsequent DNA double-strand breakage at the Protospacer Adjacent Motif (PAM) sequence, followed by depiction of the two principal DNA repair pathways (gene editing methods).
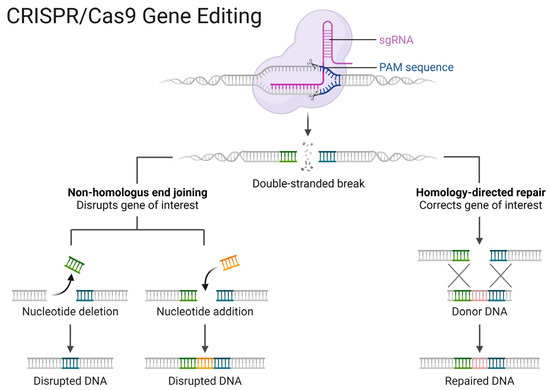
Figure 5.
Basic steps in gene editing using CRISPR/Cas9. [credit: Esmée Dragt (creator) and Louis Ngai (BioRender.com (2020). Retrieved from https://app.biorender.com/biorender-templates/figures/all/t-5f873df466346900a43c6db1-crisprcas9-gene-editing, accessed on 16 January 2024)].

Table 2.
Usual strategies for genetic transformation of filamentous fungi.
Table 2.
Usual strategies for genetic transformation of filamentous fungi.
| Method | Principle | Advantages | Disadvantages | Reference |
|---|---|---|---|---|
| TMP | Use of cell wall degrading enzymes to get protoplasts. DNA transfer occurs by adding PEG and CaCl2. | Spores, spores in the process of germination, and hyphae are usable. | The transformation rate depends on the efficiency of the lytic enzyme used to get the protoplasts. Requires cell regeneration procedure. High number of DNA copies is inserted | [66,67] |
| TMA | A. tumefaciens carries two vectors (the binary vector with the DNA of interest between the left and right repeats and the T vector containing the virulence region important for DNA transfer). During the co-cultivation of A. tumefaciens with the fungus, they achieve DNA transfer. | Spores, spores in the process of germination, and hyphae are usable. Low copy number of inserted DNA improves targeted integration. | Several parameters during the co-cultivation of A. tumefaciens and the filamentous fungus to be transformed affect the transformation rate. Time-consuming technique | [68] |
| EP | DNA transfer is mediated by reversible permeabilization of the membrane induced by local application of electrical pulses. | Spores, spores in the process of germination, and hyphae are usable. A simple and inexpensive technique | The formation of protoplasts is often necessary to perform the technique | [69] |
| TB | The DNA is coated with tungsten or gold and inserted into the cell to be transformed using a microparticle accelerator. | No pre-treatment of recipient cells | Special equipment is necessary. | [70] |
| CRISPR (RNPs) | Cas9 and guide RNAs can be delivered in form of DNA, RNA/mRNA, or ribonucleoprotein (RNP). The delivery methods are usually divided into physical (electroporation and microinjection), viral (lentiviral, adenoviral, and AAV vectors), and non-viral (plasmids, lipid and polymeric nanoparticles, and extracellular vesicles) ones. | The delivery of CRISPR ribonucleoproteins (RNPs) for genome editing in vitro and in vivo has important advantages over other delivery methods, including reduced off-target and immunogenic effects | Effective delivery of RNPs remains challenging in certain cell types due to low efficiency and cell toxicity | [71,72,73] |
Adapted from [59], (TMP) Transformation mediated by protoplasts, (TMA) Transformation mediated by Agrobacterium, (EP) Electroporation, (TB) Transformation by Biobalistic, (PEG) Polyethylene glycol, (RNPs) Ribonucleoprotein particles.
In recent years, researchers have introduced the CRISPR/Cas9 system into filamentous fungi to leverage its potential for modulating the production of secondary metabolites. For species such as Aspergillus oryzae, Trichoderma reesei, Aspergillus niger, and Aspergillus nidulans, CRISPR/Cas9-based systems have emerged as versatile platforms for precise genome editing, leading to significant advancements in the production of valuable secondary metabolites [74]. A pioneering study demonstrated the modification of Trichoderma harzianum using the CRISPR/Cas9 marker-free system, targeting the albA (pks4) and ku70 genes. The study successfully developed two strains of T. harzianum using a recyclable CRISPR/Cas9 marker-free system based on the AMA1 plasmid vector. This achievement marks the first successful application of this recyclable system for constructing fungal strains with agricultural applications [33].
The first reports on CRISPR/Cas9-mediated genome editing in filamentous fungi were published in 2015 [75,76,77,78]. Subsequently, researchers have rapidly adopted this technology, establishing genome manipulation capabilities in over 60 fungal species, including those where such manipulation was previously unattainable [74]. This swift progress underscores the adaptability and robustness of the CRISPR/Cas9 system, laying a solid foundation for the development of genetic engineering systems in numerous other species in the future.
5. Protein Secretion Pathways in Filamentous fungi
The conventional secretory pathway, also known as the endoplasmic reticulum (ER) secretory pathway, represents the classic protein secretion pathway in filamentous fungi. This pathway is like the protein secretion pathway in other eukaryotes, such as animals and plants. In this pathway, ER-associated ribosomes synthesize proteins, which are then translocated into the ER lumen. Afterward, cells transport them to the Golgi complex, where they undergo processing, modification, and selection for transport to other organelles or outside the cell. The secretory transport vesicle (STV) transports proteins from the Golgi complex to the plasma membrane, where they will undergo the process of exocytosis and released into the extracellular environment [79].
However, besides the conventional secretory pathway, filamentous fungi also have alternative secretion pathways that play important roles in specific conditions. These include the septal secretion pathway [80,81], the Golgi-independent pathway, the multivesicular body (MVB) secretion pathway [82] and autophagy or phagocytosis [83].
The classical protein secretion pathway in filamentous fungi involves three main steps, including: (1) Polypeptide transfer from the ribosome to the endoplasmic reticulum (ER), (2) Protein folding and modification in the ER, (3) Transport of protein vesicles to the Golgi apparatus and extracellular environment [4]. Figure 6 shows the protein secretion pathways of a hypothetical filamentous fungus in a representative way.
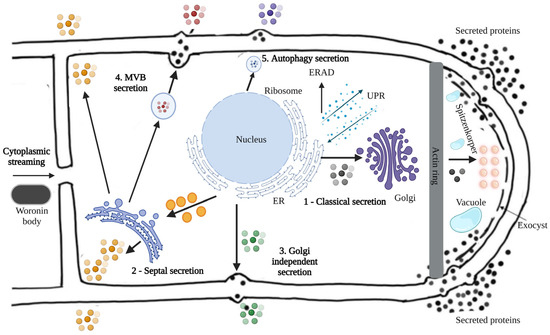
Figure 6.
Protein secretion pathways in filamentous fungi. The secretion pathways of filamentous fungi exemplify the following modes: (1) classical secretion through the ER, (2) septal secretion, (3) Golgi-independent secretion, (4) multivesicular body (MVB) secretion, and (5) autophagic secretion, adapted from [84]. The black balls represent proteins secreted by the classical secretion pathway through the ER; The orange balls represent the proteins secreted by the septal secretion; The green ones represent the proteins secreted by the Golgi-independent secretion; The red ones represent the proteins secreted by multivesicular body (MVB) secretion and the blue ones represent the proteins secreted by autophagic secretion.
In the first stage of the conventional protein secretion pathway, the co- or post-translational transport pathway handles polypeptide transfer from the ribosome to the Endoplasmic Reticulum (ER). In the co-translational transport pathway, the signal peptide recognition particle (SRP) initially binds to the signal peptide sequence, which serves to block translation [85]. The SRP then directs the nascent ribosome-mRNA-peptide complex to reach the ER membrane and binds to the SRP receptor. Subsequently, the SRP is released from the complex, translation recovered, and the nascent polypeptide enters the ER lumen via the Sec61p transport complex [79].
In the post-translational transport pathway, the nascent polypeptide undergoes translation in the cytosol and remains unfolded through interaction with Hsp70 chaperones and co-chaperones. This complex can reach the ER through interaction with the membrane receptor Sec62p-Sec72p-Sec73p [79]. The immunoglobulin protein (BiP) binds to the luminal chaperone of the endoplasmic reticulum and the membrane protein Sec63p helps this complex to enter the ER [86].
The second stage is the folding and modification of proteins in the ER, which requires the help of a series of folding enzymes, including calnexin (ClxA), immunoglobulin (BiP) and protein disulfide isomerase (PDI) [87]. Nascent peptides with correct folding undergo modifications such as glycosylation, which is one of the most common and important post-translational modifications and can significantly affect the stability, localization, and secretion of proteins [88]. After properly folding and undergoing glycosylation, cells transport secreted proteins extracellularly. The unfolded protein response (UPR) and ER-associated protein degradation (ERAD) deal with misfolded nascent peptides [89,90]. The UPR detects misfolded proteins in the ER and induces the biosynthesis of chaperones and folding enzymes, while ERAD degrades misfolded proteins.
The third stage is to transport the coiled protein vesicles to the Golgi apparatus by fusion with the target membrane and secrete them in the extracellular medium [91]. Apical vesicle clusters in Spitzenkörper in filamentous fungi transmit Golgi-derived secretory vesicles to the apical plasma membrane [85]. Vesicle formation, transport and fusion are mediated by several proteins, including GTP-binding proteins (e.g. Sar, ARF) for vesicle binding and RabGTPases for fusion with the Golgi complex [92]. The specific fusion of vesicles with the target membrane is the critical process, which is mediated by soluble N-ethylmaleimide binding receptors (SNARE). Based on localization, researchers have divided SNARE into two categories: vesicle SNARE (v-SNARE) and target membrane SNARE (t-SNARE) [93]. In filamentous fungi, the v-SNARE protein SNC1 and the t-SNARE proteins SSO1 and SSO2 are involved in vesicle fusion [94].
Increasing protein secretion in filamentous fungi by increasing intracellular protein production by optimizing the transcription and codon of the target protein is an effective strategy, as shown in some reviews [95,96,97,98]. However, researchers could use other genetic engineering strategies, such as replacing the original signal peptide with a more efficient one by regulating the UPR and ERAD, optimizing the intracellular transport process, constructing a protease-deficient strain, regulating the morphology of the mycelium, and optimizing the sterol regulatory element binding protein (SREBP). Table 3 shows some examples of improved protein secretion in some filamentous fungi.

Table 3.
Examples of genetic engineering of filamentous fungi to improve protein secretion.
6. Conclusions
Due to their robust protein secretion pathways, filamentous fungi serve as compelling candidates as cell factories for protein secretion. This review highlights select examples of filamentous fungi utilized for the production of industrially relevant enzymes; alongside genetic engineering applications aimed at enhancing enzyme yields. Leveraging various gene editing technologies such as the CRISPR-Cas system, genome engineering strategies enable the introduction of deletions, insertions, and/or point mutations throughout the genome, utilizing traceable techniques to expedite isolate modification.
As prospects, harnessing the substantial potential of genomic engineering holds promise for further augmenting protein secretion in filamentous fungi, particularly those exhibiting low efficiency in gene transformation and homologous recombination. Consequently, researchers stand to advance protein secretion in filamentous fungi, including those with limited genetic manipulation capabilities, thereby facilitating the development of fourth-generation biological products derived from microorganisms subjected to genetic engineering techniques.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/applmicrobiol4020055/s1, Table S1: Biological assets registered and used to control insects and diseases and their respective dosage per hectare according to the biological target.
Author Contributions
Conceptualization, M.C.C.d.F., L.C. and L.R.O.; formal analysis, M.V.d.C.B.C. and M.C.C.d.F.; investigation, L.R.O.; data curation, A.R.G.; writing—original draft preparation, L.R.O.; writing—review and editing, M.V.d.C.B.C., E.D.Q. and M.C.C.d.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
Author Leandro Colognese was employed by the company Bio-Input Company SoluBio Agricultural Technologies. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Kumar, A.; Chadha, S.; Rath, D. Chapter 17—CRISPR-Cas9 system for functional genomics of filamentous fungi: Applications and challenges. In Fungi Bio-Prospects in Sustainable Agriculture, Environment and Nano-Technology; Sharma, V.K., Shah, M.P., Parmar, S., Kumar, A., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 541–576. [Google Scholar] [CrossRef]
- Nevalainen, H.; Peterson, R. Making recombinant proteins in filamentous fungi-are we expecting too much? Front. Microbiol. 2014, 5, 75. [Google Scholar] [CrossRef] [PubMed]
- Demain, A.L. Small bugs, big business: The economic power of the microbe. Biotechnol. Adv. 2000, 18, 499–514. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhong, C.; Xiao, H. Genetic Engineering of Filamentous Fungi for Efficient Protein Expression and Secretion. Front. Bioeng. Biotechnol. 2020, 8, 293. [Google Scholar] [CrossRef] [PubMed]
- Brasil. Instrução Normativa Conjunta SDA/SDC nº 2, de 12 de Julho de 2013. Estabelece as Especificações de Referência de Produtos Fitossanitários com uso Aprovado para a Agricultura Orgânica; União, D.O.D., Ed.; 2013; pp. 6–8. Available online: https://www.gov.br/agricultura/pt-br/assuntos/insumos-agropecuarios/insumos-agricolas/agrotoxicos/produtos-fitossanitarios/arquivos-especificacao-de-referencia/in-conjunta-sda-sdc-no-2-de-12-de-julho-de-2013.pdf/view (accessed on 16 January 2024).
- Quintela, E.D. Lalguard Java: Produto Biológico a Base Do Fungo Cordyceps Javanica Para o Controle de Pragas—Parceria Embrapa e Lallemand Plant Care; Embrapa Rice and Beans: Santo Antônio de Goiás, Brasil, 2022. Available online: https://www.embrapa.br/busca-de-solucoes-tecnologicas/-/produto-servico/11043/bioinseticida-a-base-de-cordyceps-javanica-para-controle-biologico-de-mosca-branca (accessed on 1 May 2024).
- Corrêa, B.; da Silveira Duarte, V.; Silva, D.M.; Mascarin, G.M.; Júnior, I.D. Comparative analysis of blastospore production and virulence of Beauveria bassiana and Cordyceps fumosorosea against soybean pests. BioControl 2020, 65, 323–337. [Google Scholar] [CrossRef]
- Woo, S.L.; Hermosa, R.; Lorito, M.; Monte, E. Trichoderma: A multipurpose, plant-beneficial microorganism for eco-sustainable agriculture. Nat. Rev. Microbiol. 2023, 21, 312–326. [Google Scholar] [CrossRef] [PubMed]
- MAPA. Consulta de Produtos Formulados. 2024. Available online: https://agrofit.agricultura.gov.br/agrofit_cons/principal_agrofit_cons (accessed on 3 February 2024).
- Joop, G.; Vilcinskas, A. Coevolution of parasitic fungi and insect hosts. Zoology 2016, 119, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Molnár, I.; Gibson, D.M.; Krasnoff, S.B. Secondary metabolites from entomopathogenic Hypocrealean fungi. Nat. Prod. Rep. 2010, 27, 1241–1275. [Google Scholar] [CrossRef]
- Rohlfs, M.; Churchill, A.C. Fungal secondary metabolites as modulators of interactions with insects and other arthropods. Fungal Genet. Biol. 2011, 48, 23–34. [Google Scholar] [CrossRef]
- Wang, H.; Peng, H.; Li, W.; Cheng, P.; Gong, M. The Toxins of Beauveria bassiana and the Strategies to Improve Their Virulence to Insects. Front. Microbiol. 2021, 12, 705343. [Google Scholar] [CrossRef]
- Rukachaisirikul, V.; Chantaruk, S.; Tansakul, C.; Saithong, S.; Chaicharernwimonkoon, L.; Pakawatchai, C.; Isaka, M.; Intereya, K. A cyclopeptide from the Insect pathogenic fungus Cordyceps sp. BCC 1788. J. Nat. Prod. 2006, 69, 305–307. [Google Scholar] [CrossRef]
- Kumar, S.; Dahiya, R.; Khokra, S.L.; Mourya, R.; Chennupati, S.V.; Maharaj, S. Total Synthesis and Pharmacological Investigation of Cordyheptapeptide A. Molecules 2017, 22, 682. [Google Scholar] [CrossRef]
- Meyer, V. Genetic engineering of filamentous fungi—Progress, obstacles and future trends. Biotechnol. Adv. 2008, 26, 177–185. [Google Scholar] [CrossRef]
- Vanegas, K.G.; Rendsvig, J.K.H.; Jarczynska, Z.D.; Cortes, M.V.D.C.B.; van Esch, A.P.; Morera-Gómez, M.; Contesini, F.J.; Mortensen, U.H. A Mad7 System for Genetic Engineering of Filamentous Fungi. J. Fungi 2023, 9, 16. [Google Scholar] [CrossRef]
- Unkles, S.E.; Valiante, V.; Mattern, D.J.; Brakhage, A.A. Synthetic biology tools for bioprospecting of natural products in eukaryotes. Chem. Biol. 2014, 21, 502–508. [Google Scholar] [CrossRef]
- Schuetze, T.; Meyer, V. Polycistronic gene expression in Aspergillus niger. Microb. Cell Factories 2017, 16, 162. [Google Scholar] [CrossRef]
- Geib, E.; Brock, M. ATNT: An enhanced system for expression of polycistronic secondary metabolite gene clusters in Aspergillus niger. Fungal Biol. Biotechnol. 2017, 4, 13. [Google Scholar] [CrossRef]
- Mattern, D.J.; Valiante, V.; Horn, F.; Petzke, L.; Brakhage, A.A. Rewiring of the Austinoid Biosynthetic Pathway in Filamentous Fungi. ACS Chem. Biol. 2017, 12, 2927–2933. [Google Scholar] [CrossRef]
- MAPA. Bio-Inputs Enhance Sustainability of Agricultural Production in Brazil. 2024. Available online: https://www.gov.br/agricultura/pt-br/assuntos/inovacao/bioinsumos/material-para-imprensa/en/bio-inputs-enhance-sustainability-of-agricultural-production-in-brazil.pdf (accessed on 18 February 2024).
- Ziolkowska, J.R. Chapter 1—Biofuels Technologies. In Biofuels for a More Sustainable Future; Ren, J., Scipioni, A., Manzardo, A., Liang, H., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 1–19. [Google Scholar] [CrossRef]
- Brasil. Projeto de Lei nº 3668, 2021. Dispõe Sobre a Produção, o Registro, Comercialização, uso, Destino dos Resíduos e Embalagens, o Registro, Inspeção e Fiscalização, a Pesquisa e Experimentação, e os Incentivos à Produção de Bioinsumos para Agricultura e dá Outras Providências. Deputados, C.D., Ed.; Brasília, 2021. Available online: https://www25.senado.leg.br/web/atividade/materias/-/materia/150351 (accessed on 16 January 2024).
- Brasil. Projeto de Lei nº 658: Dispõe Sobre a Classificação, Tratamento e Produção de Bioinsumos por meio do Manejo Biológico on Farm; Ratifica o Programa Nacional de Bioinsumos e dá Outras Providências. Brasilia, 2021. Available online: https://www.camara.leg.br/propostas-legislativas/2271161 (accessed on 16 January 2024).
- Brasil. Portaria Conjunta SDA/MAPA—IBAMA—ANVISA n.º 1. Estabelece Procedimentos a Serem Adotados para o Registro de Produtos Microbiológicos Empregados no Controle de Pragas ou como Desfolhantes, Dessecantes, Estimuladores, Inibidores de Crescimento, Além de Revogar os atos Normativos Vigentes, Pertinentes à esta Matéria: Instrução Normativa Conjunta Ministério da Agricultura Pecuária e Abastecimento/Anvisa/Ibama nº 03 de 10 de Março de 2006 e o Ato CGAA/DSV/SDA nº 06. Brasil, D.O.d.R.F.d., Ed.; Brasília-DF, 2023; Vol. n.º 84. Available online: http://pesquisa.in.gov.br/imprensa/jsp/visualiza/index.jsp?data=04/05/2023&jornal=515&pagina=7 (accessed on 17 February 2024).
- Ajanovic, A. Biofuels versus food production: Does biofuels production increase food prices? Energy 2021, 36, 2070–2076. [Google Scholar] [CrossRef]
- Harvey, M.; Pilgrim, S. The new competition for land: Food, energy, and climate change. Food Policy 2011, 36, S40–S51. [Google Scholar] [CrossRef]
- Méndez-González, F.; Loera-Corral, O.; Saucedo-Castañeda, G.; Favela-Torres, E. Chapter 7—Bioreactors for the Production of Biological Control Agents Produced by Solid-State Fermentation. In Current Developments in Biotechnology and Bioengineering; Pandey, A., Larroche, C., Soccol, C.R., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 109–121. [Google Scholar] [CrossRef]
- Faria, M.R.D.; Wraight, S.P. Mycoinsecticides and Mycoacaricides: A comprehensive list with worldwide coverage and international classification of formulation types. Biol. Control 2007, 43, 237–256. [Google Scholar] [CrossRef]
- Li, Z.; Alves, S.B.; Roberts, D.W.; Fan, M.; Delalibera, I., Jr.; Tang, J.; Lopes, R.B.; Faria, M.; Rangel, D.E.N. Biological control of insects in Brazil and China: History, current programs and reasons for their successes using entomopathogenic fungi. Biocontrol Sci. Technol. 2010, 20, 117–136. [Google Scholar] [CrossRef]
- Kirst, H.A. The spinosyn family of insecticides: Realizing the potential of natural products research. J. Antibiot. 2010, 63, 101–111. [Google Scholar] [CrossRef]
- Cortes, M.V.D.C.B.; Barbosa, E.T.; Oliveira, M.I.D.S.; Maciel, L.H.R.; Lobo Junior, M.; Contesini, F.J.; Filippi, M.C.C.D.; Silva-Lobo, V.L.D. Trichoderma harzianum marker-free strain construction based on efficient CRISPR/Cas9 recyclable system: A helpful tool for the study of biological control agents. Biol. Control 2023, 184, 105281. [Google Scholar] [CrossRef]
- Schueffler, A.; Anke, T. Fungal natural products in research and development. Nat. Prod. Rep. 2014, 31, 1425–1448. [Google Scholar] [CrossRef]
- Bérdy, J. Bioactive Microbial Metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef]
- Liaud, N.; Giniés, C.; Navarro, D.; Fabre, N.; Crapart, S.; Gimbert, I.H.; Levasseur, A.; Raouche, S.; Sigoillot, J.-C. Exploring fungal biodiversity: Organic acid production by 66 strains of filamentous fungi. Fungal Biol. Biotechnol. 2014, 1, 1. [Google Scholar] [CrossRef]
- Wösten, H.A.B. Filamentous fungi for the production of enzymes, chemicals and materials. Curr. Opin. Biotechnol. 2019, 59, 65–70. [Google Scholar] [CrossRef]
- Awad, H.M.; EL-Shahed, K.Y.I.; Aziz, R.; Sarmidi, M.R.; El-Enshasy, H.A. Antibiotics as Microbial Secondary Metabolites: Production and Application. J. Teknol. 2012, 59, 101–111. [Google Scholar] [CrossRef]
- Trail, F.; Mahanti, N.; Rarick, M.; Mehigh, R.; Liang, S.H.; Zhou, R.; Linz, J.E. Physical and transcriptional map of an aflatoxin gene cluster in Aspergillus parasiticus and functional disruption of a gene involved early in the aflatoxin pathway. Appl. Environ. Microbiol. 1995, 61, 2665–2673. [Google Scholar] [CrossRef]
- Lysøe, E.; Seong, K.Y.; Kistler, H.C. The transcriptome of Fusarium graminearum during the infection of wheat. Mol. Plant Microbe Interact. 2011, 24, 995–1000. [Google Scholar] [CrossRef]
- Lind, A.L.; Lim, F.Y.; Soukup, A.A.; Keller, N.P.; Rokas, A. An LaeA- and BrlA-Dependent Cellular Network Governs Tissue-Specific Secondary Metabolism in the Human Pathogen Aspergillus fumigatus. mSphere 2018, 3, 10–1128. [Google Scholar] [CrossRef]
- Spraker, J.E.; Wiemann, P.; Baccile, J.A.; Venkatesh, N.; Schumacher, J.; Schroeder, F.C.; Sanchez, L.M.; Keller, N.P. Conserved Responses in a War of Small Molecules between a Plant-Pathogenic Bacterium and Fungi. mBio 2018, 9, 1–14. [Google Scholar] [CrossRef]
- Tkacz, J.S.; Lange, L. Advances in Fungal Biotechnology for Industry, Agriculture, and Medicine; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2004. [Google Scholar]
- Keller, N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef]
- Nielsen, J.C.; Prigent, S.; Grijseels, S.; Workman, M.; Ji, B.; Nielsen, J. Comparative Transcriptome Analysis Shows Conserved Metabolic Regulation during Production of Secondary Metabolites in Filamentous Fungi. mSystems 2019, 4, 1–14. [Google Scholar] [CrossRef]
- Mosunova, O.V.; Navarro-Muñoz, J.C.; Collemare, J. The Biosynthesis of Fungal Secondary Metabolites: From Fundamentals to Biotechnological Applications. In Encyclopedia of Mycology; Elsevier: Amsterdam, The Netherlands, 2021. [Google Scholar]
- Niu, X.; Thaochan, N.; Hu, Q. Diversity of Linear Non-Ribosomal Peptide in Biocontrol Fungi. J. Fungi 2020, 6, 61. [Google Scholar] [CrossRef]
- Collemare, J.; Billard, A.; Böhnert, H.U.; Lebrun, M.-H. Biosynthesis of secondary metabolites in the rice blast fungus Magnaporthe grisea: The role of hybrid PKS-NRPS in pathogenicity. Mycol. Res. 2008, 112, 207–215. [Google Scholar] [CrossRef]
- Herbst, D.A.; Townsend, C.A.; Maier, T. The architectures of iterative type I PKS and FAS. Nat. Prod. Rep. 2018, 35, 1046–1069. [Google Scholar] [CrossRef]
- Yu, D.; Xu, F.; Zeng, J.; Zhan, J. Type III polyketide synthases in natural product biosynthesis. IUBMB Life 2012, 64, 285–295. [Google Scholar] [CrossRef]
- Hashimoto, M.; Seshime, Y.; Kitamoto, K.; Uchiyama, N.; Goda, Y.; Fujii, I. Identification of csypyrone B2 and B3 as the minor products of Aspergillus oryzae type III polyketide synthase CsyB. Bioorg. Med. Chem. Lett. 2013, 23, 650–653. [Google Scholar] [CrossRef]
- Sun, L.; Wang, S.; Zhang, S.; Yu, D.; Qin, Y.; Huang, H.; Wang, W.; Zhan, J. Identification of a type III polyketide synthase involved in the biosynthesis of spirolaxine. Appl. Microbiol. Biotechnol. 2016, 100, 7103–7113. [Google Scholar] [CrossRef]
- Ramakrishnan, D.; Tiwari, M.K.; Manoharan, G.; Sairam, T.; Thangamani, R.; Lee, J.K.; Marimuthu, J. Molecular characterization of two alkylresorcylic acid synthases from Sordariomycetes fungi. Enzym. Microb. Technol. 2018, 115, 16–22. [Google Scholar] [CrossRef]
- Yan, H.; Sun, L.; Huang, J.; Qiu, Y.; Xu, F.; Yan, R.; Zhu, D.; Wang, W.; Zhan, J. Identification and heterologous reconstitution of a 5-alk(en)ylresorcinol synthase from endophytic fungus Shiraia sp. Slf14. J. Microbiol. 2018, 56, 805–812. [Google Scholar] [CrossRef]
- Kaneko, A.; Morishita, Y.; Tsukada, K.; Taniguchi, T.; Asai, T. Post-genomic approach based discovery of alkylresorcinols from a cricket-associated fungus, Penicillium soppi. Org. Biomol. Chem. 2019, 17, 5239–5243. [Google Scholar] [CrossRef]
- Manoharan, G.; Sairam, T.; Thangamani, R.; Ramakrishnan, D.; Tiwari, M.K.; Lee, J.K.; Marimuthu, J. Identification and characterization of type III polyketide synthase genes from culturable endophytes of ethnomedicinal plants. Enzym. Microb. Technol. 2019, 131, 109396. [Google Scholar] [CrossRef]
- Minami, A.; Ugai, T.; Ozaki, T.; Oikawa, H. Predicting the chemical space of fungal polyketides by phylogeny-based bioinformatics analysis of polyketide synthase-nonribosomal peptide synthetase and its modification enzymes. Sci. Rep. 2020, 10, 13556. [Google Scholar] [CrossRef]
- Rusu, A.V.; Trif, M.; Rocha, J.M. Microbial Secondary Metabolites via Fermentation Approaches for Dietary Supplementation Formulations. Molecules 2023, 28, 6020. [Google Scholar] [CrossRef]
- Wang, S.; Chen, H.; Tang, X.; Zhang, H.; Chen, W.; Chen, Y.Q. Molecular tools for gene manipulation in filamentous fungi. Appl. Microbiol. Biotechnol. 2017, 101, 8063–8075. [Google Scholar] [CrossRef]
- Wang, Q.; Coleman, J.J. Progress and Challenges: Development and Implementation of CRISPR/Cas9 Technology in Filamentous Fungi. Comput. Struct. Biotechnol. J. 2019, 17, 761–769. [Google Scholar] [CrossRef]
- Mojica, F.J.M.; Díez-Villaseñor, C.; García-Martínez, J.; Almendros, C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 2009, 155, 733–740. [Google Scholar] [CrossRef]
- Sternberg, S.H.; Redding, S.; Jinek, M.; Greene, E.C.; Doudna, J.A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 2014, 507, 62–67. [Google Scholar] [CrossRef]
- Kim, H.; Kim, J.-S. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014, 15, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Boettcher, M.; McManus, M.T. Choosing the Right Tool for the Job: RNAi, TALEN, or CRISPR. Mol. Cell 2015, 58, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.Z.; Zhu, Y.L.; Huang, P.W.; Yang, Q.; Dai, C.C. Strategies for gene disruption and expression in filamentous fungi. Appl. Microbiol. Biotechnol. 2019, 103, 6041–6059. [Google Scholar] [CrossRef] [PubMed]
- Case, M.E.; Schweizer, M.; Kushner, S.R.; Giles, N.H. Efficient transformation of Neurospora crassa by utilizing hybrid plasmid DNA. Proc. Natl. Acad. Sci. USA 1979, 76, 5259–5263. [Google Scholar] [CrossRef] [PubMed]
- Turgeon, B.G.; Condon, B.; Liu, J.; Zhang, N. Protoplast transformation of filamentous fungi. Methods Mol. Biol. 2010, 638, 3–19. [Google Scholar] [CrossRef] [PubMed]
- de Groot, M.J.A.; Bundock, P.; Hooykaas, P.J.J.; Beijersbergen, A.G.M. Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol. 1998, 16, 839–842. [Google Scholar] [CrossRef] [PubMed]
- Richey, M.G.; Marek, E.T.; Schardl, C.L.; Smith, D.A. Transformation of filamentous fungi with plasmid DNA by electroporation. Phytopathology 1989, 79, 844–847. [Google Scholar] [CrossRef]
- Bhairi, S.M.; Staples, R.C. Transient expression of the β-glucuronidase gene introduced into Uromyces appendiculatus uredospores by particle bombardment. Phytopathology 1992, 82, 986–989. [Google Scholar] [CrossRef]
- Chen, K.; Stahl, E.C.; Kang, M.H.; Xu, B.; Allen, R.; Trinidad, M.; Doudna, J.A. Engineering self-deliverable ribonucleoproteins for genome editing in the brain. Nat. Commun. 2024, 15, 1727. [Google Scholar] [CrossRef]
- Zhang, Y.; Iaffaldano, B.; Qi, Y. CRISPR ribonucleoprotein-mediated genetic engineering in plants. Plant Commun. 2021, 2, 100168. [Google Scholar] [CrossRef]
- Horodecka, K.; Düchler, M. CRISPR/Cas9: Principle, Applications, and Delivery through Extracellular Vesicles. Int. J. Mol. Sci. 2021, 22, 6072. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Lv, G.; Tu, Y.; Cheng, X.; Duan, Y.; Zeng, B.; He, B. Applications of CRISPR/Cas9 in the Synthesis of Secondary Metabolites in Filamentous Fungi. Front. Microbiol. 2021, 12, 638096. [Google Scholar] [CrossRef] [PubMed]
- Fuller, K.K.; Chen, S.; Loros, J.J.; Dunlap, J.C. Development of the CRISPR/Cas9 System for Targeted Gene Disruption in Aspergillus fumigatus. Eukaryot. Cell 2015, 14, 1073–1080. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Chen, L.; Jiang, Y.; Zhou, Z.; Zou, G. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov. 2015, 1, 15007. [Google Scholar] [CrossRef] [PubMed]
- Matsu-Ura, T.; Baek, M.; Kwon, J.; Hong, C. Efficient gene editing in Neurospora crassa with CRISPR technology. Fungal Biol. Biotechnol. 2015, 2, 4. [Google Scholar] [CrossRef] [PubMed]
- Nødvig, C.S.; Nielsen, J.B.; Kogle, M.E.; Mortensen, U.H. A CRISPR-Cas9 System for Genetic Engineering of Filamentous Fungi. PLoS ONE 2015, 10, e0133085. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Punt, P.J.; van Luijk, N.; van den Hondel, C.A. The secretion pathway in filamentous fungi: A biotechnological view. Fungal Genet. Biol. 2001, 33, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, Y.; Ishikawa, E.; Shoji, J.Y.; Nakano, H.; Kitamoto, K. Septum-directed secretion in the filamentous fungus Aspergillus oryzae. Mol. Microbiol. 2011, 81, 40–55. [Google Scholar] [CrossRef]
- Fiedler, M.R.M.; Cairns, T.C.; Koch, O.; Kubisch, C.; Meyer, V. Conditional Expression of the Small GTPase ArfA Impacts Secretion, Morphology, Growth, and Actin Ring Position in Aspergillus niger. Front. Microbiol. 2018, 9, 878. [Google Scholar] [CrossRef]
- Shoji, J.Y.; Kikuma, T.; Kitamoto, K. Vesicle trafficking, organelle functions, and unconventional secretion in fungal physiology and pathogenicity. Curr. Opin. Microbiol. 2014, 20, 1–9. [Google Scholar] [CrossRef]
- Miura, N.; Ueda, M. Evaluation of Unconventional Protein Secretion by Saccharomyces cerevisiae and other Fungi. Cells 2018, 7, 128. [Google Scholar] [CrossRef] [PubMed]
- Sakekar, A.A.; Gaikwad, S.R.; Punekar, N.S. Protein expression and secretion by filamentous fungi. J. Biosci. 2021, 46, 5. [Google Scholar] [CrossRef]
- Virag, A.; Harris, S.D. The Spitzenkörper: A molecular perspective. Mycol. Res. 2006, 110, 4–13. [Google Scholar] [CrossRef]
- Haßdenteufel, S.; Johnson, N.; Paton, A.W.; Paton, J.C.; High, S.; Zimmermann, R. Chaperone-Mediated Sec61 Channel Gating during ER Import of Small Precursor Proteins Overcomes Sec61 Inhibitor-Reinforced Energy Barrier. Cell Rep. 2018, 23, 1373–1386. [Google Scholar] [CrossRef]
- Saloheimo, M.; Pakula, T.M. The cargo and the transport system: Secreted proteins and protein secretion in Trichoderma reesei (Hypocrea jecorina). Microbiology 2012, 158, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Mitra, N.; Sinha, S.; Ramya, T.N.; Surolia, A. N-linked oligosaccharides as outfitters for glycoprotein folding, form and function. Trends Biochem. Sci. 2006, 31, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Bernasconi, R.; Molinari, M. ERAD and ERAD tuning: Disposal of cargo and of ERAD regulators from the mammalian ER. Curr. Opin. Cell Biol. 2011, 23, 176–183. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, D.; Chen, S. Effect of earlier unfolded protein response and efficient protein disposal system on cellulase production in Rut C30. World J. Microbiol. Biotechnol. 2014, 30, 2587–2595. [Google Scholar] [CrossRef]
- Spang, A. Membrane traffic in the secretory pathway. Cell. Mol. Life Sci. 2008, 65, 2781–2789. [Google Scholar] [CrossRef]
- Hutagalung, A.H.; Novick, P.J. Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 2011, 91, 119–149. [Google Scholar] [CrossRef]
- Söllner, T.; Whiteheart, S.W.; Brunner, M.; Erdjument-Bromage, H.; Geromanos, S.; Tempst, P.; Rothman, J.E. SNAP receptors implicated in vesicle targeting and fusion. Nature 1993, 362, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Valkonen, M.; Kalkman, E.R.; Saloheimo, M.; Penttilaö, M.; Read, N.D.; Duncan, R.R. Spatially Segregated SNARE Protein Interactions in Living Fungal Cells. J. Biol. Chem. 2007, 282, 22775–22785. [Google Scholar] [CrossRef] [PubMed]
- Saunders, G.; Picknett, T.M.; Tuite, M.F.; Ward, M. Heterologous gene expression in filamentous fungi. Trends Biotechnol. 1989, 7, 283–287. [Google Scholar] [CrossRef]
- Jeenes, D.J.; Mackenzie, D.A.; Roberts, I.N.; Archer, D.B. Heterologous protein production by filamentous fungi. Biotechnol. Genet. Eng. Rev. 1991, 9, 327–367. [Google Scholar]
- Nevalainen, K.M.; Te’o, V.S.; Bergquist, P.L. Heterologous protein expression in filamentous fungi. Trends Biotechnol. 2005, 23, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Schmitz, G.; Zhang, M.; Mackie, R.I.; Cann, I.K. Heterologous gene expression in filamentous fungi. Adv. Appl. Microbiol. 2012, 81, 1–61. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, Y.H.; Liu, T.Q.; Zhang, H.; Zhang, H.; Li, J. The GlaA signal peptide substantially increases the expression and secretion of α-galactosidase in Aspergillus niger. Biotechnol. Lett. 2018, 40, 949–955. [Google Scholar] [CrossRef] [PubMed]
- Ohno, A.; Maruyama, J.; Nemoto, T.; Arioka, M.; Kitamoto, K. A carrier fusion significantly induces unfolded protein response in heterologous protein production by Aspergillus oryzae. Appl. Microbiol. Biotechnol. 2011, 92, 1197–1206. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, D.I.; Olsthoorn, M.M.; Maillet, I.; Akeroyd, M.; Breestraat, S.; Donkers, S.; van der Hoeven, R.A.; van den Hondel, C.A.; Kooistra, R.; Lapointe, T.; et al. Effective lead selection for improved protein production in Aspergillus niger based on integrated genomics. Fungal Genet. Biol. 2009, 46 (Suppl. 1), S141–S152. [Google Scholar] [CrossRef]
- Wu, Y.; Sun, X.; Xue, X.; Luo, H.; Yao, B.; Xie, X.; Su, X. Overexpressing key component genes of the secretion pathway for enhanced secretion of an Aspergillus niger glucose oxidase in Trichoderma reesei. Enzym. Microb. Technol. 2017, 106, 83–87. [Google Scholar] [CrossRef]
- Hoang, H.D.; Maruyama, J.; Kitamoto, K. Modulating endoplasmic reticulum-Golgi cargo receptors for improving secretion of carrier-fused heterologous proteins in the filamentous fungus Aspergillus oryzae. Appl. Environ. Microbiol. 2015, 81, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Garst, A.D.; Bassalo, M.C.; Pines, G.; Lynch, S.A.; Halweg-Edwards, A.L.; Liu, R.; Liang, L.; Wang, Z.; Zeitoun, R.; Alexander, W.G.; et al. Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering. Nat. Biotechnol. 2017, 35, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xue, W.; Sims, A.H.; Zhao, C.; Wang, A.; Tang, G.; Qin, J.; Wang, H. Isolation of four pepsin-like protease genes from Aspergillus niger and analysis of the effect of disruptions on heterologous laccase expression. Fungal Genet. Biol. 2008, 45, 17–27. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).