Bioprospection of Bacterial Strains from Chromite Process Industry Residues from Mexico for Potential Remediation
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Site
2.2. Biodiversity Analysis
2.3. Colony Isolation Procedure
2.4. Screening by Formation of Chromium Reduction Halos
2.5. Determination of Chromium Resistance
2.6. Chromium VI Quantification
2.7. Phylogenetic Analyses of Isolated Strains
3. Results
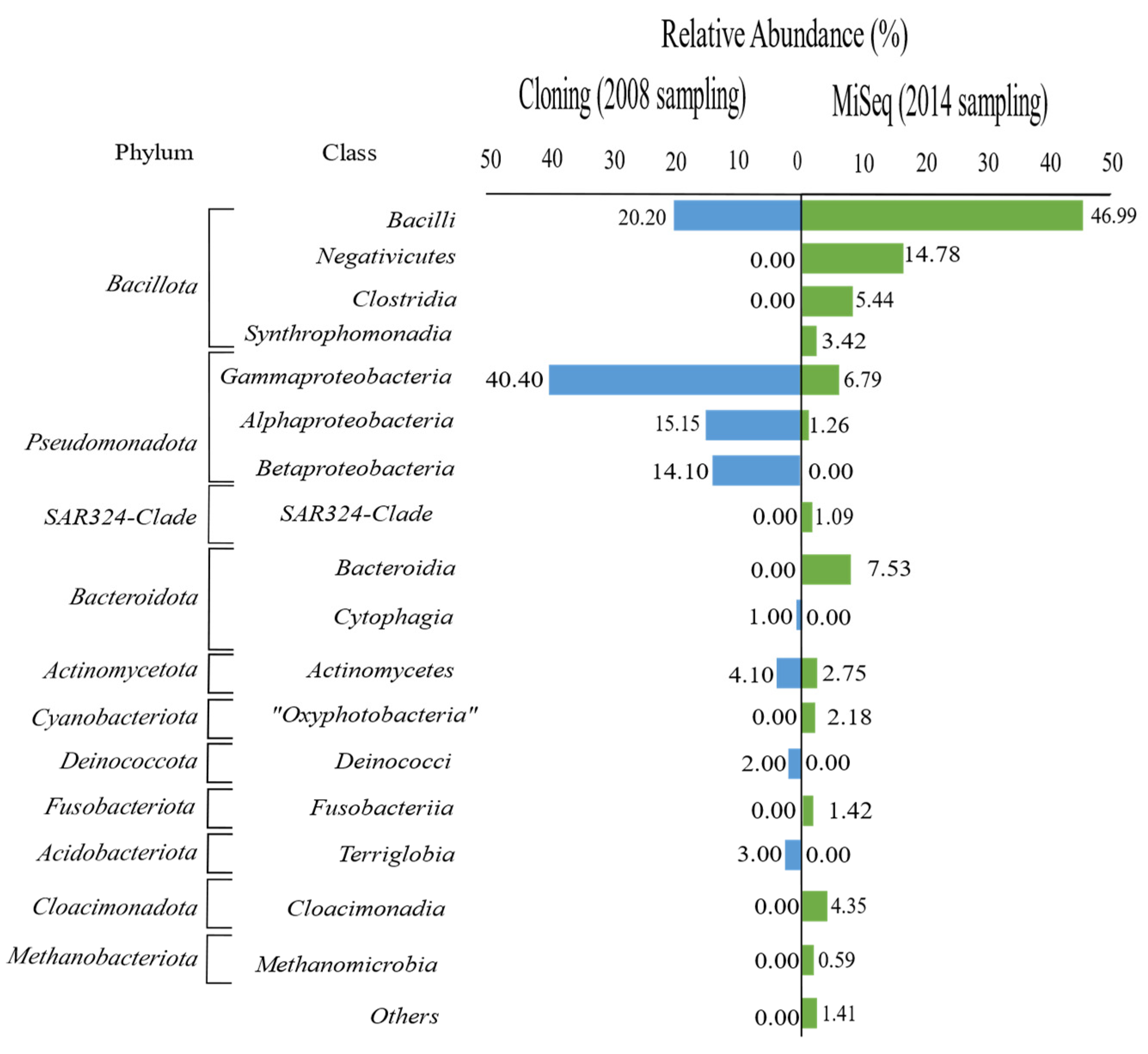
3.1. Diversity
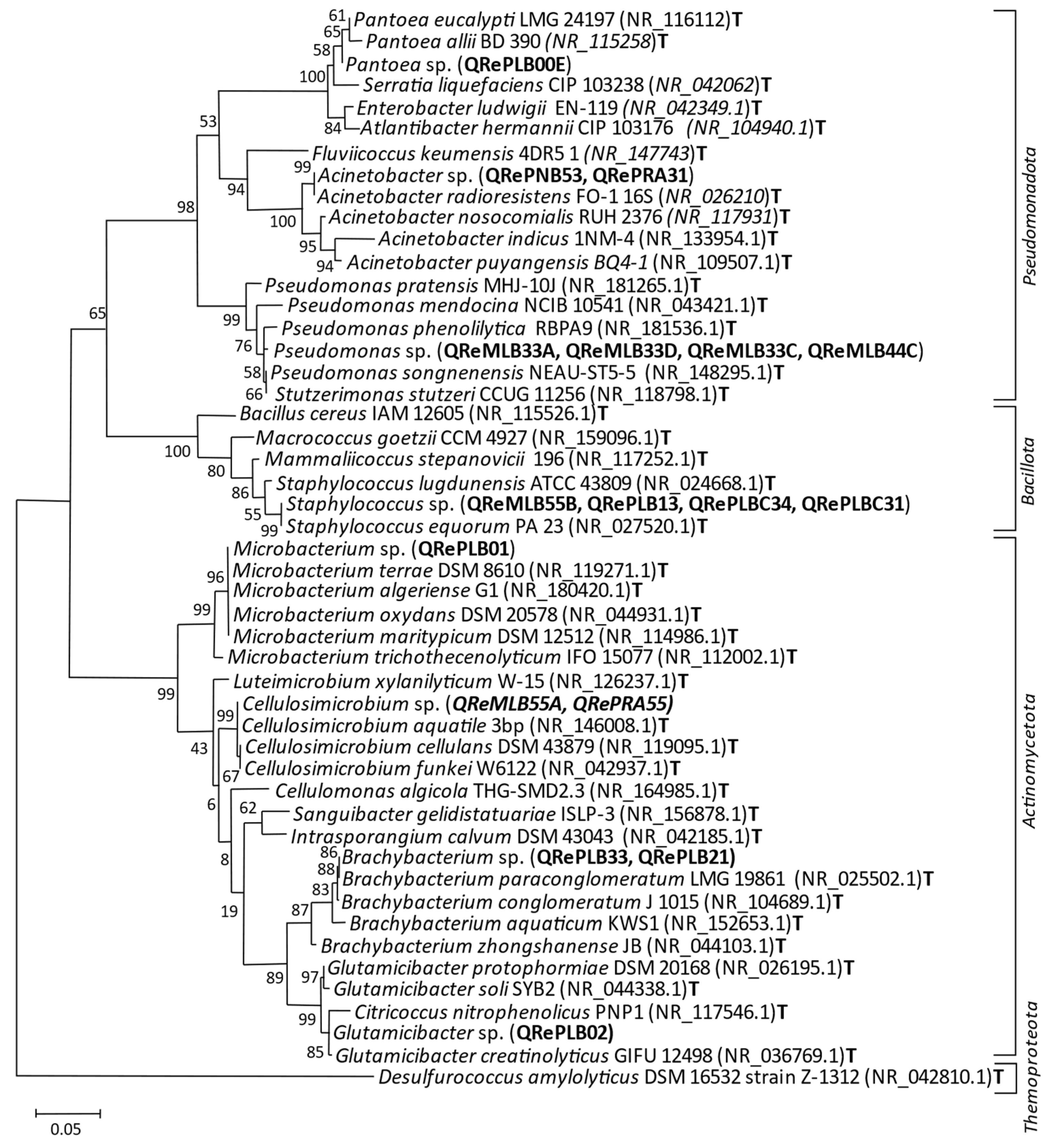
3.2. Colonies and Sequenced Strains
3.3. Chromium Reduction Halos
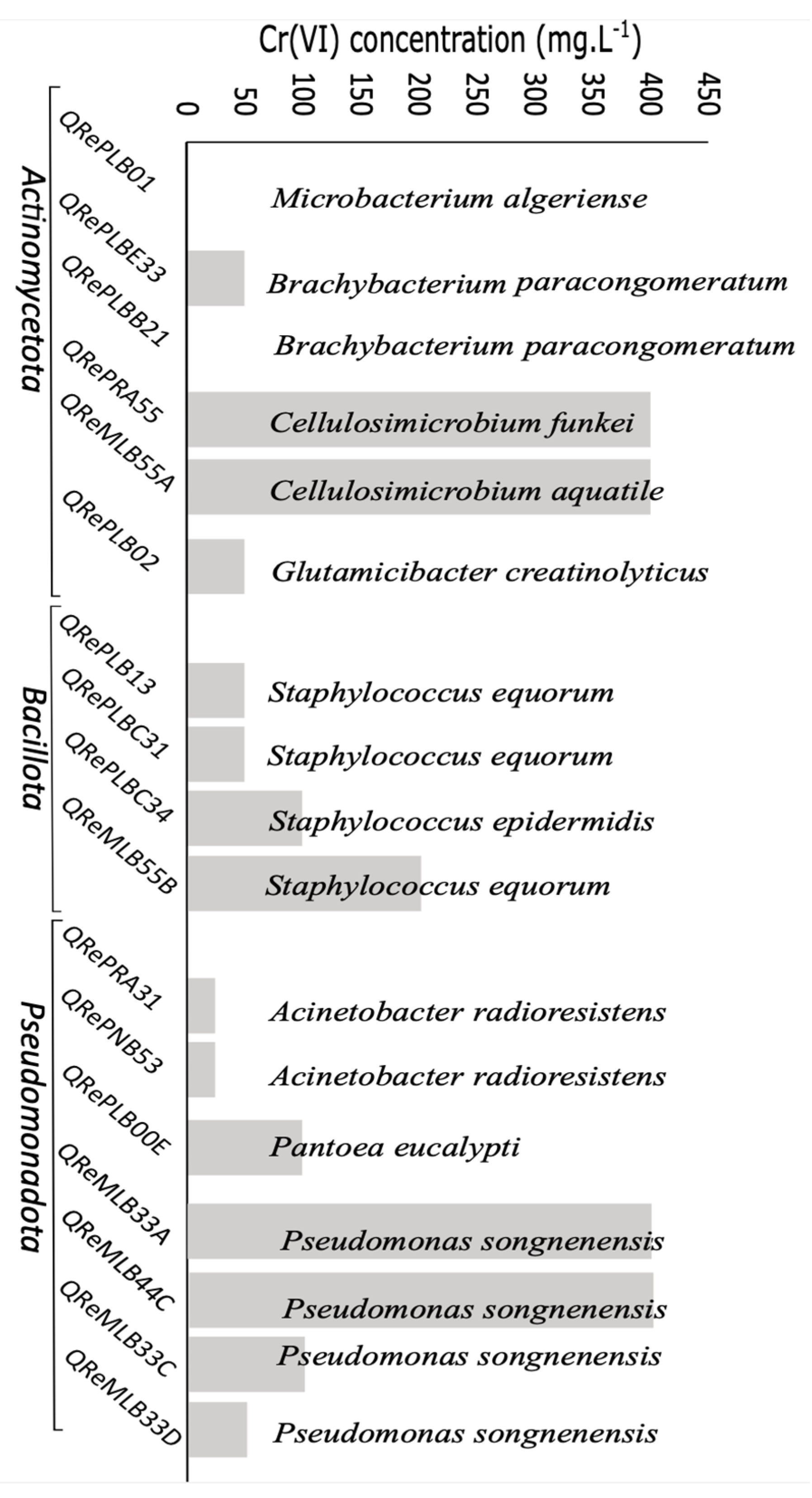
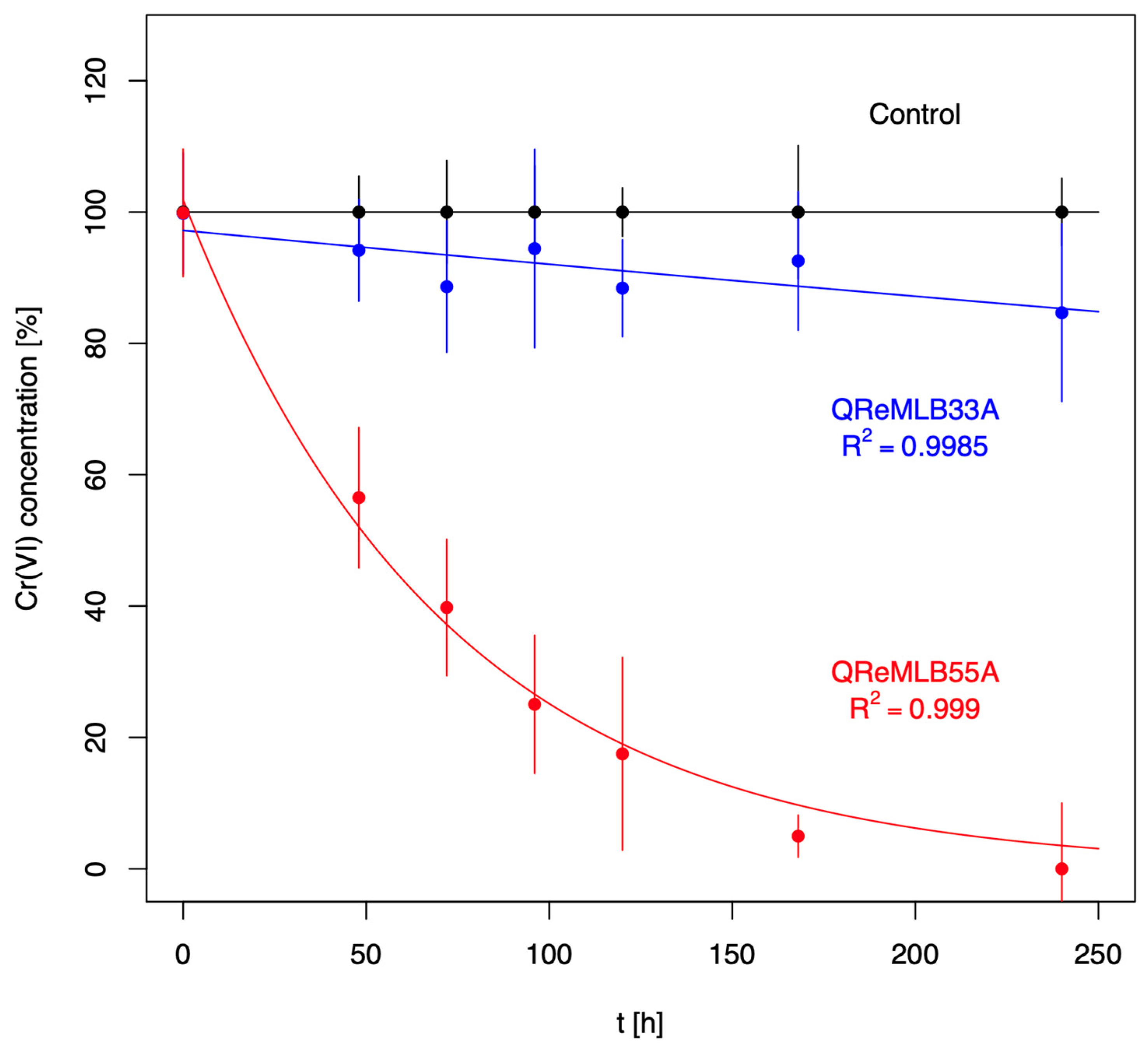
3.4. Chromium Resistance and Depletion
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thatoi, H.; Das, S.; Mishra, J.; Rath, B.P.; Das, N. Bacterial chromate reductase, a potential enzyme for bioremediation of hexavalent chromium: A review. J. Environ. Manag. 2014, 146, 383–399. [Google Scholar] [CrossRef] [PubMed]
- Guan, C.-Y.; Tseng, Y.-H.; Tsang, D.C.W.; Hu, A.; Yu, C.-P. Wetland plant microbial fuel cells for remediation of hexavalent chromium contaminated soils and electricity production. J. Hazard. Mater. 2019, 365, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Patra, R.C.; Malik, S.; Beer, M.; Megharaj, M.; Naidu, R. Molecular characterization of chromium (VI) reducing potential in Gram positive bacteria isolated from contaminated sites. Soil Biol. Biochem. 2010, 42, 1857–1863. [Google Scholar] [CrossRef]
- Fu, F.; Wang, Q. Removal of heavy metal ions from wastewaters: A review. J. Environ. Manag. 2011, 92, 407–418. [Google Scholar] [CrossRef]
- Vidales, L.E.; Cárdenas, L.C.; Robleto, E.; Yasbin, R.E.; Pedraza-Reyes, M. Defects in the Error Prevention Oxidized Guanine System Potentiate Stationary-Phase Mutagenesis in Bacillus subtilis. J. Bacteriol. 2009, 191, 506–513. [Google Scholar] [CrossRef]
- Vendruscolo, F.; da Ferreira, G.L.; Antoniosi Filho, N.R. Biosorption of hexavalent chromium by microorganisms. Int. Biodeterior. Biodegrad. 2017, 119, 87–95. [Google Scholar] [CrossRef]
- Buscaróns, F.; Artigas, J. Detection and colorimetric determination of the cr04-2 ion with o-dianisidine. Anal. Chim. Acta 1957, 16, 452–454. [Google Scholar] [CrossRef]
- Farhan, S.N.; Khadom, A.A. Biosorption of heavy metals from aqueous solutions by Saccharomyces Cerevisiae. Int. J. Ind. Chem. 2015, 6, 119–130. [Google Scholar] [CrossRef]
- Long, D.; Hashmi, M.Z.; Su, X.; Pongpiachan, S. Cr(VI) reduction by an extracellular polymeric substance (EPS) produced from a strain of Pseudochrobactrum saccharolyticum. 3 Biotech 2019, 9, 111. [Google Scholar] [CrossRef]
- Armienta, M.A.; Rodríguez, R.; Queré, A.; Juárez, F.; Ceniceros, N.; Aguayo, A. Ground Water Pollution with Chromium in Leon Valley, Mexico. Int. J. Environ. Anal. Chem. 1993, 54, 1–13. [Google Scholar] [CrossRef]
- Piñón-Castillo, H.; Brito, E.; Goñi-Urriza, M.; Guyoneaud, R.; Duran, R.; Nevarez-Moorillon, G.; Gutiérrez-Corona, J.; Caretta, C.; Reyna-López, G. Hexavalent chromium reduction by bacterial consortia and pure strains from an alkaline industrial effluent. J. Appl. Microbiol. 2010, 109, 2173–2182. [Google Scholar] [CrossRef] [PubMed]
- Brito, E.M.S.; Piñón-Castillo, H.A.; Guyoneaud, R.; Caretta, C.A.; Gutiérrez-Corona, J.F.; Duran, R.; Reyna-López, G.E.; Nevárez-Moorillón, G.V.; Fahy, A.; Goñi-Urriza, M. Bacterial biodiversity from anthropogenic extreme environments: A hyper-alkaline and hyper-saline industrial residue contaminated by chromium and iron. Appl. Microbiol. Biotechnol. 2012, 97, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Escudié, F.; Auer, L.; Bernard, M.; Mariadassou, M.; Cauquil, L.; Vidal, K.; Maman, S.; Hernandez-Raquet, G.; Combes, S.; Pascal, G. FROGS: Find, Rapidly, OTUs with Galaxy Solution. Bioinformatics 2018, 34, 1287–1294. [Google Scholar] [CrossRef] [PubMed]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [PubMed]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef] [PubMed]
- Reasoner, D.J.; E Geldreich, E. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 1985, 49, 1–7. [Google Scholar] [CrossRef]
- Kneebone, B.M.; Freiser, H. Determination of chromium(VI) in industrial atmospheres by a catalytic method. Anal. Chem. 1975, 47, 595–598. [Google Scholar] [CrossRef] [PubMed]
- Adams, V.D. Water and Wastewater Examination Manual; Lewis Publishers: Lakeland, FL, USA, 2017. [Google Scholar]
- Brito, E.M.S.; Romero-Núñez, V.M.; Caretta, C.A.; Bertin, P.; Valerdi-Negreros, J.C.; Guyoneaud, R.; Goñi-Urriza, M. The bacterial diversity on steam vents from Paricutín and Sapichu volcanoes. Extremophiles 2019, 23, 249–263. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Frank, K.L.; del Pozo, J.L.; Patel, R. From Clinical Microbiology to Infection Pathogenesis: How Daring to Be Different Works for Staphylococcus lugdunensis. Clin. Microbiol. Rev. 2008, 21, 111–133. [Google Scholar] [CrossRef] [PubMed]
- Freney, J.; Brun, Y.; Bes, M.; Meugnier, H.; Grimont, F.; Grimont, P.A.D.; Nervi, C.; Fleurette, J. Staphylococcus lugdunensis sp. nov. and Staphylococcus schleiferi sp. nov., Two Species from Human Clinical Specimens. Int. J. Syst. Evol. Microbiol. 1988, 38, 168–172. [Google Scholar] [CrossRef]
- Nishimura, Y.; Ino, T.; Iizuka, H. Acinetobacter radioresistens sp. nov. Isolated from Cotton and Soil. Int. J. Syst. Evol. Microbiol. 1988, 38, 209–211. [Google Scholar] [CrossRef]
- Ji, J.; Yuan, D.; Jin, C.; Wang, G.; Li, X.; Guan, C. Enhancement of growth and salt tolerance of rice seedlings (Oryza sativa L.) by regulating ethylene production with a novel halotolerant PGPR strain Glutamicibacter sp. YD01 containing ACC deaminase activity. Acta Physiol. Plant. 2020, 42, 42. [Google Scholar] [CrossRef]
- Acinas, S.G.; Sarma-Rupavtarm, R.; Klepac-Ceraj, V.; Polz, M.F. PCR-Induced Sequence Artifacts and Bias: Insights from Comparison of Two 16S rRNA Clone Libraries Constructed from the Same Sample. Appl. Environ. Microbiol. 2005, 71, 8966–8969. [Google Scholar] [CrossRef]
- Kennedy, K.; Hall, M.W.; Lynch, M.D.J.; Moreno-Hagelsieb, G.; Neufeld, J.D. Evaluating Bias of Illumina-Based Bacterial 16S rRNA Gene Profiles. Appl. Environ. Microbiol. 2014, 80, 5717–5722. [Google Scholar] [CrossRef]
- Tremblay, J.; Singh, K.; Fern, A.; Kirton, E.S.; He, S.; Woyke, T.; Lee, J.; Chen, F.; Dangl, J.L.; Tringe, S.G. Primer and platform effects on 16S rRNA tag sequencing. Front. Microbiol. 2015, 6, 771. [Google Scholar] [CrossRef] [PubMed]
- Green, J.; Bohannan, B.J. Spatial scaling of microbial biodiversity. Trends Ecol. Evol. 2006, 21, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Gai, X.; Zhong, Z.; Bian, F.; Yang, C.; Li, Y.; Wen, X. Understanding variations in soil properties and microbial communities in bamboo plantation soils along a chromium pollution gradient. Ecotoxicol. Environ. Saf. 2021, 222, 112507. [Google Scholar] [CrossRef]
- Liu, B.; Su, G.; Yang, Y.; Yao, Y.; Huang, Y.; Hu, L.; Zhong, H.; He, Z. Vertical distribution of microbial communities in chromium-contaminated soil and isolation of Cr(Ⅵ)-Reducing strains. Ecotoxicol. Environ. Saf. 2019, 180, 242–251. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Singh, N.R.; Kumar, U.; Mishra, S.R.; Perumal, R.C.; Benny, J.; Thatoi, H. Illumina MiSeq based assessment of bacterial community structure and diversity along the heavy metal concentration gradient in Sukinda chromite mine area soils, India. Ecol. Genet. Genom. 2020, 15, 100054. [Google Scholar] [CrossRef]
- Smith, D.D.N.; Kirzinger, M.W.B.; Stavrinides, J. Draft Genome Sequence of the Antibiotic-Producing Epiphytic Isolate Pantoea ananatis BRT175. Genome Announc. 2013, 1. [Google Scholar] [CrossRef] [PubMed]
- Cruz, A.T.; Cazacu, A.C.; Allen, C.H. Pantoea agglomerans, a Plant Pathogen Causing Human Disease. J. Clin. Microbiol. 2007, 45, 1989–1992. [Google Scholar] [CrossRef] [PubMed]
- Ozdemir, G.; Ceyhan, N.; Ozturk, T.; Akirmak, F.; Cosar, T. Biosorption of chromium(VI), cadmium(II) and copper(II) by Pantoea sp. TEM18. Chem. Eng. J. 2004, 102, 249–253. [Google Scholar] [CrossRef]
- Luziatelli, F.; Gatti, L.; Ficca, A.G.; Medori, G.; Silvestri, C.; Melini, F.; Muleo, R.; Ruzzi, M. Metabolites Secreted by a Plant-Growth-Promoting Pantoea agglomerans Strain Improved Rooting of Pyrus communis L. cv Dar Gazi Cuttings. Front. Microbiol. 2020, 11, 539359. [Google Scholar] [CrossRef] [PubMed]
- Castagno, L.; Estrella, M.; Sannazzaro, A.; Grassano, A.; Ruiz, O. Phosphate-solubilization mechanism and in vitro plant growth promotion activity mediated by Pantoea eucalypti isolated from Lotus tenuis rhizosphere in the Salado River Basin (Argentina). J. Appl. Microbiol. 2011, 110, 1151–1165. [Google Scholar] [CrossRef] [PubMed]
- Cumpa-Velásquez, L.M.; Moriconi, J.I.; Dip, D.P.; Castagno, L.N.; Puig, M.L.; Maiale, S.J.; Santa-María, G.E.; Sannazzaro, A.I.; Estrella, M.J. Prospecting phosphate solubilizing bacteria in alkaline-sodic environments reveals intra-specific variability in Pantoea eucalypti affecting nutrient acquisition and rhizobial nodulation in Lotus tenuis. Appl. Soil Ecol. 2021, 168, 104125. [Google Scholar] [CrossRef]
- Vahling-Armstrong, C.; Dung, J.K.S.; Humann, J.L.; Schroeder, B.K. Effects of postharvest onion curing parameters on bulb rot caused by Pantoea agglomerans, Pantoea ananatis and Pantoea allii in storage. Plant Pathol. 2015, 65, 536–544. [Google Scholar] [CrossRef]
- Navon-Venezia, S.; Zosim, Z.; Gottlieb, A.; Legmann, R.; Carmeli, S.; Ron, E.Z.; Rosenberg, E. Alasan, a new bioemulsifier from Acinetobacter radioresistens. Appl. Environ. Microbiol. 1995, 61, 3240–3244. [Google Scholar] [CrossRef]
- Chen, S.-J.; Cheng, C.-Y.; Chen, T.-L. Production of an alkaline lipase by Acinetobacter radioresistens. J. Ferment. Bioeng. 1998, 86, 308–312. [Google Scholar] [CrossRef]
- Liu, F.-Y.; Hong, M.-Z.; Liu, D.-M.; Li, Y.-W.; Shou, P.-S.; Yan, H.; Shi, G.-Q. Biodegradation of methyl parathion by Acinetobacter radioresistens USTB-04. J. Environ. Sci. 2007, 19, 1257–1260. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, W.; Shah, S.B.; Zanaroli, G.; Xu, P.; Tang, H. Phenol biodegradation by Acinetobacter radioresistens APH1 and its application in soil bioremediation. Appl. Microbiol. Biotechnol. 2019, 104, 427–437. [Google Scholar] [CrossRef]
- Mazzoli, R.; Pessione, E.; Giuffrida, M.G.; Fattori, P.; Barello, C.; Giunta, C.; Lindley, N.D. Degradation of aromatic compounds by Acinetobacter radioresistens S13: Growth characteristics on single substrates and mixtures. Arch. Microbiol. 2007, 188, 55–68. [Google Scholar] [CrossRef]
- Jawad, A.; Snelling, A.; Heritage, J.; Hawkey, P. Exceptional desiccation tolerance of Acinetobacter radioresistens. J. Hosp. Infect. 1998, 39, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Spiers, A.J.; Buckling, A.; Rainey, P.B. MINI REVIEW The Causes of Pseudomonas Diversity. 2016. Available online: www.microbiologyresearch.org (accessed on 20 January 2024).
- Poblete-Castro, I.; Becker, J.; Dohnt, K.; Dos Santos, V.M.; Wittmann, C. Industrial biotechnology of Pseudomonas putida and related species. Appl. Microbiol. Biotechnol. 2012, 93, 2279–2290. [Google Scholar] [CrossRef] [PubMed]
- Nikel, P.I.; de Lorenzo, V. Metabolic Engineering of Pseudomonas. In Metabolic Engineering; Wiley: Hoboken, NJ, USA, 2021; pp. 519–550. [Google Scholar] [CrossRef]
- Zhao, F.; Wu, Y.; Wang, Q.; Zheng, M.; Cui, Q. Glycerol or crude glycerol as substrates make Pseudomonas aeruginosa achieve anaerobic production of rhamnolipids. Microb. Cell Factories 2021, 20, 185. [Google Scholar] [CrossRef] [PubMed]
- Fakhar, A.; Gul, B.; Gurmani, A.R.; Khan, S.M.; Ali, S.; Sultan, T.; Chaudhary, H.J.; Rafique, M.; Rizwan, M. Heavy metal remediation and resistance mechanism of Aeromonas, Bacillus, and Pseudomonas: A review. Crit. Rev. Environ. Sci. Technol. 2020, 52, 1868–1914. [Google Scholar] [CrossRef]
- Zhang, L.; Pan, Y.; Wang, K.; Zhang, X.; Zhang, S.; Fu, X.; Zhang, C.; Jiang, J. Pseudomonas songnenensis sp. nov., isolated from saline and alkaline soils in Songnen Plain, China. Antonie Van Leeuwenhoek 2014, 107, 711–721. [Google Scholar] [CrossRef]
- Alvarez, A.; Saez, J.M.; Costa, J.S.D.; Colin, V.L.; Fuentes, M.S.; Cuozzo, S.A.; Benimeli, C.S.; Polti, M.A.; Amoroso, M.J. Actinobacteria: Current research and perspectives for bioremediation of pesticides and heavy metals. Chemosphere 2017, 166, 41–62. [Google Scholar] [CrossRef]
- Elahi, A.; Ajaz, M.; Rehman, A.; Vuilleumier, S.; Khan, Z.; Hussain, S.Z. Isolation, characterization, and multiple heavy metal-resistant and hexavalent chromium-reducing Microbacterium testaceum B-HS2 from tannery effluent. J. King Saud Univ.-Sci. 2019, 31, 1437–1444. [Google Scholar] [CrossRef]
- Ouertani, R.; Ouertani, A.; Mahjoubi, M.; Bousselmi, Y.; Najjari, A.; Cherif, H.; Chamkhi, A.; Mosbah, A.; Khdhira, H.; Sghaier, H.; et al. New Plant Growth-Promoting, Chromium-Detoxifying Microbacterium Species Isolated From a Tannery Wastewater: Performance and Genomic Insights. Front. Bioeng. Biotechnol. 2020, 8, 521. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Chen, S.; Saratale, G.D.; Saratale, R.G.; Ferreira, L.F.R.; Bilal, M.; Bharagava, R.N. Reduction of hexavalent chromium by Microbacterium paraoxydans isolated from tannery wastewater and characterization of its reduced products. J. Water Process Eng. 2020, 39, 101748. [Google Scholar] [CrossRef]
- Busse, H.-J. Review of the taxonomy of the genus Arthrobacter, emendation of the genus Arthrobacter sensu lato, proposal to reclassify selected species of the genus Arthrobacter in the novel genera Glutamicibacter gen. nov., Paeniglutamicibacter gen. nov., Pseudoglutamicibacter gen. nov., Paenarthrobacter gen. nov. and Pseudarthrobacter gen. nov., and emended description of Arthrobacter roseus. Int. J. Syst. Evol. Microbiol. 2016, 66, 9–37. [Google Scholar] [CrossRef]
- Wang, X.S. Cd (II) removal by marine Arthrobacter protophormiae biomass: Mechanism characterization and adsorption performance. Desalin. Water Treat. 2013, 51, 7710–7720. [Google Scholar] [CrossRef]
- Wan, Y.-S.; Chen, Y.-Q.; Hang, L. Interaction of Pb (II) and Cd (II) with Arthrobacter protophormiae biomass: Mechanism and characterization. Desalin. Water Treat. 2016, 57, 25773–25782. [Google Scholar] [CrossRef]
- Arshad, Q.; Ahmed, A. Chromium-resistant PGPB: Growth stimulatory impact on Vigna radiata L. under chromium stress. Rom. Biotechnol. Lett. 2017, 22, 12988. [Google Scholar]
- Duraisamy, P.; Sekar, J.; Arunkumar, A.D.; Ramalingam, P.V. Kinetics of Phenol Biodegradation by Heavy Metal Tolerant Rhizobacteria Glutamicibacter nicotianae MSSRFPD35 from Distillery Effluent Contaminated Soils. Front. Microbiol. 2020, 11, 1573. [Google Scholar] [CrossRef] [PubMed]
- Kiran, G.S.; Sabarathnam, B.; Thajuddin, N.; Selvin, J. Production of Glycolipid Biosurfactant from Sponge-Associated Marine Actinobacterium Brachybacterium paraconglomeratum MSA21. J. Surfactants Deterg. 2014, 17, 531–542. [Google Scholar] [CrossRef]
- Harboul, K.; Alouiz, I.; Hammani, K.; El-Karkouri, A. Isotherm and kinetics modeling of biosorption and bioreduction of the Cr(VI) by Brachybacterium paraconglomeratum ER41. Extremophiles 2022, 26, 30. [Google Scholar] [CrossRef]
- Bharagava, R.N.; Mishra, S. Hexavalent chromium reduction potential of Cellulosimicrobium sp. isolated from common effluent treatment plant of tannery industries. Ecotoxicol. Environ. Saf. 2018, 147, 102–109. [Google Scholar] [CrossRef]
- Rehman, F.; Faisal, M. Toxic hexavalent chromium reduction by Bacillus pumilis, Cellulosimicrobium cellulans and Exiguobacterium. Chin. J. Oceanol. Limnol. 2014, 33, 585–589. [Google Scholar] [CrossRef]
- Karthik, C.; Barathi, S.; Pugazhendhi, A.; Ramkumar, V.S.; Thi, N.B.D.; Arulselvi, P.I. Evaluation of Cr(VI) reduction mechanism and removal by Cellulosimicrobium funkei strain AR8, a novel haloalkaliphilic bacterium. J. Hazard. Mater. 2017, 333, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Karthik, C.; Arulselvi, P.I. Biotoxic Effect of Chromium (VI) on Plant Growth-Promoting Traits of Novel Cellulosimicrobium funkei Strain AR8 Isolated from Phaseolus vulgaris Rhizosphere. Geomicrobiol. J. 2016, 34, 434–442. [Google Scholar] [CrossRef]
- Karthik, C.; Ramkumar, V.S.; Pugazhendhi, A.; Gopalakrishnan, K.; Arulselvi, P.I. Biosorption and biotransformation of Cr(VI) by novel Cellulosimicrobium funkei strain AR6. J. Taiwan Inst. Chem. Eng. 2017, 70, 282–290. [Google Scholar] [CrossRef]
- Karthik, C.; Kadirvelu, K.; Bruno, B.; Maharajan, K.; Rajkumar, M.; Manoj, S.R.; Arulselvi, P.I. Cellulosimicrobium funkei strain AR6 alleviate Cr(VI) toxicity in Lycopersicon esculentum by regulating the expression of growth responsible, stress tolerant and metal transporter genes. Rhizosphere 2021, 18, 100351. [Google Scholar] [CrossRef]
- Roohi, I.; Ahmed, N.; Khalid, M.I.; Jamil, M. Isolation and Phylogenetic Identification of Halotolerant/Halophilic Bacteria from the Salt Mines of Karak, Pakistan. Int. J. Agric. Biol. 2014, 16, 564–570. Available online: http://www.fspublishers.org (accessed on 20 January 2024).
- Wadood, H.Z.; Latif, A.; Mukhtar, H.; Javed, M.; Mukhtar, H.; Rehman, Y. Planktonic cells of Staphylococcus and Bacillus species capable of faster chromium reduction in short incubation times as compared to their biofilms. Arab. J. Geosci. 2021, 14, 1797. [Google Scholar] [CrossRef]
- Rahman, Z.; Thomas, L.; Chetri, S.P.K.; Bodhankar, S.; Kumar, V.; Naidu, R. A comprehensive review on chromium (Cr) contamination and Cr(VI)-resistant extremophiles in diverse extreme environments. Environ. Sci. Pollut. Res. 2023, 30, 59163–59193. [Google Scholar] [CrossRef]

| Isolates | ) | ) | % Diminution after 8 Days |
|---|---|---|---|
| QReMLB33A | 105.71 ± 15.32 232.42 ± 30.60 294.43 ± 26.97 368 ± 21.34 | 58.9 ± 10.17 143.45 ± 16.68 238.31 ± 26.92 313.06 ± 23.70 | 44.8 38.28 19.06 14.98 |
| QReMLB55A | 135.37 ± 52.0 215.76 ± 11.86 283.41 ± 24.74 406.69 ± 18.51 | 53.50 ± 14.79 136.59 ± 18.10 207.18 ± 13.57 269.44 ± 23.50 | 60.47 36.69 26.90 33.75 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Aldape, P.A.; Sandoval-Vergara, M.E.; Padilla-Hernández, R.E.; Caretta, C.A.; Valerdi-Negreros, J.C.; Casanova, P.; Monteiro, M.M.; Gassie, C.; Goñi-Urriza, M.; Brito, E.M.S.; et al. Bioprospection of Bacterial Strains from Chromite Process Industry Residues from Mexico for Potential Remediation. Appl. Microbiol. 2024, 4, 665-681. https://doi.org/10.3390/applmicrobiol4020046
Martínez-Aldape PA, Sandoval-Vergara ME, Padilla-Hernández RE, Caretta CA, Valerdi-Negreros JC, Casanova P, Monteiro MM, Gassie C, Goñi-Urriza M, Brito EMS, et al. Bioprospection of Bacterial Strains from Chromite Process Industry Residues from Mexico for Potential Remediation. Applied Microbiology. 2024; 4(2):665-681. https://doi.org/10.3390/applmicrobiol4020046
Chicago/Turabian StyleMartínez-Aldape, Paola Abigail, Mario Enrique Sandoval-Vergara, Reyna Edith Padilla-Hernández, César Augusto Caretta, Julio César Valerdi-Negreros, Pablo Casanova, Magna Maria Monteiro, Claire Gassie, Marisol Goñi-Urriza, Elcia Margareth Souza Brito, and et al. 2024. "Bioprospection of Bacterial Strains from Chromite Process Industry Residues from Mexico for Potential Remediation" Applied Microbiology 4, no. 2: 665-681. https://doi.org/10.3390/applmicrobiol4020046
APA StyleMartínez-Aldape, P. A., Sandoval-Vergara, M. E., Padilla-Hernández, R. E., Caretta, C. A., Valerdi-Negreros, J. C., Casanova, P., Monteiro, M. M., Gassie, C., Goñi-Urriza, M., Brito, E. M. S., & Guyoneaud, R. (2024). Bioprospection of Bacterial Strains from Chromite Process Industry Residues from Mexico for Potential Remediation. Applied Microbiology, 4(2), 665-681. https://doi.org/10.3390/applmicrobiol4020046






