Customizing Sanitization Protocols for Food-Borne Pathogens Based on Biofilm Formation, Surfaces and Disinfectants—Their Two- and Three-Way Interactions
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Surfaces, and Disinfectants
2.2. Bacterial Biofilm Formation on Eight Surfaces
2.3. Statistical Analyses
3. Results
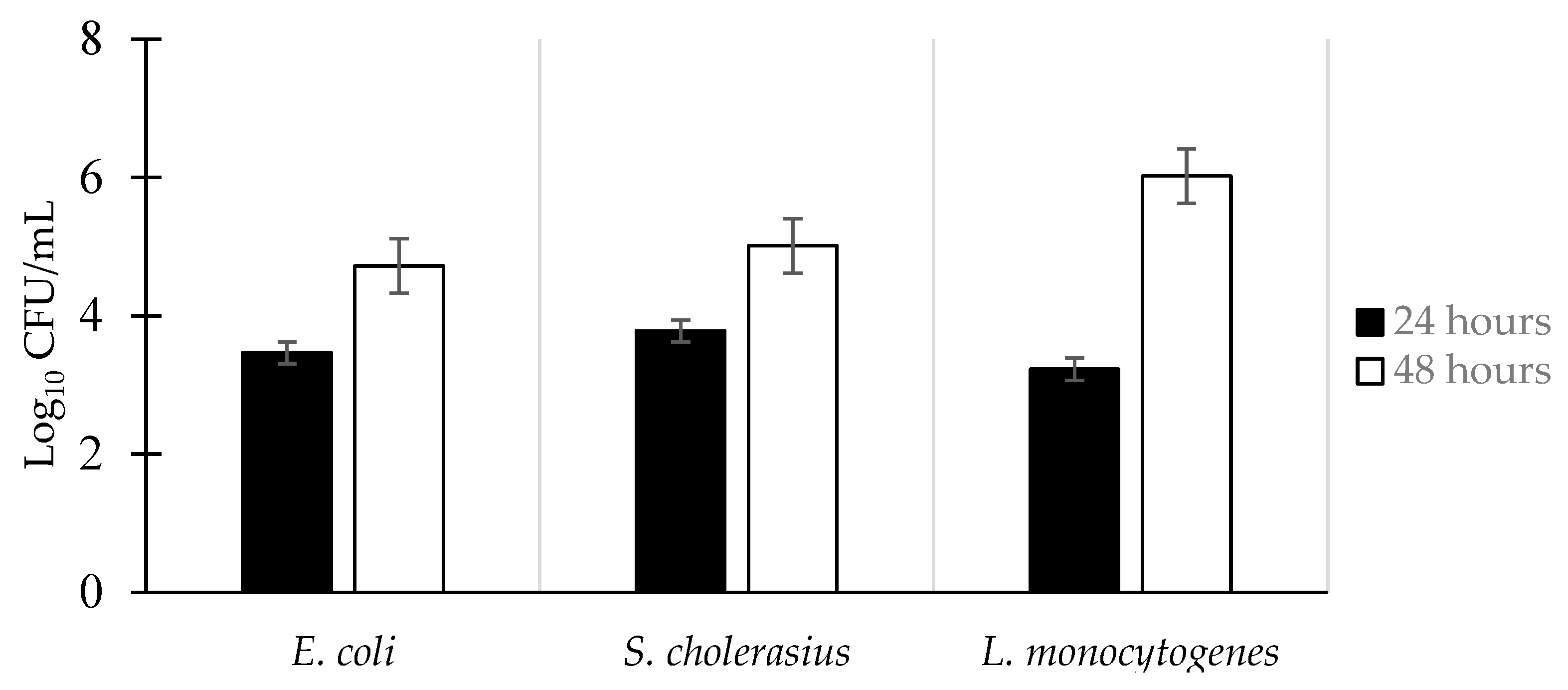
3.1. Biofilm Formation of E. coli, S. choleraesuis, and L. monocytogenes
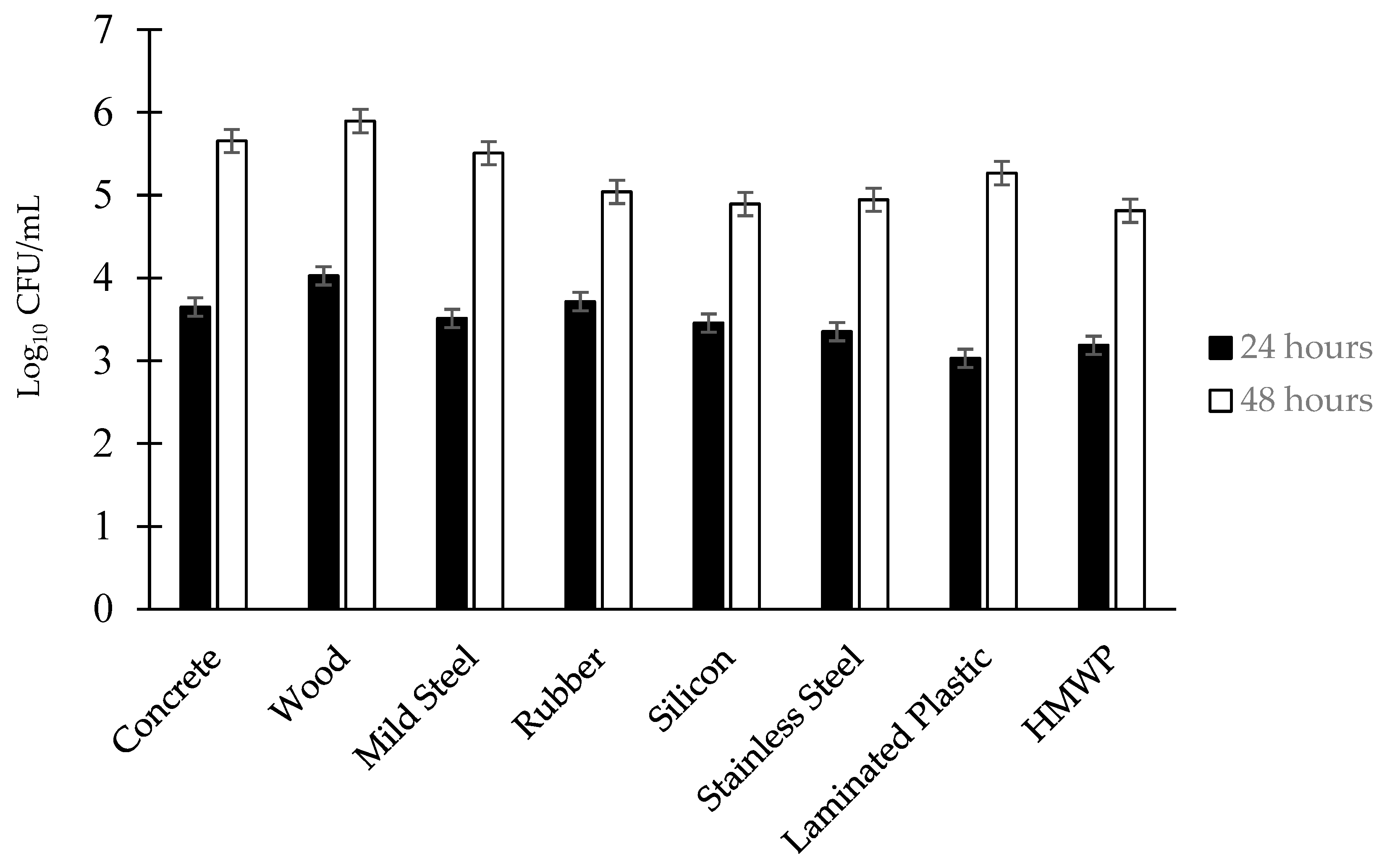
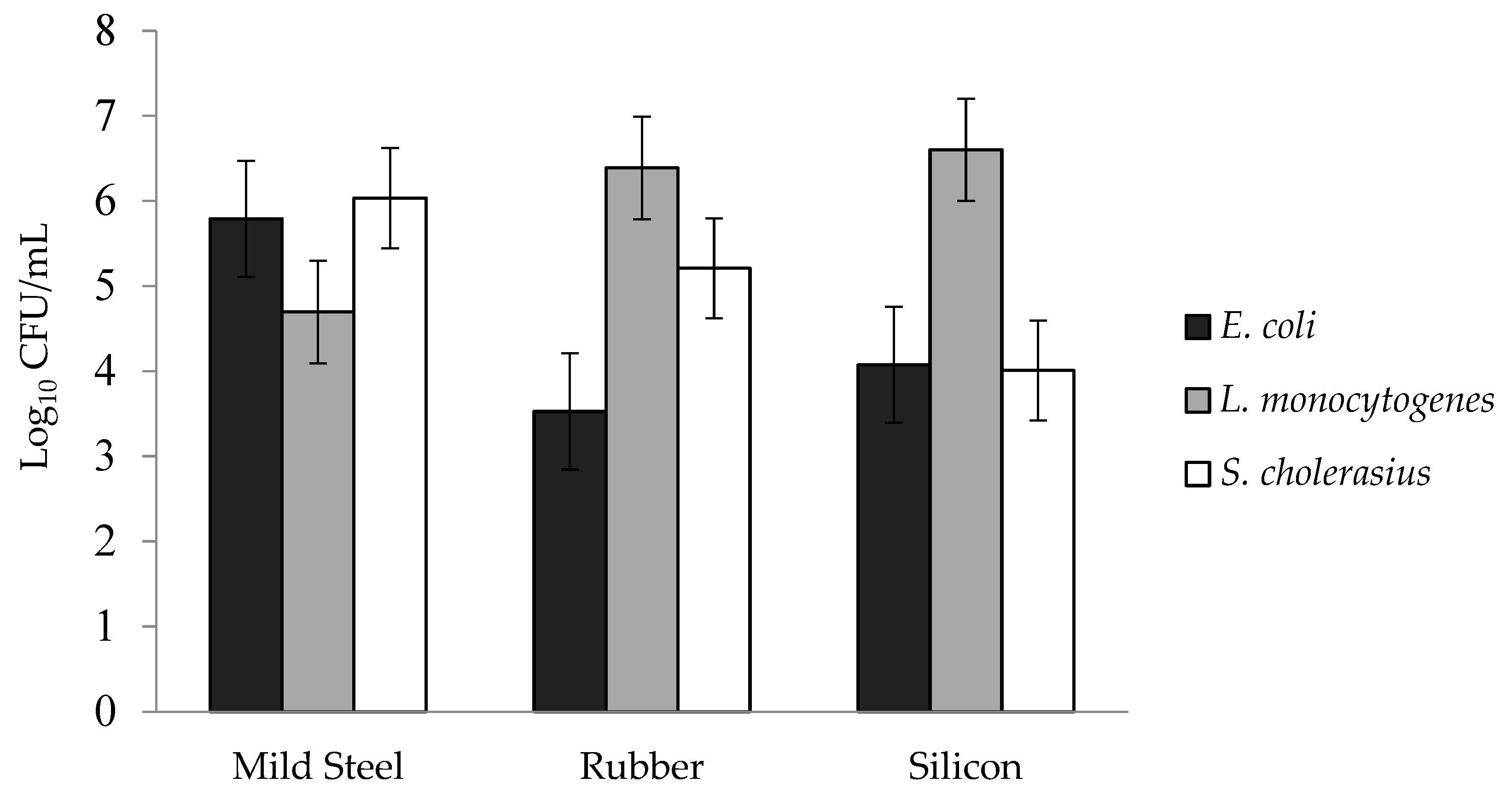
3.2. Effect of the Surface on Biofilm Population Density
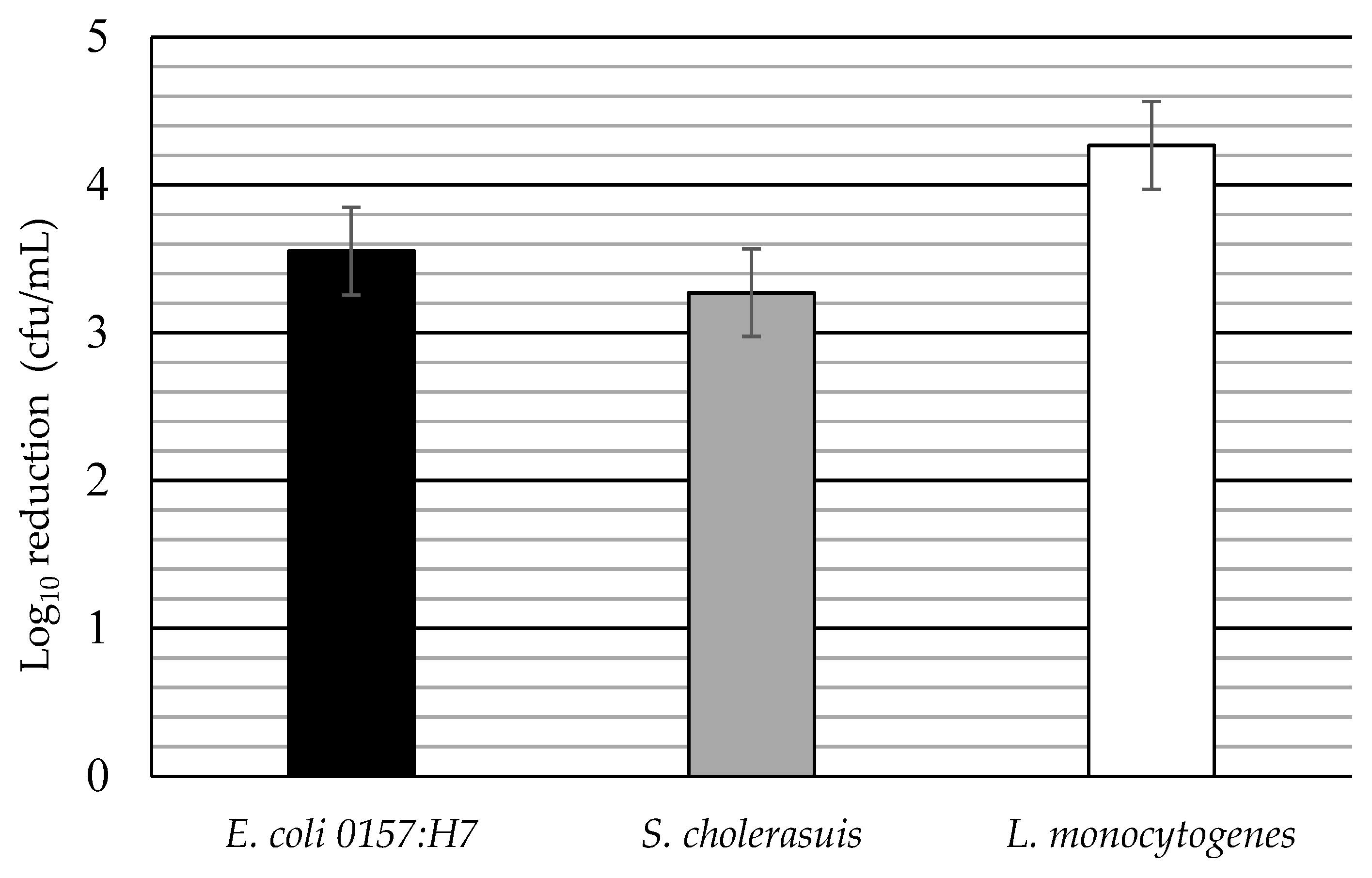
3.3. Log Reductions of Food-Borne Pathogen Biofilms by Disinfectant: Single Factors
3.4. Log Reductions of Food-Borne Pathogen Biofilms: Two-Way Interactions
3.5. Three-Way Interactions of Bacterial Species × Surfaces × Disinfectants
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Josephs-Spaulding, J.; Beeler, E.; Singh, O.V. Human microbiome versus food-borne pathogens: Friend or foe. Appl. Microbiol. Biotechnol. 2016, 100, 4845–4863. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.H.; Dalmasso, M.; Ingmer, H.; Langsrud, S.; Malakauskas, M.; Mader, A.; Møretrø, T.; Možina, S.S.; Rychli, K.; Wagner, M.; et al. Persistence of foodborne pathogens and their control in primary and secondary food production chains. Food Control 2014, 44, 92–109. [Google Scholar] [CrossRef]
- Butucel, E.; Balta, I.; Ahmadi, M.; Dumitrescu, G.; Morariu, F.; Pet, I.; Stef, L.; Corcionivoschi, N. Biocides as biomedicines against foodborne pathogenic bacteria. Biomedicines 2022, 10, 379. [Google Scholar] [CrossRef] [PubMed]
- Eppinger, M.; Cebula, T.A. Future perspectives, applications and challenges of genomic epidemiology studies for food-borne pathogens: A case study of Enterohemorrhagic Escherichia coli (EHEC) of the O157: H7 serotype. Gut Microbes 2015, 6, 194–201. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yu, Q.; Zhai, L.; Bie, X.; Lu, Z.; Zhang, C.; Tao, T.; Li, J.; Lv, F.; Zhao, H. Survey of five food-borne pathogens in commercial cold food dishes and their detection by multiplex PCR. Food Control 2016, 59, 862–869. [Google Scholar] [CrossRef]
- Aryal, M.; Muriana, P.M. Efficacy of commercial sanitizers used in food processing facilities for inactivation of Listeria monocytogenes, E. coli O157: H7, and Salmonella biofilms. Foods 2019, 8, 639. [Google Scholar] [CrossRef] [PubMed]
- Moerman, F.; Mager, K. Cleaning and disinfection in dry food processing facilities. In Handbook of Hygiene Control in the Food Industry; Woodhead Publishing: Sawston, UK, 2016; pp. 521–554. [Google Scholar]
- Holah, J.T. Cleaning and disinfection practices in food processing. In Hygiene in Food Processing, 2nd ed.; Woodhead Publishing: Cambridge, UK, 2014; pp. 259–304. [Google Scholar]
- Midelet, G.; Carpentier, B. Impact of cleaning and disinfection agents on biofilm structure and on microbial transfer to a solid model food. J. Appl. Microbiol. 2004, 97, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Simões, M.; Simões, L.C.; Machado, I.; Pereira, M.O.; Vieira, M.J. Control of flow-generated biofilms with surfactants: Evidence of resistance and recovery. Food Bioprod. Process. 2006, 84, 338–345. [Google Scholar] [CrossRef]
- Cadnum, J.L.; Pearlmutter, B.S.; Haq, M.F.; Jencson, A.L.; Donskey, C.J. Effectiveness and real-world materials compatibility of a novel hydrogen peroxide disinfectant cleaner. Am. J. Infect. Control 2021, 49, 1572–1574. [Google Scholar] [CrossRef]
- Aranke, M.; Moheimani, R.; Phuphanich, M.; Kaye, A.D.; Ngo, A.L.; Viswanath, O.; Herman, J. Disinfectants in interventional practices. Curr. Pain Headache Rep. 2021, 25, 1–10. [Google Scholar] [CrossRef]
- Abdallah, M.; Benoliel, C.; Drider, D.; Dhulster, P.; Chihib, N.E. Biofilm formation and persistence on abiotic surfaces in the context of food and medical environments. Arch. Microbiol. 2014, 196, 453–472. [Google Scholar] [CrossRef] [PubMed]
- Srey, S.; Jahid, I.K.; Ha, S.D. Biofilm formation in food industries: A food safety concern. Food Control 2013, 31, 572–585. [Google Scholar] [CrossRef]
- Bridier, A.; Sanchez-Vizuete, P.; Guilbaud, M.; Piard, J.C.; Naïtali, M.; Briandet, R. Biofilm-associated persistence of food-borne pathogens. Food Microbiol. 2015, 45, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Lewandowski, Z.; Caldwell, D.E.; Korber, D.R.; Lappin-Scott, H.M. Microbial biofilms. Annu. Rev. Microbiol. 1995, 49, 711–745. [Google Scholar] [CrossRef] [PubMed]
- Iturriaga, M.H.; Tamplin, M.L.; Escartin, E.F. Colonization of tomatoes by Salmonella montevideo is affected by relative humidity and storage temperature. J. Food Prot. 2007, 70, 30–34. [Google Scholar] [CrossRef] [PubMed]
- Zobell, C.E. The effect of solid surfaces upon bacterial activity. J. Bacteriol. 1943, 46, 39–56. [Google Scholar] [CrossRef] [PubMed]
- Chitlapilly Dass, S.; Wang, R. Biofilm through the looking glass: A microbial food safety perspective. Pathogens 2022, 11, 346. [Google Scholar] [CrossRef]
- Ghosh, S.; Sarkar, T.; Chakraborty, R. Formation and development of biofilm-an alarming concern in food safety perspectives. Biocatal. Agric. Biotechnol. 2021, 38, 102210. [Google Scholar] [CrossRef]
- Bai, X.; Nakatsu, C.H.; Bhunia, A.K. Bacterial biofilms and their implications in pathogenesis and food safety. Foods 2021, 10, 2117. [Google Scholar] [CrossRef]
- Raposo, A.; Ramos, F.; Raheem, D.; Saraiva, A.; Carrascosa, C. Food safety, security, sustainability and nutrition as priority objectives of the food sector. Int. J. Environ. Res. Public Health 2021, 18, 8073. [Google Scholar] [CrossRef]
- Mevo, S.I.U.; Ashrafudoulla, M.; Furkanur Rahaman Mizan, M.; Park, S.H.; Ha, S.D. Promising strategies to control persistent enemies: Some new technologies to combat biofilm in the food industry—A review. Compr. Rev. Food Sci. Food Saf. 2021, 20, 5938–5964. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Yang, C.; Bao, X.; Chen, F.; Guo, X. Strategies for controlling biofilm formation in food industry. Grain Oil Sci. Technol. 2022, 5, 179–186. [Google Scholar] [CrossRef]
- Abebe, G.M. The role of bacterial biofilm in antibiotic resistance and food contamination. Int. J. Microbiol. 2020, 2020, 1705814. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, M.; Beito, B.; Oliveira, M.; Tudela Martins, M.; Gallas, B.; Salmain, M.; Boujday, S.; Humblot, V. Strategies for antimicrobial peptides immobilization on surfaces to prevent biofilm growth on biomedical devices. Antibiotics 2021, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Amod, A.; Pandey, P.; Bose, P.; Pingali, M.S.; Shivalkar, S.; Varadwaj, P.K.; Sahoo, A.K.; Samanta, S.K. Bacterial biofilm infections, their resistance to antibiotics therapy and current treatment strategies. Biomed. Mater. 2022, 17, 022003. [Google Scholar] [CrossRef] [PubMed]
- Rather, M.A.; Gupta, K.; Mandal, M. Impact of microbial biofilm on crop productivity and agricultural sustainability. In Microbes in Land Use Change Management; Elsevier: Amsterdam, The Netherlands, 2021; pp. 451–469. [Google Scholar]
- Rayanoothala, P.; Divya, M.; Mahapatra, S.; Das, S. Microbial Biofilm: Formation, Quorum Sensing, and Its Applications in Plant Disease Management. In Emerging Trends in Plant Pathology; Springer: Singapore, 2021; pp. 385–397. [Google Scholar]
- Fazeli-Nasab, B.; Sayyed, R.Z.; Mojahed, L.S.; Rahmani, A.F.; Ghafari, M.; Antonius, S. Biofilm production: A strategic mechanism for survival of microbes under stress conditions. Biocatal. Agric. Biotechnol. 2022, 42, 102337. [Google Scholar] [CrossRef]
- Beier, R.C.; Byrd, J.A.; Andrews, K.; Caldwell, D.; Crippen, T.L.; Anderson, R.C.; Nisbet, D.J. Disinfectant and antimicrobial susceptibility studies of the foodborne pathogen Campylobacter jejuni isolated from the litter of broiler chicken houses. Poult. Sci. 2021, 100, 1024–1033. [Google Scholar] [CrossRef]
- Trubenová, B.; Roizman, D.; Moter, A.; Rolff, J.; Regoes, R.R. Population genetics, biofilm recalcitrance, and antibiotic resistance evolution. Trends Microbiol. 2022, 30, 841–852. [Google Scholar] [CrossRef]
- Nunez, C.; Kostoulias, X.; Peleg, A.Y.; Short, F.; Qu, Y. A comprehensive comparison of biofilm formation and capsule production for bacterial survival on hospital surfaces. Biofilm 2023, 5, 100105. [Google Scholar] [CrossRef]
- Thaarup, I.C.; Iversen, A.K.S.; Lichtenberg, M.; Bjarnsholt, T.; Jakobsen, T.H. Biofilm survival strategies in chronic wounds. Microorganisms 2022, 10, 775. [Google Scholar] [CrossRef]
- Gloag, E.S.; Fabbri, S.; Wozniak, D.J.; Stoodley, P. Biofilm mechanics: Implications in infection and survival. Biofilm 2020, 2, 100017. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Rossman, M.L.; Pawar, D.M. Attachment of enterohemorrhagic Escherichia coli to the surface of beef and a culture medium. LWT-Food Sci. Technol. 2007, 40, 249–254. [Google Scholar] [CrossRef]
- Frank, J.F.; Koffi, R.A. Surface-adherent growth of Listeria monocytogenes is associated with increased resistance to surfactant sanitizers and heat. J. Food Prot. 1990, 53, 550–554. [Google Scholar] [CrossRef] [PubMed]
- Jessen, B.; Lammert, L. Biofilm and disinfection in meat processing plants. Int. Biodeterior. Biodegrad. 2003, 51, 265–269. [Google Scholar] [CrossRef]
- Somers, E.B.; Wong, A.C.L. Efficacy of two cleaning and sanitizing combinations on Listeria monocytogenes biofilms formed at low temperature on a variety of materials in the presence of ready-to-eat meat residue. J. Food Prot. 2004, 67, 2218–2229. [Google Scholar] [CrossRef] [PubMed]
- Duguid, J.P.; Anderson, E.S.; Campbell, I. Fimbriae and adhesive properties in Salmonellae. J. Pathol. Bacteriol. 1966, 92, 107–137. [Google Scholar] [CrossRef] [PubMed]
- Frank, J.F.; Ehlers, J.; Wicker, L. Removal of Listeria monocytogenes and poultry soil-containing biofilms using chemical cleaning and sanitizing agents under static conditions. Food Prot. Trends 2003, 23, 654–663. [Google Scholar]
- Herald, P.J.; Zottola, E.A. Attachment of Listeria monocytogenes to stainless steel surfaces at various temperatures and pH values. J. Food Sci. 1988, 53, 1549–1562. [Google Scholar] [CrossRef]
- Mafu, A.A.; Roy, D.; Goulet, J.; Magny, P. Attachment of Listeria monocytogenes to stainless steel, glass, polypropylene, and rubber surfaces after short contact times. J. Food Prot. 1990, 53, 742–746. [Google Scholar] [CrossRef]
- Duze, S.T.; Marimani, M.; Patel, M. Tolerance of Listeria monocytogenes to biocides used in food processing environments. Food Microbiol. 2021, 97, 103758. [Google Scholar] [CrossRef]
- Herald, P.J.; Zottola, E.A. Scanning electron microscopic examination of Yersinia enterocolitica attached to stainless steel at selected temperatures and pH values. J. Food Prot. 1988, 51, 445–448. [Google Scholar] [CrossRef] [PubMed]
- Kuusela, P.; Moran, A.P.; Vartio, T.; Kosunen, T.U. Interaction of Campylobacter jejuni with extracellular matrix components. Biochim. Biophys. Acta (BBA)-Gen. Subj. 1989, 993, 297–300. [Google Scholar] [CrossRef]
- Xu, J.G.; Meng, J.; Bao, W.J.; Kang, J.M.; Chen, J.Y.; Han, B.Z. Occurrence of disinfectant-resistant bacteria in a fresh-cut vegetables processing facility and their role in protecting Salmonella enteritidis. RSC Adv. 2021, 11, 10291–10299. [Google Scholar] [CrossRef] [PubMed]
- Puligundla, P.; Lim, S. Biocontrol approaches against Escherichia coli O157: H7 in foods. Foods 2022, 11, 756. [Google Scholar] [CrossRef] [PubMed]
- Dewanti, R.; Wong, A.C. Influence of culture conditions on biofilm formation by Escherichia coli O157: H7. Int. J. Food Microbiol. 1995, 26, 147–164. [Google Scholar] [CrossRef] [PubMed]
- Giaouris, E.; Simões, M.; Dubois-Brissonnet, F. The role of biofilms in the development and dissemination of microbial resistance within the food industry. Foods 2020, 9, 816. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, P.; Allison, D.G.; McBain, A.J. Biofilms in vitro and in vivo: Do singular mechanisms imply cross-resistance? J. Appl. Microbiol. 2002, 92, 98S–110S. [Google Scholar] [CrossRef]
- Fagerlund, A.; Langsrud, S.; Heir, E.; Mikkelsen, M.I.; Møretrø, T. Biofilm matrix composition affects the susceptibility of food associated staphylococci to cleaning and disinfection agents. Front. Microbiol. 2016, 7, 856. [Google Scholar] [CrossRef]
- Demeter, M.A.; Lemire, J.A.; Turner, R.J.; Harrison, J.J. Biofilm Survival Strategies in Polluted Environments. In Biofilms in Bioremediation: Current Research and Emerging Technologies; Caister Academic Press: Poole, UK, 2016; pp. 43–56. ISBN 9781910190296. [Google Scholar]
- Razdan, K.; Garcia-Lara, J.; Sinha, V.R.; Singh, K.K. Pharmaceutical strategies for the treatment of bacterial biofilms in chronic wounds. Drug Discov. Today 2022, 27, 2137–2150. [Google Scholar] [CrossRef]
- Pragman, A.A.; Berger, J.P.; Williams, B.J. Understanding persistent bacterial lung infections: Clinical implications informed by the biology of the microbiota and biofilms. Clin. Pulm. Med. 2016, 23, 57. [Google Scholar] [CrossRef]
- Harrison, J.J.; Turner, R.J.; Marques, L.L.R.; Ceri, H. Biofilms: A new understanding of these microbial communities is driving a revolution that may transform the science of microbiology. Am. Sci. 2005, 93, 508–515. [Google Scholar] [CrossRef]
- Harding, M.W.; Marques, L.L.; Howard, R.J.; Olson, M.E. Can filamentous fungi form biofilms? Trends Microbiol. 2009, 17, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.Y.E.; Chew, S.C.; Tan, S.Y.Y.; Givskov, M.; Yang, L. Emerging frontiers in detection and control of bacterial biofilms. Curr. Opin. Biotechnol. 2014, 26, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Gkana, E.N.; Doulgeraki, A.I.; Chorianopoulos, N.G.; Nychas, G.J.E. Anti-adhesion and anti-biofilm potential of organosilane nanoparticles against foodborne pathogens. Front. Microbiol. 2017, 8, 1295. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.M.; Lim, E.S.; Kim, J.S.; Paik, H.D.; Koo, O.K. Survival of foodborne pathogens on stainless steel soiled with different food residues. Food Sci. Biotechnol. 2020, 29, 729–737. [Google Scholar] [CrossRef] [PubMed]
- Harding, M.W.; Howard, R.J.; Daniels, G.D.; Mobbs, S.L.; Lisowski, S.L.I.; Allan, N.D.; Omar, A.; Olson, M.E. A multi-well plate method for rapid growth, characterization and biocide sensitivity testing of microbial biofilms on various surface materials. In Science against Microbial Pathogens: Communicating Current Research and Technological Advances; Formatex: Badajoz, Spain, 2011; pp. 872–877. [Google Scholar]
- Jang, Y.; Lee, K.; Yun, S.; Lee, M.; Song, J.; Chang, B.; Choe, N.H. Efficacy evaluation of commercial disinfectants by using Salmonella enterica serovar Typhimurium as a test organism. J. Vet. Sci. 2017, 18, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.J.; Lee, N.R.; Jo, W.; Jung, W.K.; Lim, J.S. Effects of substrates on biofilm formation observed by atomic force microscopy. Ultramicroscopy 2009, 109, 874–880. [Google Scholar] [CrossRef]
- Song, F.; Koo, H.; Ren, D. Effects of material properties on bacterial adhesion and biofilm formation. J. Dent. Res. 2015, 94, 1027–1034. [Google Scholar] [CrossRef]
- Feng, G.; Cheng, Y.; Wang, S.Y.; Borca-Tasciuc, D.A.; Worobo, R.W.; Moraru, C.I. Bacterial attachment and biofilm formation on surfaces are reduced by small-diameter nanoscale pores: How small is small enough? Npj Biofilms Microbiomes 2015, 1, 15022. [Google Scholar] [CrossRef]
- Wang, R.; Zhou, Y.; Kalchayanand, N.; Harhay, D.M.; Wheeler, T.L. Consecutive treatments with a multicomponent sanitizer inactivate biofilms formed by Escherichia coli O157: H7 and Salmonella enterica and remove biofilm matrix. J. Food Prot. 2021, 84, 408–417. [Google Scholar] [CrossRef]
- Peterson, S.B.; Irie, Y.; Borlee, B.R.; Murakami, K.; Harrison, J.J.; Colvin, K.M.; Parsek, M.R. Different methods for culturing biofilms in vitro. In Biofilm Infections; Springer: New York, NY, USA, 2011; pp. 251–266. [Google Scholar]
- Harding, M.W.; Olson, M.E.; Marques, L.L.R.; Howard, R.J. Biology and management of microbial biofilms on plant surfaces. In Proceedings of the International Symposium on Biological Control of Postharvest Diseases: Challenges and Opportunities, Leesburg, VA, USA, 25–28 October 2010; 905, pp. 43–55. [Google Scholar]
- Harding, M.W.; Daniels, G.C. In Vitro Assessment of Biofilm Formation by Soil-and Plant-Associated Microorganisms. In Biofilms in Plant and Soil Health; Wiley: Hoboken, NJ, USA, 2017; pp. 253–273. [Google Scholar]
- Omar, A.; Nadworny, P. Antimicrobial efficacy validation using in vitro and in vivo testing methods. Adv. Drug Deliv. Rev. 2017, 112, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Ceri, H.; Olson, M.E.; Stremick, C.; Read, R.R.; Morck, D.; Buret, A. The Calgary Biofilm Device: New technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 1999, 37, 1771–1776. [Google Scholar] [CrossRef] [PubMed]
- Mulla, S.; Kumar, A.; Rajdev, S. Comparison of MIC with MBEC assay for in vitro antimicrobial susceptibility testing in biofilm forming clinical bacterial isolates. Adv. Microbiol. 2016, 6, 73. [Google Scholar] [CrossRef]
- Parker, A.E.; Walker, D.K.; Goeres, D.M.; Allan, N.; Olson, M.E.; Omar, A. Ruggedness and reproducibility of the MBEC biofilm disinfectant efficacy test. J. Microbiol. Methods 2014, 102, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Harding, M.; Nadworny, P.; Buziak, B.; Omar, A.; Daniels, G.; Feng, J. Improved methods for treatment of phytopathogenic biofilms: Metallic compounds as anti-bacterial coatings and fungicide tank-mix partners. Molecules 2019, 24, 2312. [Google Scholar] [CrossRef] [PubMed]
- Howard, R.J.; Harding, M.W.; Daniels, G.C.; Mobbs, S.L.; Lisowski, S.L.I.; De Boer, S.H. Efficacy of agricultural disinfectants on biofilms of the bacterial ring rot pathogen, Clavibacter michiganensis subsp. sepedonicus. Can. J. Plant Pathol. 2015, 37, 273–284. [Google Scholar] [CrossRef]
- Harding, M.W.; Daniels, G.D.; Howard, R.J.; Marques, L.L.R.; Allan, N.D.; Omar, A.; Olson, M.E. In vitro evaluations of microbial biofilms and their responses to chemical disinfectants. Acta Hortic. 2014, 1053, 245–255. [Google Scholar] [CrossRef]
- Allan, N.D.; Omar, A.; Harding, M.W.; Olson, M.E. A rapid, high-throughput method for culturing, characterizing and biocide efficacy testing of both planktonic cells and biofilms. In Science against Microbial Pathogens: Communicating Current Research and Technological Advances; Microbiology Book Series; Mendez-Vilas, A., Ed.; Formatex: Badajoz, Spain, 2011; pp. 864–871. [Google Scholar]
- Corcoran, M.; Morris, D.; De Lappe, N.; O’connor, J.; Lalor, P.; Dockery, P.; Cormican, M. Commonly used disinfectants fail to eradicate Salmonella enterica biofilms from food contact surface materials. Appl. Environ. Microbiol. 2014, 80, 1507–1514. [Google Scholar] [CrossRef]
- Giaouris, E.; Heir, E.; Desvaux, M.; Hébraud, M.; Møretrø, T.; Langsrud, S.; Doulgeraki, A.; Nychas, G.J.; Kačániová, M.; Czaczyk, K.; et al. Intra-and inter-species interactions within biofilms of important foodborne bacterial pathogens. Front. Microbiol. 2015, 6, 841. [Google Scholar] [CrossRef]
- Zheng, S.; Bawazir, M.; Dhall, A.; Kim, H.E.; He, L.; Heo, J.; Hwang, G. Implication of surface properties, bacterial motility, and hydrodynamic conditions on bacterial surface sensing and their initial adhesion. Front. Bioeng. Biotechnol. 2021, 9, 643722. [Google Scholar] [CrossRef]
- Alonso, V.P.P.; Gonçalves, M.P.M.; de Brito, F.A.E.; Barboza, G.R.; Rocha, L.D.O.; Silva, N.C.C. Dry surface biofilms in the food processing industry: An overview on surface characteristics, adhesion and biofilm formation, detection of biofilms, and dry sanitization methods. Compr. Rev. Food Sci. Food Saf. 2023, 22, 688–713. [Google Scholar] [CrossRef] [PubMed]
- Bryers, J.D. Biologically active surfaces: Processes governing the formation and persistence of biofilms. Biotechnol. Prog. 1987, 3, 57–68. [Google Scholar] [CrossRef]
- Boulange-Petermann, L.; Baroux, B.; Bellon-Fontaine, M.N. The influence of metallic surface wettability on bacterial adhesion. J. Adhes. Sci. Technol. 1993, 7, 221–230. [Google Scholar] [CrossRef]
- McEldowney, S.; Fletcher, M. Adhesion of bacteria from mixed cell suspension to solid surfaces. Arch. Microbiol. 1987, 148, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Baker, J.H. Factors affecting the bacterial colonization of various surfaces in a river. Can. J. Microbiol. 1984, 30, 511–515. [Google Scholar] [CrossRef]
- Bos, R.; van der Mei, H.; Busscher, H. Physico-chemistry of initial microbial adhesive interactions—Its mechanisms and methods for study. FEMS Microbiol. Rev. 1999, 23, 179–230. [Google Scholar] [CrossRef]
- Characklis, W.G.; McFeters, G.A.; Marshall, K.C. Physiological ecology in biofilm systems. In Biofilms; John Wiley & Sons: New York, NY, USA, 1990; pp. 341–394. [Google Scholar]
- Jones, C.R.; Adams, M.R.; Zhdan, P.A.; Chamberlain, A.H.L. The role of surface physicochemical properties in determining the distribution of the autochthonous microflora in mineral water bottles. J. Appl. Microbiol. 1999, 86, 917–927. [Google Scholar] [CrossRef]
- Sasahara, K.C.; Zottola, E.A. Biofilm formation by Listeria monocytogenes utilizes a primary colonizing microorganism in flowing systems. J. Food Prot. 1993, 56, 1022–1028. [Google Scholar] [CrossRef]
- EPA Technical Guidance Manual. Disinfection Profiling and Benchmarking. Office of Water (4606M) EPA 815-R-20-003, June. 2020. Available online: https://www.epa.gov/system/files/documents/2022-02/disprof_bench_3rules_final_508.pdf (accessed on 15 August 2023).
- Rutala, W.A.; Weber, D.J. CDC Guideline for Disinfection and Sterilization in Healthcare Facilities. 2019. Available online: https://www.cdc.gov/infectioncontrol/pdf/guidelines/disinfection-guidelines-H.pdf (accessed on 15 August 2023).
- FSIS Cooking Guideline for Meat and Poultry Products (Revised Appendix A); Document ID: FSIS-GD-2021-14; USDA: Washington DC, USA, 2021. Available online: https://www.fsis.usda.gov/sites/default/files/media_file/2021-12/Appendix-A.pdf (accessed on 15 August 2023).
- Aviat, F.; Gerhards, C.; Rodriguez-Jerez, J.J.; Michel, V.; Bayon, I.L.; Ismail, R.; Federighi, M. Microbial safety of wood in contact with food: A review. Compr. Rev. Food Sci. Food Saf. 2016, 15, 491–505. [Google Scholar] [CrossRef]


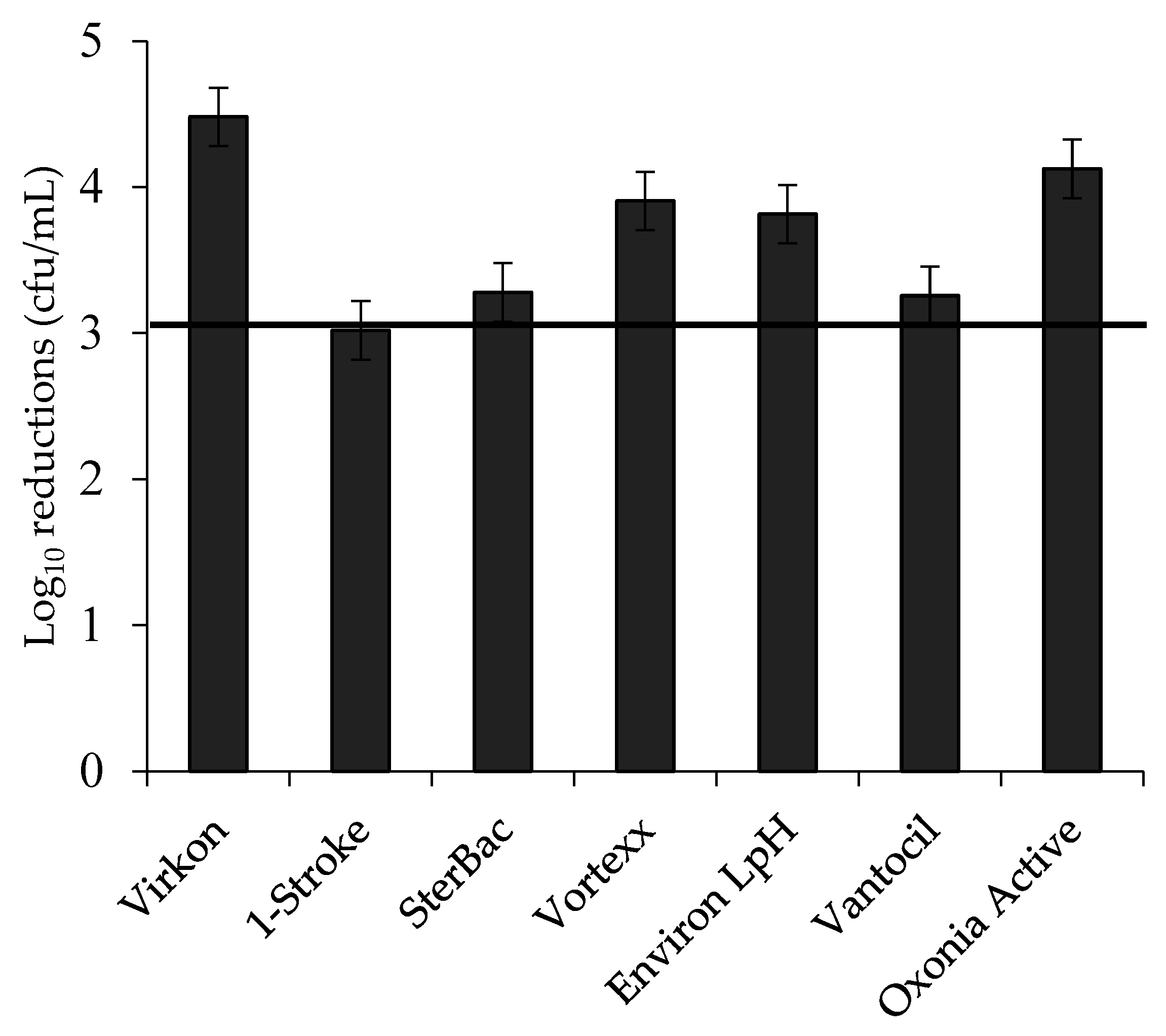
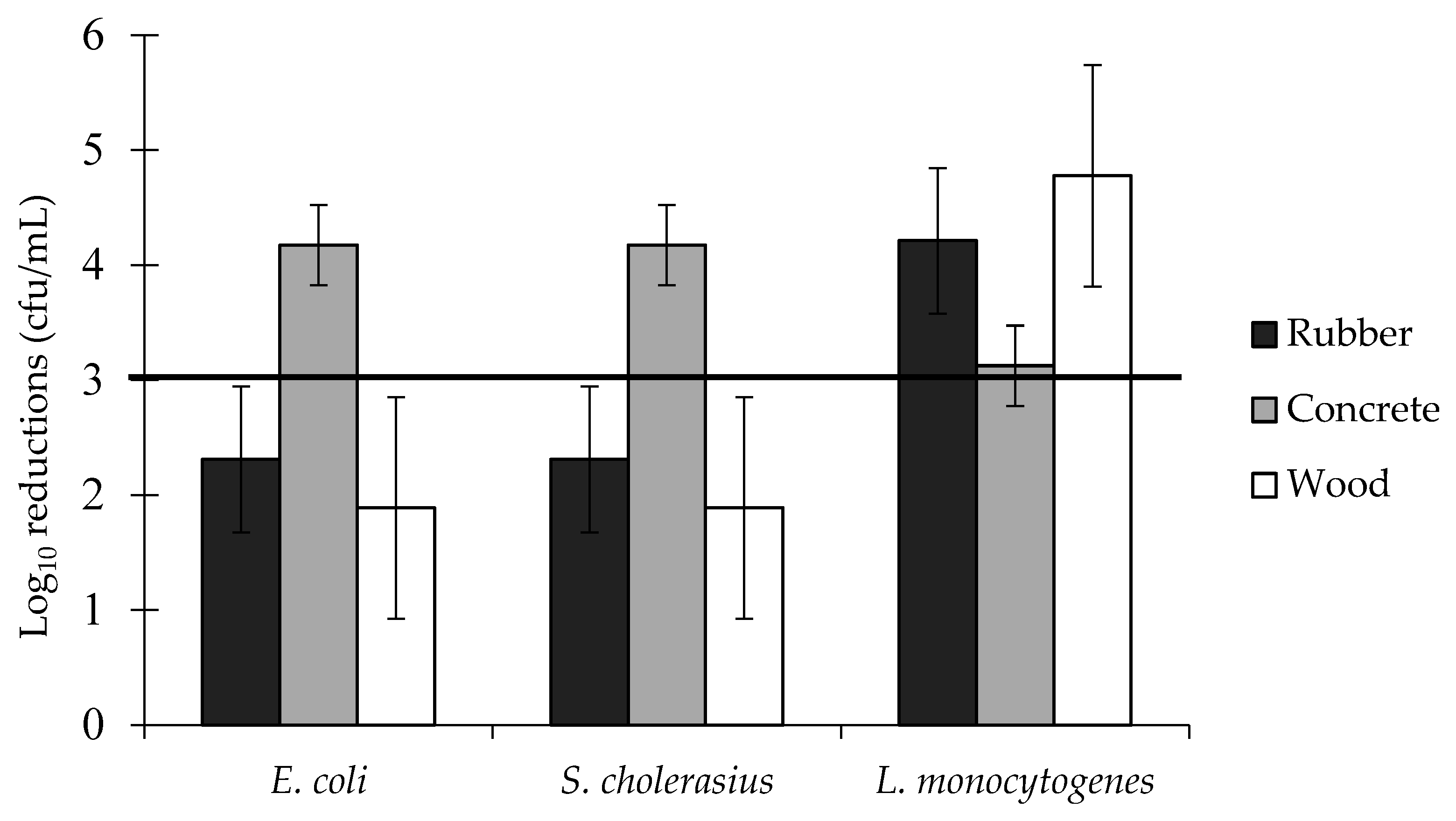

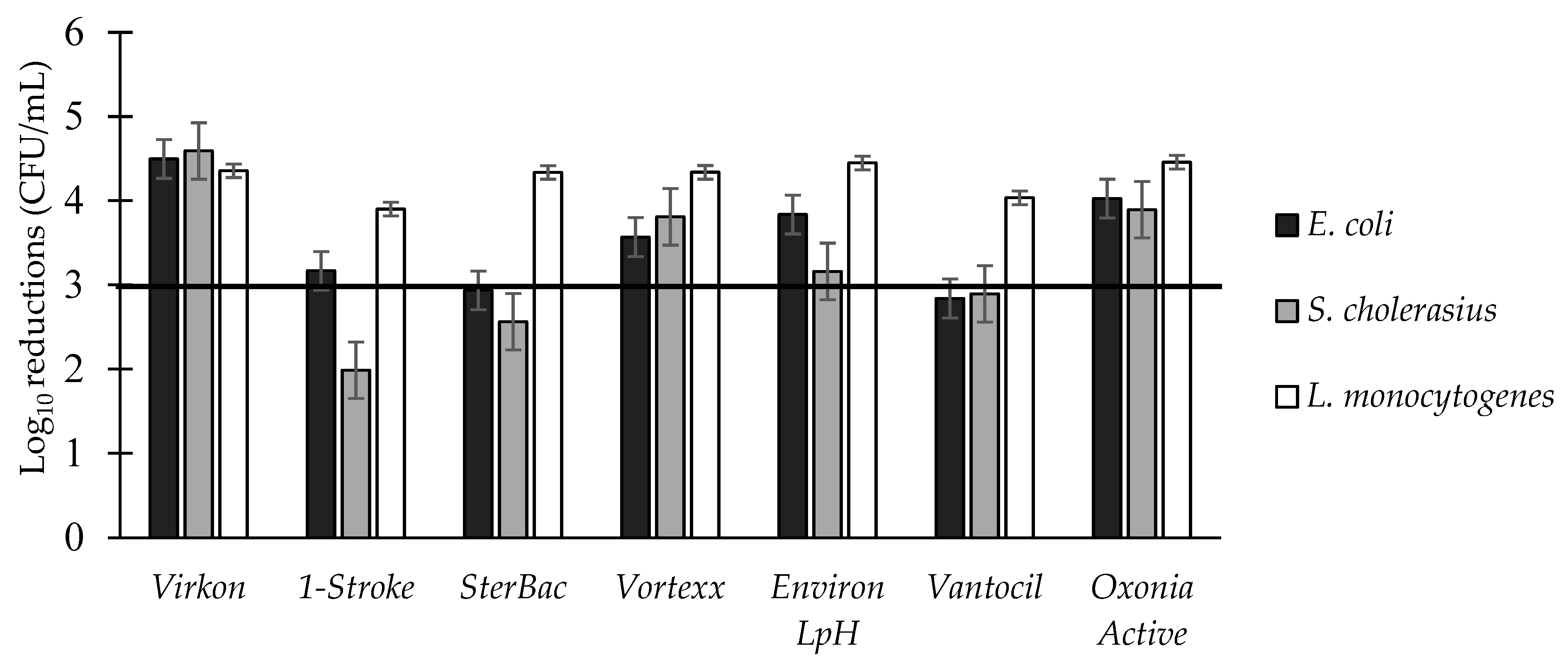
| Disinfectant | Family | Active Ingredients | Conc. Used | Conc. on Label |
|---|---|---|---|---|
| Virkon™ | Oxidizing agents | Potassium peroxymonosulfate 21.4% | 1.5% w/v | 1.5% w/v |
| 1-Stroke | Phenols | ortho-phenylphenol 10%, ortho bezyl-para-cholorophenol 8.5% | 0.8% v/v | 0.4% v/v |
| Environ® LpH | Phenols | o-Benzyl-p-chlorophenol 6.4%, p-tertiary-amylphenol 3.0%, o-phenyl phenol 0.5%, hexylene glycol 4.0%, glycolic acid (hydroxyacetic) 12.6%, isopropanol 8.0% | 1.56% v/v | 0.78% v/v |
| Vantocil™ | PolyHydrochloride | Polyhexamethylene biguanide | 0.6% v/v | 0.3% v/v |
| Vortexx | Peroxides | Caprylic acid 3%, hydrogen peroxide 6.9%, peroxyacetic acid 4.4% | 0.5% v/v | 0.25% v/v |
| Oxonia Active™ | Peroxides | Hydrogen peroxide 15–40%, acetic acid 7–13%, peracetic acid 5–10% | 0.6% v/v | 0.3% v/v |
| SterBac® | Quaternary ammonium | Benzyl-c12-c16-alkyldimethyl, chlorides 7–13%, ethanol 1–5% | 0.4% v/v | 0.2% v/v |
| Surface | Bacteria Species | Mean | ||
|---|---|---|---|---|
| E. coli 0157H7 | S. cholerasius | L. monocytogenes | ||
| Stainless steel | 4.85 | 5.01 | 4.96 | 4.94 |
| Mild steel | 5.76 | 6.03 | 4.93 | 5.58 |
| Wood | 5.40 | 5.92 | 6.28 | 5.87 |
| Laminated plastic | 5.01 | 3.86 | 6.87 | 5.25 |
| UHMWP | 4.23 | 4.65 | 5.49 | 4.79 |
| Rubber | 5.21 | 5.20 | 6.56 | 5.66 |
| Concrete | 4.90 | 5.26 | 6.75 | 5.64 |
| Silicon | 4.07 | 4.01 | 6.59 | 4.89 |
| Mean | 4.93 | 4.99 | 6.05 | 5.32 |
| p value | LSD (p = 0.05) | |||
| Surface | 0.000 *** | 0.14 | ||
| Bacteria | 0.000 *** | 0.08 | ||
| Surface × Bacterial species interaction | 0.000 *** | 0.24 | ||
| CV% = 2.7 | ||||
| Surface | Biocide | Bacteria Species | Mean | ||
|---|---|---|---|---|---|
| E. coli 0157:H7 | S. cholerasius | L. monocytogenes | |||
| Stainless steel | Control Recovery ** | (4.47) | (3.59) | (4.51) | (4.19) |
| Virkon | 4.47 | 3.59 | 4.51 | 4.19 | |
| 1-Stroke | 4.47 | 3.59 | 4.51 | 4.19 | |
| SterBac | 4.47 | 3.59 | 4.51 | 4.19 | |
| Vortexx | 4.47 | 3.59 | 4.51 | 4.19 | |
| Environ LpH | 4.47 | 3.59 | 4.51 | 4.19 | |
| Vantocil | 4.47 | 3.59 | 4.51 | 4.19 | |
| Oxonia Active | 4.47 | 3.59 | 4.51 | 4.19 | |
| Mean | 4.47 | 3.59 | 4.51 | 4.19 | |
| Mild steel | Control Recovery ** | (4.60) | (5.07) | (4.57) | (4.75) |
| Virkon | 4.60 | 5.07 | 4.57 | 4.75 | |
| 1-Stroke | 1.35 * | 0.34 * | 4.57 | 2.09 * | |
| SterBac | 1.32 * | 2.64 * | 4.57 | 2.84 * | |
| Vortexx | 4.60 | 4.40 | 4.57 | 4.52 | |
| Environ LpH | 4.60 | 3.93 | 4.57 | 4.37 | |
| Vantocil | 4.60 | 4.34 | 4.57 | 4.50 | |
| Oxonia Active | 4.60 | 5.07 | 4.57 | 4.75 | |
| Mean | 3.67 | 3.69 | 4.57 | 3.97 | |
| Rubber | Control Recovery ** | (4.65) | (5.15) | (4.70) | (4.83) |
| Virkon | 4.08 | 5.15 | 4.70 | 4.64 | |
| 1-Stroke | 2.55 * | 0.14 * | 1.95 * | 1.55 * | |
| SterBac | 2.18 * | 1.05 * | 4.70 | 2.64 * | |
| Vortexx | 4.65 | 2.32 * | 4.70 | 3.89 | |
| Environ LpH | 4.08 | 1.23 * | 4.70 | 3.34 | |
| Vantocil | 2.95 * | 1.60 * | 4.70 | 3.08 | |
| Oxonia Active | 4.65 | 4.58 | 4.03 | 4.42 | |
| Mean | 3.59 | 2.29 * | 4.21 | 3.37 | |
| Concrete | Control Recovery ** | (5.45) | (6.29) | (4.55) | (5.43) |
| Virkon | 5.45 | 4.87 | 2.97 | 4.43 | |
| 1-Stroke | 5.45 | 2.99 | 4.55 | 4.33 | |
| SterBac | 5.45 | 3.15 | 3.31 | 3.97 | |
| Vortexx | 5.45 | 6.29 | 4.55 | 5.43 | |
| Environ LpH | 5.45 | 2.79 * | 2.41 * | 3.55 | |
| Vantocil | 5.45 | 2.83 * | 0.93 * | 3.07 | |
| Oxonia Active | 5.45 | 6.29 | 3.16 | 4.97 | |
| Mean | 5.45 | 4.17 | 3.13 | 4.25 | |
| Wood | Control Recovery ** | (5.35) | (5.90) | (5.21) | (5.49) |
| Virkon | 5.35 | 5.90 | 5.21 | 5.49 | |
| 1-Stroke | 1.43 * | 0.34 * | 5.21 | 2.33 * | |
| SterBac | 0.12 * | 0.62 * | 5.21 | 1.98 * | |
| Vortexx | 0.29 * | 1.73 * | 2.18 * | 1.40 * | |
| Environ LpH | 2.15 * | 1.60 * | 5.21 | 2.99 | |
| Vantocil | 0.02 * | 0.90 * | 5.21 | 2.05 * | |
| Oxonia Active | 2.86 * | 2.12 * | 5.21 | 3.40 | |
| Mean | 1.75 * | 1.89 * | 4.78 | 2.84 * | |
| Laminated plastic | Control Recovery ** | (4.73) | (4.61) | (4.47) | (4.60) |
| Virkon | 4.73 | 4.61 | 4.47 | 4.60 | |
| 1-Stroke | 1.42 * | 1.55 * | 3.62 | 2.20 * | |
| SterBac | 1.80 * | 1.79 * | 2.67 * | 2.09 * | |
| Vortexx | 4.16 | 4.61 | 4.47 | 4.41 | |
| Environ LpH | 2.61 * | 4.61 | 4.47 | 3.90 | |
| Vantocil | 4.06 | 2.35 * | 4.47 | 3.63 | |
| Oxonia Active | 4.73 | 1.96 * | 4.47 | 3.72 | |
| Mean | 3.36 | 3.07 | 4.09 | 3.50 | |
| UHMWP | Control Recovery ** | (3.89) | (4.15) | (4.66) | (4.23) |
| Virkon | 3.89 | 4.15 | 4.66 | 4.23 | |
| 1-Stroke | 3.89 | 4.15 | 4.66 | 4.23 | |
| SterBac | 3.32 | 4.15 | 4.66 | 4.04 | |
| Vortexx | 3.89 | 4.15 | 4.66 | 4.23 | |
| Environ LpH | 3.89 | 4.15 | 4.66 | 4.23 | |
| Vantocil | 0.26 * | 4.15 | 4.66 | 3.02 | |
| Oxonia Active | 3.89 | 4.15 | 4.66 | 4.23 | |
| Mean | 3.29 | 4.15 | 4.66 | 4.03 | |
| Silicon | Control Recovery ** | (4.78) | (3.39) | (5.06) | (4.41) |
| Virkon | 3.41 | 3.39 | 3.76 | 3.52 | |
| 1-Stroke | 4.78 | 2.82 * | 2.15 * | 3.25 | |
| SterBac | 4.78 | 3.39 | 5.06 | 4.41 | |
| Vortexx | 1.05 * | 3.39 | 5.06 | 3.17 | |
| Environ LpH | 3.44 | 3.39 | 5.06 | 3.96 | |
| Vantocil | 0.93 * | 3.39 | 3.23 | 2.52 * | |
| Oxonia Active | 1.58 * | 3.39 | 5.06 | 3.34 | |
| Mean | 2.85 * | 3.31 | 4.20 | 3.45 | |
| p value | LSD p = 0.05 | ||||
| Surface | 0.000 *** | 0.18 | |||
| Bacterial species | 0.000 *** | 0.11 | |||
| Biocide | 0.000 *** | 0.17 | |||
| Surface × Biocide interaction | 0.000 *** | 0.31 | |||
| Bacterial species × Biocide interaction | 0.000 *** | 0.47 | |||
| Surface × Bacterial species interaction | 0.000 *** | 0.29 | |||
| Surface × Bacterial species × Biocide interaction | 0.000 *** | 0.81 | |||
| CV% = 13.7% | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kundu, M.; Omar, A.; Buziak, B.; Allan, N.; Marques, L.; Olson, M.; Howard, R.; Harding, M.W. Customizing Sanitization Protocols for Food-Borne Pathogens Based on Biofilm Formation, Surfaces and Disinfectants—Their Two- and Three-Way Interactions. Appl. Microbiol. 2024, 4, 27-46. https://doi.org/10.3390/applmicrobiol4010003
Kundu M, Omar A, Buziak B, Allan N, Marques L, Olson M, Howard R, Harding MW. Customizing Sanitization Protocols for Food-Borne Pathogens Based on Biofilm Formation, Surfaces and Disinfectants—Their Two- and Three-Way Interactions. Applied Microbiology. 2024; 4(1):27-46. https://doi.org/10.3390/applmicrobiol4010003
Chicago/Turabian StyleKundu, Manju, Amin Omar, Brenton Buziak, Nick Allan, Lyriam Marques, Merle Olson, Ronald Howard, and Michael W. Harding. 2024. "Customizing Sanitization Protocols for Food-Borne Pathogens Based on Biofilm Formation, Surfaces and Disinfectants—Their Two- and Three-Way Interactions" Applied Microbiology 4, no. 1: 27-46. https://doi.org/10.3390/applmicrobiol4010003
APA StyleKundu, M., Omar, A., Buziak, B., Allan, N., Marques, L., Olson, M., Howard, R., & Harding, M. W. (2024). Customizing Sanitization Protocols for Food-Borne Pathogens Based on Biofilm Formation, Surfaces and Disinfectants—Their Two- and Three-Way Interactions. Applied Microbiology, 4(1), 27-46. https://doi.org/10.3390/applmicrobiol4010003








