Antimicrobial Resistance Profile of Planctomycetota Isolated from Oyster Shell Biofilm: Ecological Relevance within the One Health Concept
Abstract
1. Introduction
2. Material and Methods
2.1. Sampling
2.2. Isolation of Planctomycetota from Oysters
2.2.1. Isolation of Planctomycetota from Surface Biofilm
2.2.2. Isolation of Planctomycetota from the Enrichment of the Edible Content
2.3. Amplification and Sequencing Analysis of 16S rRNA Gene and Bacterial Phylogenetic Analysis
2.4. Antimicrobial Susceptibility Testing
3. Results and Discussion
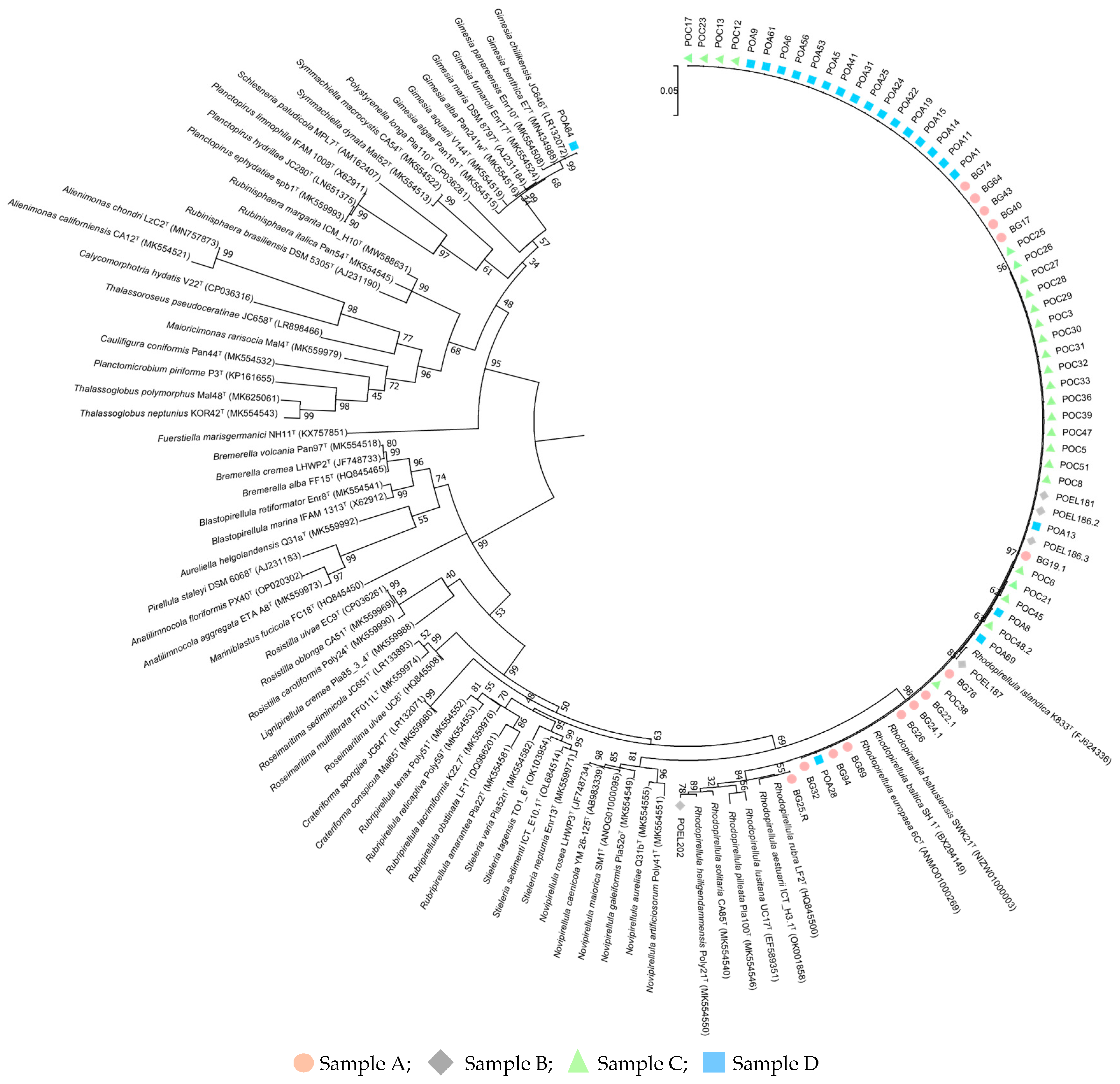
3.1. Planctomycetota Isolation
3.2. Antimicrobial Susceptibility Patterns of Planctomycetota
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wagner, M.; Horn, M. The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr. Opin. Biotechnol. 2006, 17, 241–249. [Google Scholar] [CrossRef]
- Wiegand, S.; Jogler, M.; Jogler, C. On the maverick Planctomycetes. FEMS Microbiol. Rev. 2018, 42, 739–760. [Google Scholar] [CrossRef] [PubMed]
- Lage, O.M.; van Niftrik, L.; Jogler, C.; Devos, D.P. Planctomycetes. In Encyclopedia of Microbiology, 4th ed.; Schmidt, T.M., Ed.; Academic Press: Oxford, UK, 2019; pp. 614–626. [Google Scholar]
- Mahajan, M.; Seeger, C.; Yee, B.; Andersson, S.G.E. Evolutionary remodelling of the cell envelope in bacteria of the Planctomycetes phylum. Genome Biol. Evol. 2020, 12, 1528–1548. [Google Scholar] [CrossRef] [PubMed]
- Lage, O.M.; Bondoso, J. Bringing Planctomycetes into pure culture. Front. Microbiol. 2012, 3, 405. [Google Scholar] [CrossRef] [PubMed]
- Godinho, O.; Calisto, R.; Øvreås, L.; Quinteira, S.; Lage, O.M. Antibiotic susceptibility of marine Planctomycetes. Antonie van Leeuwenhoek 2019, 112, 1273–1280. [Google Scholar] [CrossRef] [PubMed]
- Kuenen, J.G. Anammox bacteria: From discovery to application. Nat. Rev. Microbiol. 2008, 6, 320–326. [Google Scholar] [CrossRef]
- Vitorino, I.; Santos, J.D.N.; Godinho, O.; Vicente, F.; Vasconcelos, V.; Lage, O.M. Novel and conventional isolation techniques to obtain Planctomycetes from marine environments. Microorganisms 2021, 9, 2078. [Google Scholar] [CrossRef] [PubMed]
- Buckley, D.H.; Huangyutitham, V.; Nelson, T.A.; Rumberger, A.; Thies, J.E. Diversity of Planctomycetes in soil in relation to soil history and environmental heterogeneity. Appl. Environ. Microbiol. 2006, 72, 4522–4531. [Google Scholar] [CrossRef]
- Kulichevskaya, I.; Pankratov, T.; Dedysh, S. Detection of representatives of the Planctomycetes in Sphagnum peat bogs by molecular and cultivation approaches. Microbiology 2006, 75, 329–335. [Google Scholar] [CrossRef]
- Wang, J.; Jenkins, C.; Webb, R.I.; Fuerst, J.A. Isolation of Gemmata-like and Isosphaera-like planctomycete bacteria from soil and freshwater. Appl. Environ. Microbiol. 2002, 68, 417–422. [Google Scholar] [CrossRef]
- Storesund, J.E.; Øvreås, L. Diversity of Planctomycetes in iron-hydroxide deposits from the Arctic Mid Ocean Ridge (AMOR) and description of Bythopirellula goksoyri gen. nov., sp. nov., a novel Planctomycete from deep sea iron-hydroxide deposits. Antonie Van Leeuwenhoek 2013, 104, 569–584. [Google Scholar] [CrossRef] [PubMed]
- Elcheninov, A.G.; Podosokorskaya, O.A.; Kovaleva, O.L.; Novikov, A.A.; Toshchakov, S.V.; Bonch-Osmolovskaya, E.A.; Kublanov, I.V. Thermogemmata fonticola gen. nov., sp. nov., the first thermophilic planctomycete of the order Gemmatales from a Kamchatka hot spring. Syst. Appl. Microbiol. 2021, 44, 126157. [Google Scholar] [CrossRef] [PubMed]
- Andrew, D.R.; Fitak, R.R.; Munguia-Vega, A.; Racolta, A.; Martinson, V.G.; Dontsova, K. Abiotic factors shape microbial diversity in Sonoran Desert soils. Appl. Environ. Microbiol. 2012, 78, 7527–7537. [Google Scholar] [CrossRef] [PubMed]
- Brümmer, I.H.M.; Fehr, W.; Wagner-Döbler, I. Biofilm community structure in polluted rivers: Abundance of dominant phylogenetic groups over a complete annual cycle. Appl. Environ. Microbiol. 2000, 66, 3078–3082. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chouari, R.; Paslier, D.L.; Daegelen, P.; Ginestet, P.; Weissenbach, J.; Sghir, A. Molecular evidence for novel Planctomycete diversity in a municipal wastewater treatment plant. Appl. Environ. Microbiol. 2003, 69, 7354–7363. [Google Scholar] [CrossRef] [PubMed]
- Pimentel-Elardo, S.; Wehrl, M.; Friedrich, A.B.; Jensen, P.R.; Hentschel, U. Isolation of Planctomycetes from Aplysina sponges. Aquat. Microb. Ecol. 2003, 33, 239–245. [Google Scholar] [CrossRef]
- Lage, O.M.; Bondoso, J. Planctomycetes and macroalgae, a striking association. Front. Microbiol. 2014, 5, 267. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.R.; Pop, M.; Deboy, R.T.; Eckburg, P.B.; Turnbaugh, P.J.; Samuel, B.S.; Gordon, J.I.; Relman, D.A.; Fraser-Liggett, C.M.; Nelson, K.E. Metagenomic analysis of the human distal gut microbiome. Science 2006, 312, 1355–1359. [Google Scholar] [CrossRef]
- Costello, E.K.; Lauber, C.L.; Hamady, M.; Fierer, N.; Gordon, J.I.; Knight, R. Bacterial community variation in human body habitats across space and time. Science 2009, 326, 1694–1697. [Google Scholar] [CrossRef]
- Maldonado-Contreras, A.; Goldfarb, K.; Godoy-Vitorino, F.; Karaoz, U.; Contreras, M.; Blaser, M.J.; Brodie, E.L.; Dominguez-Bello, M.G. Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME J. 2011, 5, 574–579. [Google Scholar] [CrossRef]
- Cayrou, C.; Sambe, B.; Armougom, F.; Raoult, D.; Drancourt, M. Molecular diversity of the Planctomycetes in the human gut microbiota in France and Senegal. Apmis 2013, 121, 1082–1090. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Piquer, J.; Bowman, J.; Ross, T.; Tamplin, M. Molecular analysis of the bacterial communities in the live Pacific oyster (Crassostrea gigas) and the influence of postharvest temperature on its structure. J. Appl. Microbiol. 2012, 112, 1134–1143. [Google Scholar] [CrossRef] [PubMed]
- King, G.M.; Judd, C.; Kuske, C.R.; Smith, C. Analysis of stomach and gut microbiomes of the eastern oyster (Crassostrea virginica) from Coastal Louisiana, USA. PLoS ONE 2012, 7, e51475. [Google Scholar] [CrossRef] [PubMed]
- Pierce, M.L.; Ward, J.E. Gut microbiomes of the eastern oyster (Crassostrea virginica) and the blue mussel (Mytilus edulis): Temporal variation and the influence of marine aggregate-associated microbial communities. mSphere 2019, 4, e00730-19. [Google Scholar] [CrossRef] [PubMed]
- Clerissi, C.; de Lorgeril, J.; Petton, B.; Lucasson, A.; Escoubas, J.-M.; Gueguen, Y.; Dégremont, L.; Mitta, G.; Toulza, E. Microbiota composition and evenness predict survival rate of oysters confronted to Pacific Oyster mortality syndrome. Front. Microbiol. 2020, 11, 311. [Google Scholar] [CrossRef]
- Stevick, R.J.; Post, A.F.; Gómez-Chiarri, M. Functional plasticity in oyster gut microbiomes along a eutrophication gradient in an urbanized estuary. Anim. Microbiome 2021, 3, 5. [Google Scholar] [CrossRef]
- Unzueta-Martínez, A.; Welch, H.; Bowen, J.L. Determining the composition of resident and transient members of the oyster microbiome. Front. Microbiol. 2022, 12, 828692. [Google Scholar] [CrossRef]
- Liu, M.; Li, Q.; Tan, L.; Wang, L.; Wu, F.; Li, L.; Zhang, G. Host-microbiota interactions play a crucial role in oyster adaptation to rising seawater temperature in summer. Environ. Res. 2023, 216, 114585. [Google Scholar] [CrossRef]
- Pathak, A.; Stothard, P.; Chauhan, A. Comparative genomic analysis of three Pseudomonas species isolated from the eastern oyster (Crassostrea virginica) tissues, mantle fluid, and the overlying estuarine water column. Microorganisms 2021, 9, 490. [Google Scholar] [CrossRef]
- Guedes, B.; Godinho, O.; Lage, O.M.; Quinteira, S. Microbiological quality, antibiotic resistant bacteria and relevant re-sistance genes in ready-to-eat Pacific oysters (Magallana gigas). FEMS Microbiol. Lett. 2023, 370, fnad053. [Google Scholar] [CrossRef]
- Lage, O.M.; Bondoso, J. Planctomycetes diversity associated with macroalgae. FEMS Microbiol. Ecol. 2011, 78, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.J. 16S/23S rRNA Sequencing. In Nucleic Acid Techniques in Bacterial Systematic; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- DeLong, E.F.; Franks, D.G.; Alldredge, A.L. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol. Oceanogr. 1993, 38, 924–934. [Google Scholar] [CrossRef]
- Singh, P.; Williams, D.; Velez, F.J.; Nagpal, R. Comparison of the gill microbiome of retail oysters from two geographical locations exhibited distinct microbial signatures: A pilot study for potential future applications for monitoring authenticity of their origins. Appl. Microbiol. 2023, 3, 1–10. [Google Scholar] [CrossRef]
- Offret, C.; Paulino, S.; Gauthier, O.; Château, K.; Bidault, A.; Corporeau, C.; Miner, P.; Petton, B.; Pernet, F.; Fabioux, C.; et al. The marine intertidal zone shapes oyster and clam digestive bacterial microbiota. FEMS Microbiol. Ecol. 2020, 96, fiaa078. [Google Scholar] [CrossRef]
- Vitorino, I.R.; Lage, O.M. The biology of Planctomycetia: An overview of the currently largest class within the phylum Planctomycetes. Antonie Van Leeuwenhoek 2022, 115, 169–201. [Google Scholar] [CrossRef]
- Dugeny, E.; de Lorgeril, J.; Petton, B.; Toulza, E.; Gueguen, Y.; Pernet, F. Seaweeds influence oyster microbiota and disease susceptibility. J. Anim. Ecol. 2022, 91, 805–818. [Google Scholar] [CrossRef]
- Hieu, C.X.; Voigt, B.; Albrecht, D.; Becher, D.; Lombardot, T.; Glöckner, F.O.; Amann, R.; Hecker, M.; Schweder, T. Detailed proteome analysis of growing cells of the planctomycete Rhodopirellula baltica SH1T. Proteomics 2008, 8, 1608–1623. [Google Scholar] [CrossRef] [PubMed]
- Lage, O.M.; Bondoso, J.; Viana, F. Isolation and characterisation of Planctomycetes from the sediments of a fish farm wastewater treatment tank. Arch. Microbiol. 2012, 194, 879–885. [Google Scholar] [CrossRef] [PubMed]
- Kallscheuer, N.; Wiegand, S.; Jogler, M.; Boedeker, C.; Peeters, S.H.; Rast, P.; Heuer, A.; Jetten, M.S.M.; Rohde, M.; Jogler, C. Rhodopirellula heiligendammensis sp. nov., Rhodopirellula pilleata sp. nov., and Rhodopirellula solitaria sp. nov. isolated from natural or artificial marine surfaces in Northern Germany and California, USA, and emended description of the genus Rhodopirellula. Antonie van Leeuwenhoek 2020, 113, 1737–1750. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Gaurav, K.; Pk, S.; Uppada, J.; Ch., S.; Ch.V., R. Gimesia chilikensis sp. nov., a haloalkali-tolerant planctomycete isolated from Chilika lagoon and emended description of the genus Gimesia. Int. J. Syst. Evol. Microbiol. 2020, 70, 3647–3655. [Google Scholar] [CrossRef]
- Wiegand, S.; Jogler, M.; Boedeker, C.; Pinto, D.; Vollmers, J.; Rivas-Marín, E.; Kohn, T.; Peeters, S.H.; Heuer, A.; Rast, P.; et al. Cultivation and functional characterization of 79 planctomycetes uncovers their unique biology. Nat. Microbiol. 2020, 5, 126–140. [Google Scholar] [CrossRef] [PubMed]
- Cayrou, C.; Raoult, D.; Drancourt, M. Broad-spectrum antibiotic resistance of Planctomycetes organisms determined by Etest. J. Antimicrob. Chemother. 2010, 65, 2119–2122. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, A.A.; Miroshnikov, K.K.; Oshkin, I.Y. Exploring antibiotic susceptibility, resistome and mobilome structure of Planctomycetes from Gemmataceae family. Sustainability 2021, 13, 5031. [Google Scholar] [CrossRef]
- Jeske, O.; Schüler, M.; Schumann, P.; Schneider, A.; Boedeker, C.; Jogler, M.; Bollschweiler, D.; Rohde, M.; Mayer, C.; Engelhardt, H.; et al. Planctomycetes do possess a peptidoglycan cell wall. Nat. Commun. 2015, 6, 7116. [Google Scholar] [CrossRef]
- Aghnatios, R.; Drancourt, M. Gemmata species: Planctomycetes of medical interest. Future Microbiol. 2016, 11, 659–667. [Google Scholar] [CrossRef]
- Michalopoulos, A.S.; Livaditis, I.G.; Gougoutas, V. The revival of fosfomycin. Int. J. Infect. Dis. 2011, 15, e732–e739. [Google Scholar] [CrossRef]
- Blair, J.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Andrade, F.F.; Silva, D.; Rodrigues, A.; Pina-Vaz, C. Colistin update on its mechanism of action and resistance, present and future challenges. Microorganisms 2020, 8, 1716. [Google Scholar] [CrossRef] [PubMed]
- Mohapatra, S.S.; Dwibedy, S.K.; Padhy, I. Polymyxins, the last-resort antibiotics: Mode of action, resistance emergence, and potential solutions. J. Biosci. 2021, 46, 85. [Google Scholar] [CrossRef] [PubMed]
- Faria, M.; Bordin, N.; Kizina, J.; Harder, J.; Devos, D.; Lage, O.M. Planctomycetes attached to algal surfaces: Insight into their genomes. Genomics 2018, 110, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Graça, A.P.; Calisto, R.; Lage, O.M. Planctomycetes as novel source of bioactive molecules. Front. Microbiol. 2016, 7, 1241. [Google Scholar] [CrossRef]
- Gimranov, E.; Santos, J.; Vitorino, I.; Martín, J.; Reyes, F.; Moura, L.; Tavares, F.; Santos, C.; Mariz-Ponte, N.; Lage, O.M. Marine bacterial activity against phytopathogenic Pseudomonas show high efficiency of Planctomycetes extracts. Eur. J. Plant Pathol. 2022, 162, 843–854. [Google Scholar] [CrossRef]
- Vitorino, I.R.; Lobo-da-Cunha, A.; Vasconcelos, V.; Vicente, F.; Lage, O.M. Isolation, diversity and antimicrobial activity of Planctomycetes from the Tejo river estuary (Portugal). FEMS Microbiol. Ecol. 2022, 98, fiaa078. [Google Scholar] [CrossRef]
- Park, J.W.; Park, S.R.; Nepal, K.K.; Han, A.R.; Ban, Y.H.; Yoo, Y.J.; Kim, E.J.; Kim, E.M.; Kim, D.; Sohng, J.K.; et al. Discovery of parallel pathways of kanamycin biosynthesis allows antibiotic manipulation. Nat. Chem. Biol. 2011, 7, 843–852. [Google Scholar] [CrossRef]
| Species | No of Isolates | Origin of Production | Colony Color | Cell Shape |
|---|---|---|---|---|
| Rhodopirellula baltica | 62 | Portuguese/French | Pink to red | Ovoid, ellipsoidal or pear-shaped |
| Rhodopirellula rubra | 1 | French | Red | Pear- or club-shaped |
| Rhodopirellula heiligendammensis | 1 | Portuguese | Coral pink | Ovoid to pear-shaped |
| Gimesia chilikensis | 1 | Portuguese | Orange | Ovoid to pear-shaped |
| Target | Class | Antibiotic | Disk Content | Diameter (mm) of Inhibition Zone | |
|---|---|---|---|---|---|
| Gimesia chilikensis POA64 | Rhodopirellula heiligendammensis POEL202 | ||||
| Cell wall biosynthesis | Beta-lactams | Amoxycillin | 10 µg | 0 | 0 |
| Amoxycillin/Clavulanic acid | 30 µg | 0 | 0 | ||
| Aztreonam | 30 µg | 0 | 0 | ||
| Cefotaxime | 30 µg | 0 | 0 | ||
| Cefoxitin | 30 µg | 0 | 0 | ||
| Ceftazidime | 30 µg | 0 | 0 | ||
| Imipenem | 10 µg | 0 | 0 | ||
| Meropenem | 10 µg | 0 | 0 | ||
| Piperacillin | 100 µg | 0 | 0 | ||
| Piperacillin/Tazobactam | 110 µg | 0 | 0 | ||
| Fosfomycin | Fosfomycin | 50 µg | 0 | 0 | |
| Glycopeptides | Teicoplanin | 30 µg | ND | 0 | |
| Vancomycin | 30 µg | ND | 0 | ||
| Structure of cell membrane | Polymyxins | Polymyxin B | 300 IU | 27 | 18 |
| Protein synthesis | Aminoglycosides | Amikacin | 30 µg | 0 | 0 |
| Gentamicin | 10 µg | 0 | 0 | ||
| Kanamycin | 30 µg | 0 | 0 | ||
| Tobramycin | 10 µg | 0 | 0 | ||
| Amphenicol | Chloramphenicol | 30 µg | 10 | 33 | |
| Lincosamide | Clindamycin | 2 µg | 38 | 53 | |
| Macrolides | Erythromycin | 15 μg | >50 | 40 | |
| Tetracyclines | Doxycycline | 30 µg | >50 | 26 | |
| Tetracycline | 30 µg | >50 | 12 | ||
| DNA replication/Protein synthesis | Nitrofuran | Nitrofurantoin | 300 μg | 0 | 35 |
| DNA replication | Quinolones | Ciprofloxacin | 5 µg | 33 | 47 |
| Nalidixic acid | 30 µg | 0 | 0 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guedes, B.; Godinho, O.; Quinteira, S.; Lage, O.M. Antimicrobial Resistance Profile of Planctomycetota Isolated from Oyster Shell Biofilm: Ecological Relevance within the One Health Concept. Appl. Microbiol. 2024, 4, 16-26. https://doi.org/10.3390/applmicrobiol4010002
Guedes B, Godinho O, Quinteira S, Lage OM. Antimicrobial Resistance Profile of Planctomycetota Isolated from Oyster Shell Biofilm: Ecological Relevance within the One Health Concept. Applied Microbiology. 2024; 4(1):16-26. https://doi.org/10.3390/applmicrobiol4010002
Chicago/Turabian StyleGuedes, Bárbara, Ofélia Godinho, Sandra Quinteira, and Olga Maria Lage. 2024. "Antimicrobial Resistance Profile of Planctomycetota Isolated from Oyster Shell Biofilm: Ecological Relevance within the One Health Concept" Applied Microbiology 4, no. 1: 16-26. https://doi.org/10.3390/applmicrobiol4010002
APA StyleGuedes, B., Godinho, O., Quinteira, S., & Lage, O. M. (2024). Antimicrobial Resistance Profile of Planctomycetota Isolated from Oyster Shell Biofilm: Ecological Relevance within the One Health Concept. Applied Microbiology, 4(1), 16-26. https://doi.org/10.3390/applmicrobiol4010002








