Soil Bacteriome Resilience and Reduced Nitrogen Toxicity in Tomato by Controlled Release Nitrogen Fertilizer Compared to Urea
Abstract
1. Introduction
2. Materials and Methods
2.1. Soil and Plant Selection
2.2. Fertilizer Selection
2.3. Plant Biomass Sampling
2.4. DNA Extraction and 16S Amplicon Sequencing with MinION Flow Cells
| Gene | Process | Reaction | KEGG Entry |
|---|---|---|---|
| nifH | N-fixation | N2− > NH3 | K02588 |
| pmoA-amoA | Nitrification | NH3− > NH2OH | K10944 |
| hao | Nitrification | NH2OH− > NO2− | K10535 |
| nirK | Denitrification | NO2− > NO− | K00368 |
| nosZ | Denitrification | N2O− > N2 | K00376 |
2.5. Statistical Analysis
3. Results
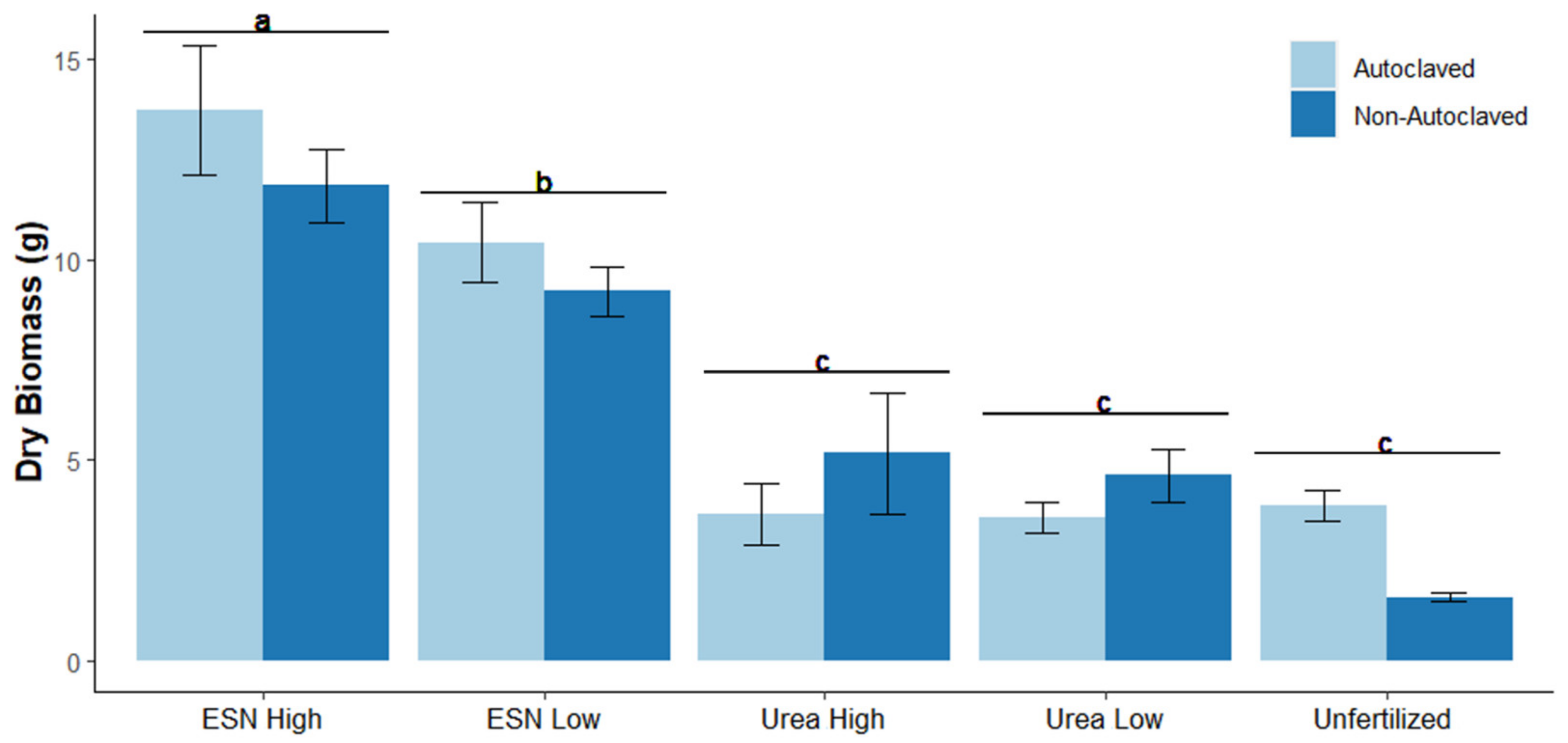
3.1. Total Plant Biomass
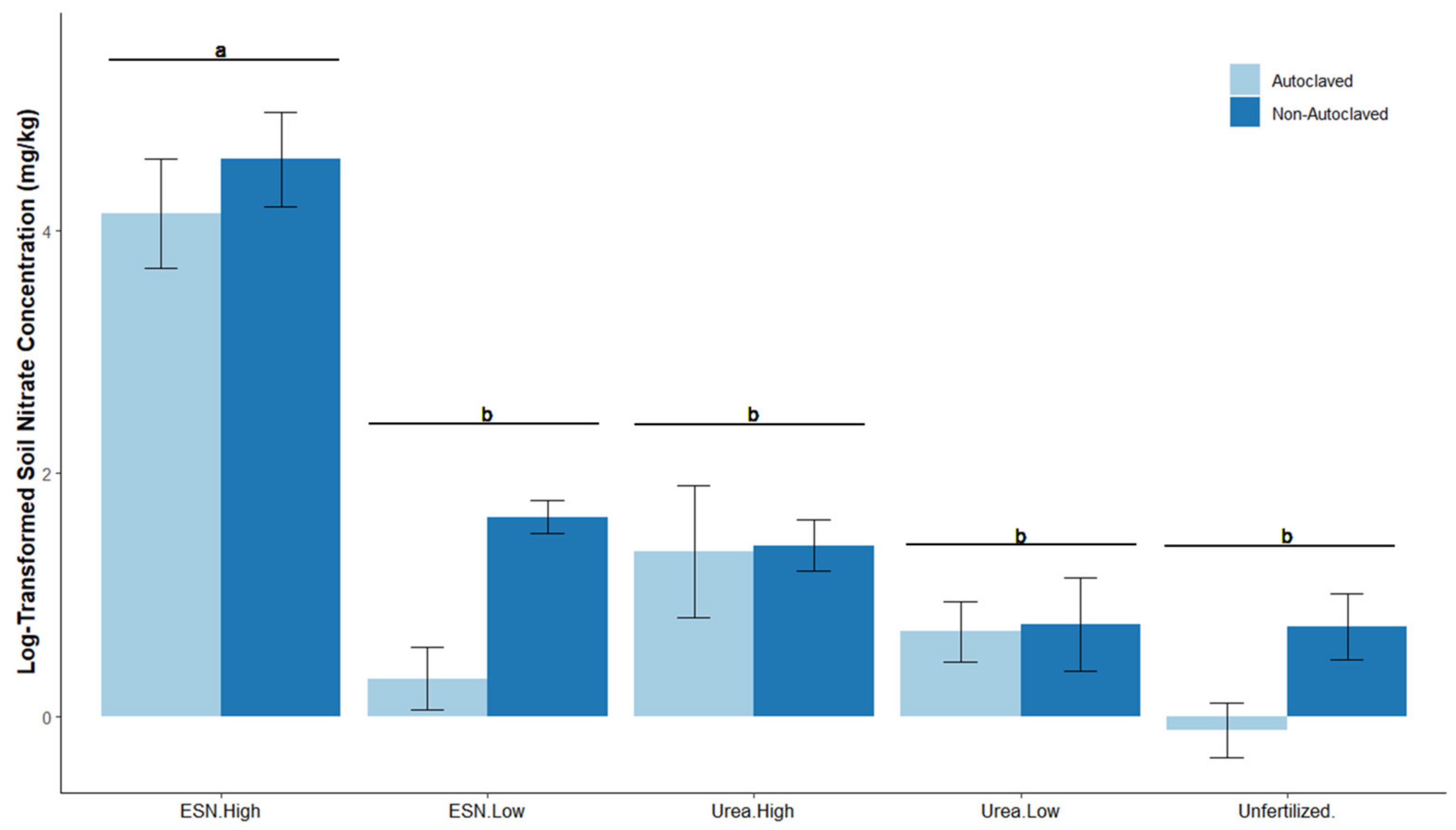
3.2. Soil Nitrate Analysis
3.3. Soil Bacterial Community Composition
3.4. Bacteria Abundance Changes with Fertilizer
3.5. Nitrogen-Cycling Bacteria Abundance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kraiser, T.; Gras, D.E.; Gutiérrez, A.G.; Gonzalez, B.; Gutiérrez, R.A. A holistic view of nitrogen acquisition in plants. J. Exp. Bot. 2011, 62, 1455–1466. [Google Scholar] [CrossRef]
- Galloway, J.N.; Dentener, F.J.; Capone, D.G.; Boyer, E.W.; Howarth, R.W.; Seitzinger, S.P.; Asner, G.P.; Cleveland, C.C.; Green, P.; Holland, E.A. Nitrogen cycles: Past, present, and future. Biogeochemistry 2004, 70, 153–226. [Google Scholar] [CrossRef]
- Cameron, K.C.; Di, H.J.; Moir, J.L. Nitrogen losses from the soil/plant system: A review. Ann. Appl. Biol. 2013, 162, 145–173. [Google Scholar] [CrossRef]
- Zhao, Z.-B.; He, J.-Z.; Geisen, S.; Han, L.-L.; Wang, J.-T.; Shen, J.-P.; Wei, W.-X.; Fang, Y.-T.; Li, P.-P.; Zhang, L.-M. Protist communities are more sensitive to nitrogen fertilization than other microorganisms in diverse agricultural soils. Microbiome 2019, 7, 33. [Google Scholar] [CrossRef]
- Yang, T.; Lupwayi, N.; Marc, S.-A.; Siddique, K.H.; Bainard, L.D. Anthropogenic drivers of soil microbial communities and impacts on soil biological functions in agroecosystems. Glob. Ecol. Conserv. 2021, 27, e01521. [Google Scholar] [CrossRef]
- Zeng, J.; Liu, X.; Song, L.; Lin, X.; Zhang, H.; Shen, C.; Chu, H. Nitrogen fertilization directly affects soil bacterial diversity and indirectly affects bacterial community composition. Soil Biol. Biochem. 2016, 92, 41–49. [Google Scholar] [CrossRef]
- Kavamura, V.N.; Hayat, R.; Clark, I.M.; Rossmann, M.; Mendes, R.; Hirsch, P.R.; Mauchline, T.H. Inorganic nitrogen application affects both taxonomical and predicted functional structure of wheat rhizosphere bacterial communities. Front. Microbiol. 2018, 9, 1074. [Google Scholar] [CrossRef]
- Dixon, M.; Rohrbaugh, C.; Afkairin, A.; Vivanco, J. Impacts of the Green Revolution on Rhizosphere Microbiology Related to Nutrient Acquisition. Appl. Microbiol. 2022, 2, 992–1003. [Google Scholar] [CrossRef]
- Kaye, J.P.; Hart, S.C. Competition for nitrogen between plants and soil microorganisms. Trends Ecol. Evol. 1997, 12, 139–143. [Google Scholar] [CrossRef]
- Kuzyakov, Y.; Xu, X. Competition between roots and microorganisms for nitrogen: Mechanisms and ecological relevance. New Phytol. 2013, 198, 656–669. [Google Scholar] [CrossRef]
- Liu, Q.; Qiao, N.; Xu, X.; Xin, X.; Han, J.Y.; Tian, Y.; Ouyang, H.; Kuzyakov, Y. Nitrogen acquisition by plants and microorganisms in a temperate grassland. Sci. Rep. 2016, 6, 22642. [Google Scholar] [CrossRef]
- Inselsbacher, E.; Umana, N.H.-N.; Stange, F.C.; Gorfer, M.; Schüller, E.; Ripka, K.; Zechmeister-Boltenstern, S.; Hood-Novotny, R.; Strauss, J.; Wanek, W. Short-term competition between crop plants and soil microbes for inorganic N fertilizer. Soil Biol. Biochem. 2010, 42, 360–372. [Google Scholar] [CrossRef]
- Kastl, E.-M.; Schloter-Hai, B.; Buegger, F.; Schloter, M. Impact of fertilization on the abundance of nitrifiers and denitrifiers at the root–soil interface of plants with different uptake strategies for nitrogen. Biol. Fertil. Soils 2015, 51, 57–64. [Google Scholar] [CrossRef]
- Zhu, Q.; Riley, W.J.; Tang, J.; Koven, C.D. Multiple soil nutrient competition between plants, microbes, and mineral surfaces: Model development, parameterization, and example applications in several tropical forests. Biogeosciences 2016, 13, 341–363. [Google Scholar] [CrossRef]
- Norton, J.; Ouyang, Y. Controls and adaptive management of nitrification in agricultural soils. Front. Microbiol. 2019, 10, 1931. [Google Scholar] [CrossRef]
- Alphei, J.; Scheu, S. Effects of biocidal treatments on biological and nutritional properties of a mull-structured woodland soil. In Soil Structure/Soil Biota Interrelationships; Elsevier: Amsterdam, The Netherlands, 1993; pp. 435–448. [Google Scholar]
- Dimkpa, C.O.; Fugice, J.; Singh, U.; Lewis, T.D. Development of fertilizers for enhanced nitrogen use efficiency–Trends and perspectives. Sci. Total Environ. 2020, 731, 139113. [Google Scholar] [CrossRef]
- Timilsena, Y.P.; Adhikari, R.; Casey, P.; Muster, T.; Gill, H.; Adhikari, B. Enhanced efficiency fertilisers: A review of formulation and nutrient release patterns. J. Sci. Food Agric. 2015, 95, 1131–1142. [Google Scholar] [CrossRef]
- Delgado, J.; Mosier, A. Mitigation Alternatives to Decrease Nitrous Oxides Emissions and Urea-Nitrogen Loss and Their Effect on Methane Flux; Wiley Online Library: Hoboken, NJ, USA, 1996. [Google Scholar]
- Rathnappriya, R.; Sakai, K.; Okamoto, K.; Kimura, S.; Haraguchi, T.; Nakandakari, T.; Setouchi, H.; Bandara, W. Examination of the effectiveness of controlled release fertilizer to balance sugarcane yield and reduce nitrate leaching to groundwater. Agronomy 2022, 12, 695. [Google Scholar] [CrossRef]
- Thapa, R.; Chatterjee, A.; Awale, R.; McGranahan, D.A.; Daigh, A. Effect of enhanced efficiency fertilizers on nitrous oxide emissions and crop yields: A meta-analysis. Soil Sci. Soc. Am. J. 2016, 80, 1121–1134. [Google Scholar] [CrossRef]
- Akiyama, H.; Yan, X.; Yagi, K. Evaluation of effectiveness of enhanced-efficiency fertilizers as mitigation options for N2O and NO emissions from agricultural soils: Meta-analysis. Glob. Chang. Biol. 2010, 16, 1837–1846. [Google Scholar] [CrossRef]
- Sempeho, S.I.; Kim, H.T.; Mubofu, E.; Hilonga, A. Meticulous overview on the controlled release fertilizers. Adv. Chem. 2014, 2014, 363071. [Google Scholar] [CrossRef]
- Lawrencia, D.; Wong, S.K.; Low, D.Y.S.; Goh, B.H.; Goh, J.K.; Ruktanonchai, U.R.; Soottitantawat, A.; Lee, L.H.; Tang, S.Y. Controlled release fertilizers: A review on coating materials and mechanism of release. Plants 2021, 10, 238. [Google Scholar] [CrossRef] [PubMed]
- Houser, M. Farmer Motivations for Excess Nitrogen Use in the US Corn Belt. Case Stud. Environ. 2022, 6, 1688823. [Google Scholar] [CrossRef]
- Golden, B.; Slaton, N.; Norman, R.; Gbur, E.; Wilson, C. Nitrogen release from environmentally smart nitrogen fertilizer as influenced by soil series, temperature, moisture, and incubation method. Commun. Soil Sci. Plant Anal. 2011, 42, 1809–1824. [Google Scholar] [CrossRef]
- Liu, H.; Qiu, Z.; Ye, J.; Verma, J.P.; Li, J.; Singh, B.K. Effective colonisation by a bacterial synthetic community promotes plant growth and alters soil microbial community. J. Sustain. Agric. Environ. 2022, 1, 30–42. [Google Scholar] [CrossRef]
- Li, K.; DiLegge, M.J.; Minas, I.S.; Hamm, A.; Manter, D.; Vivanco, J.M. Soil sterilization leads to re-colonization of a healthier rhizosphere microbiome. Rhizosphere 2019, 12, 100176. [Google Scholar] [CrossRef]
- Newberger, D.R.; Minas, I.S.; Manter, D.K.; Vivanco, J.M. A Microbiological Approach to Alleviate Soil Replant Syndrome in Peaches. Microorganisms 2023, 11, 1448. [Google Scholar] [CrossRef]
- O’sullivan, L.A.; Roussel, E.G.; Weightman, A.J.; Webster, G.; Hubert, C.R.; Bell, E.; Head, I.; Sass, H.; Parkes, R.J. Survival of Desulfotomaculum spores from estuarine sediments after serial autoclaving and high-temperature exposure. ISME J. 2015, 9, 922–933. [Google Scholar] [CrossRef]
- Urbaniak, G.C.; Plous, S. Research Randomizer (Version 4.0). Available online: https://www.randomizer.org/ (accessed on 1 September 2021).
- Zhang, S.; Liu, Y.; Du, M.; Shou, G.; Wang, Z.; Xu, G. Nitrogen as a regulator for flowering time in plant. Plant Soil 2022, 480, 1–29. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Badri, D.V.; Vivanco, J.M. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014, 8, 790–803. [Google Scholar] [CrossRef]
- Wick, R.; Menzel, P. rrwick/Filtlong: Quality Filtering Tool for Long Reads. 2017. Available online: https://github.com/rrwick/Filtlong/ (accessed on 1 December 2021).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Curry, K.D.; Wang, Q.; Nute, M.G.; Tyshaieva, A.; Reeves, E.; Soriano, S.; Wu, Q.; Graeber, E.; Finzer, P.; Mendling, W. Emu: Species-level microbial community profiling of full-length 16S rRNA Oxford Nanopore sequencing data. Nat. Methods 2022, 19, 845–853. [Google Scholar] [CrossRef] [PubMed]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Manter, D.K.; Hamm, A.K.; Deel, H.L. Community structure and abundance of ACC deaminase containing bacteria in soils with 16S-PICRUSt2 inference or direct acdS gene sequencing. J. Microbiol. Methods 2023, 211, 106740. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Barbera, P.; Kozlov, A.M.; Czech, L.; Morel, B.; Darriba, D.; Flouri, T.; Stamatakis, A. EPA-ng: Massively parallel evolutionary placement of genetic sequences. Syst. Biol. 2019, 68, 365–369. [Google Scholar] [CrossRef]
- Louca, S.; Doebeli, M. Efficient comparative phylogenetics on large trees. Bioinformatics 2018, 34, 1053–1055. [Google Scholar] [CrossRef]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.-Y.; Dillies, M.-A. SARTools: A DESeq2-and EdgeR-based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef]
- Barreto, R.; Prado, R.; Leal, A.; Troleis, M.; Junior, G.S.; Monteiro, C.; Santos, L.; Carvalho, R. Mitigation of ammonium toxicity by silicon in tomato depends on the ammonium concentration. Acta Agric. Scand. Sect. B—Soil Plant Sci. 2016, 66, 483–488. [Google Scholar] [CrossRef][Green Version]
- Mo, X.; Peng, H.; Xin, J.; Wang, S. Analysis of urea nitrogen leaching under high-intensity rainfall using HYDRUS-1D. J. Environ. Manag. 2022, 312, 114900. [Google Scholar] [CrossRef] [PubMed]
- Beeckman, F.; Motte, H.; Beeckman, T. Nitrification in agricultural soils: Impact, actors and mitigation. Curr. Opin. Biotechnol. 2018, 50, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Goyal, S.S.; Huffaker, R.C. Nitrogen toxicity in plants. Nitrogen Crop Prod. 1984, 97–118. [Google Scholar] [CrossRef]
- Hoque, M.M.; Ajwa, H.A.; Smith, R. Nitrite and ammonium toxicity on lettuce grown under hydroponics. Commun. Soil Sci. Plant Anal. 2007, 39, 207–216. [Google Scholar] [CrossRef]
- Shedley, E.; Dell, B.; Grove, T. Effects of inorganic nitrogen forms on growth of Eucalyptus globulus seedlings. In Plant Nutrition—From Genetic Engineering to Field Practice: Proceedings of the Twelfth International Plant Nutrition Colloquium, 21–26 September 1993, Perth, Western Australia; Springer Netherlands: Cham, The Netherlands, 1993; pp. 595–598. [Google Scholar]
- Castellano-Hinojosa, A.; Strauss, S.L.; González-López, J.; Bedmar, E.J. Changes in the diversity and predicted functional composition of the bulk and rhizosphere soil bacterial microbiomes of tomato and common bean after inorganic N-fertilization. Rhizosphere 2021, 18, 100362. [Google Scholar] [CrossRef]
- Rehman, S.; Fayaz, H.; Rehmaan, I.U.; Rashid, K.; Rashid, S. Nitrogen Toxicity in Plants, Symptoms, and Safeguards. In Advances in Plant Nitrogen Metabolism; CRC Press: Boca Raton, FL, USA, 2022; pp. 213–225. [Google Scholar]
- Sánchez, E.; Soto, J.M.; García, P.C.; López-Lefebre, L.R.; Rivero, R.M.; Ruiz, J.M.; Romero, L. Phenolic compounds and oxidative metabolism in green bean plants under nitrogen toxicity. Funct. Plant Biol. 2000, 27, 973–978. [Google Scholar] [CrossRef]
- Kamnqa, U.; Etsassala, N.G.; Akinpelu, E.A.; Nchu, F. Effects of Varying Nitrogen Fertilization on Growth, Yield and Flowering of Capsicum annuum (California wonder). In Proceedings of the 18th SOUTH AFRICA Int’l Conference on Agricultural, Chemical, Biological & Environmental Sciences (ACBES-20), Johannesburg, South Africa, 16–17 November 2020; pp. 16–17. [Google Scholar]
- Ozores-Hampton, M.; Simonne, E.; Morgan, K.; Cushman, K.; Sato, S.; Albright, C.; Waldo, E.; Polak, A. Can we use controlled release fertilizers (CRF) in tomato production. Proc. Fla. Tomato Inst. PRO526 2009, 10–13. [Google Scholar] [CrossRef]
- Shrestha, R.C.; Ghazaryan, L.; Poodiack, B.; Zorin, B.; Gross, A.; Gillor, O.; Khozin-Goldberg, I.; Gelfand, I. The effects of microalgae-based fertilization of wheat on yield, soil microbiome and nitrogen oxides emissions. Sci. Total Environ. 2022, 806, 151320. [Google Scholar] [CrossRef]
- Lourenço, K.S.; Suleiman, A.K.; Pijl, A.; Van Veen, J.; Cantarella, H.; Kuramae, E. Resilience of the resident soil microbiome to organic and inorganic amendment disturbances and to temporary bacterial invasion. Microbiome 2018, 6, 142. [Google Scholar] [CrossRef]
- Mahmood, T.; Mehnaz, S.; Fleischmann, F.; Ali, R.; Hashmi, Z.; Iqbal, Z. Soil sterilization effects on root growth and formation of rhizosheaths in wheat seedlings. Pedobiologia 2014, 57, 123–130. [Google Scholar] [CrossRef]
- Jurburg, S.D.; Salles, J.F. Functional redundancy and ecosystem function—The soil microbiota as a case study. In Biodiversity in Ecosystems-Linking Structure and Function; IntechOpen Limited: London, UK, 2015; pp. 29–49. [Google Scholar] [CrossRef]
- Han, F.; Zhou, W. Nitrogen recovery from wastewater by microbial assimilation—A review. Bioresour. Technol. 2022, 363, 127933. [Google Scholar] [CrossRef] [PubMed]
- Harrison, K.A.; Bol, R.; Bardgett, R.D. Preferences for different nitrogen forms by coexisting plant species and soil microbes. Ecology 2007, 88, 989–999. [Google Scholar] [CrossRef] [PubMed]
- Jackson, L.E.; Schimel, J.P.; Firestone, M.K. Short-term partitioning of ammonium and nitrate between plants and microbes in an annual grassland. Soil Biol. Biochem. 1989, 21, 409–415. [Google Scholar] [CrossRef]
- Poretsky, R.; Rodriguez-R, L.M.; Luo, C.; Tsementzi, D.; Konstantinidis, K.T. Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS ONE 2014, 9, e93827. [Google Scholar] [CrossRef]
- Naz, M.Y.; Sulaiman, S.A. Slow release coating remedy for nitrogen loss from conventional urea: A review. J. Control. Release 2016, 225, 109–120. [Google Scholar] [CrossRef]
- Zhu, Q.; Zhang, M.; Ma, Q. Copper-based foliar fertilizer and controlled release urea improved soil chemical properties, plant growth and yield of tomato. Sci. Hortic. 2012, 143, 109–114. [Google Scholar] [CrossRef]
- Carson, L.C.; Ozores-Hampton, M.; Morgan, K.T.; Sargent, S.A. Effects of controlled-release fertilizer nitrogen rate, placement, source, and release duration on tomato grown with seepage irrigation in Florida. HortScience 2014, 49, 798–806. [Google Scholar] [CrossRef]
- Shaviv, A. Advances in controlled-release fertilizers. Adv. Agron. 2001, 71, 1–49. [Google Scholar]
- Torralbo, F.; Boardman, D.; Houx III, J.H.; Fritschi, F.B. Distinct enhanced efficiency urea fertilizers differentially influence ammonia volatilization losses and maize yield. Plant Soil 2022, 475, 551–563. [Google Scholar] [CrossRef]
- Ma, Q.; Qian, Y.; Yu, Q.; Cao, Y.; Tao, R.; Zhu, M.; Ding, J.; Li, C.; Guo, W.; Zhu, X. Controlled-release nitrogen fertilizer application mitigated N losses and modified microbial community while improving wheat yield and N use efficiency. Agric. Ecosyst. Environ. 2023, 349, 108445. [Google Scholar] [CrossRef]
- Tkacz, A.; Bestion, E.; Bo, Z.; Hortala, M.; Poole, P.S. Influence of plant fraction, soil, and plant species on microbiota: A multikingdom comparison. MBio 2020, 11, 10–128. [Google Scholar] [CrossRef] [PubMed]
- DiLegge, M.J.; Manter, D.K.; Vivanco, J.M. Soil microbiome disruption reveals specific and general plant-bacterial relationships in three agroecosystem soils. PLoS ONE 2022, 17, e0277529. [Google Scholar] [CrossRef] [PubMed]

| Fertilizer Type | Fertilizer Rate | Fertilizer Quantity (g N/Pot) |
|---|---|---|
| Unfertilized Control | Unfertilized Control | Unfertilized Control |
| Quick Release Urea | High | 0.92 |
| Quick Release Urea | Low | 0.23 |
| Controlled Release Environmentally Smart Nitrogen (ESN) | High | 0.90 |
| Controlled Release Environmentally Smart Nitrogen (ESN) | Low | 0.24 |
| Taxa | Predicted Function | N-Cycling Gene | Enriched Group | Contrast | Fold Change | Adjusted p-Value |
|---|---|---|---|---|---|---|
| Nitrobacter winogradskyi | Denitrification | narG; narH; nirK | High ESN | Unfertilized | 4.997 | 7.51 × 10−7 |
| High Urea | Unfertilized | 4.531 | 1.98 × 10−5 | |||
| Nitrosomonas communis | Nitrification | pmoA; hao | High ESN | Unfertilized | 4.587 | 7.22 × 10−8 |
| High Urea | Unfertilized | 4.158 | 3.74 × 10−6 | |||
| Nitrosospira lacus | Nitrification; Denitrification | pmoA; nirK | High ESN | Unfertilized | 3.586 | 2.22 × 10−2 |
| High ESN | Low Urea | 4.073 | 2.05 × 10−2 | |||
| Nitrosospira multiformis | Nitrification; Denitrification | pmoA; hao; norB; norC; nirK | High ESN | Unfertilized | 2.342 | 6.50 × 10−12 |
| High ESN | Low ESN | 1.806 | 2.11 × 10−6 | |||
| High ESN | Low Urea | 1.793 | 1.22 × 10−5 | |||
| High Urea | Unfertilized | 1.402 | 4.32 × 10−4 | |||
| Nitrospira defluvii | Denitrification | narG; narH; nirK | High ESN | Unfertilized | 7.539 | 3.37 × 10−4 |
| High ESN | Low Urea | 7.567 | 2.96 × 10−3 | |||
| Sporosarcina koreensis | Denitrification | narG; narH | High Urea | High ESN | 30.508 | 1.97 × 10−8 |
| High Urea | Unfertilized | 23.058 | 2.70 × 10−5 | |||
| High Urea | Low ESN | 30.578 | 1.18 × 10−8 | |||
| High Urea | Low Urea | 29.502 | 3.66 × 10−7 | |||
| Sporosarcina luteola | Denitrification | narG; narH | High Urea | High ESN | 28.600 | 6.11 × 10−11 |
| High Urea | Unfertilized | 24.850 | 4.71 × 10−9 | |||
| High Urea | Low ESN | 28.759 | 2.22 × 10−11 | |||
| High Urea | Low Urea | 28.978 | 2.00 × 10−10 | |||
| Sporosarcina soli | Denitrification | narG; narH | High Urea | High ESN | 27.218 | 1.28 × 10−6 |
| High Urea | Unfertilized | 24.313 | 8.93 × 10−6 | |||
| High Urea | Low ESN | 27.255 | 9.17 × 10−7 | |||
| High Urea | Low Urea | 27.039 | 6.04 × 10−6 | |||
| Archangium gephyra | NA | NA | Unfertilized | High Urea | 1.444 | 1.30 × 10−2 |
| Bacillus lentus | NA | NA | High ESN | Unfertilized | 18.649 | 1.20 × 10−6 |
| High ESN | Low Urea | 37.483 | 8.68 × 10−24 | |||
| High Urea | Unfertilized | 22.440 | 3.02 × 10−9 | |||
| High Urea | Low Urea | 41.275 | 7.21 × 10−28 | |||
| Low ESN | Unfertilized | 16.784 | 1.23 × 10−4 | |||
| Low ESN | Low Urea | 35.619 | 3.37 × 10−21 | |||
| Bacillus sp. OxB-1 | NA | NA | High ESN | Unfertilized | 19.415 | 6.50 × 10−12 |
| High Urea | Unfertilized | 24.892 | 3.58 × 10−19 | |||
| Low ESN | Unfertilized | 17.102 | 1.59 × 10−8 | |||
| Low Urea | Unfertilized | 17.378 | 1.04 × 10−7 | |||
| Brevendimonas naejangsanensis | NA | NA | High ESN | Unfertilized | 18.330 | 5.31 × 10−7 |
| High ESN | Low ESN | 24.960 | 2.13 × 10−12 | |||
| High ESN | Low Urea | 24.007 | 2.89 × 10−10 | |||
| High Urea | Unfertilized | 22.296 | 4.82 × 10−10 | |||
| High Urea | Low ESN | 28.926 | 2.22 × 10−16 | |||
| High Urea | Low Urea | 27.972 | 8.63 × 10−14 | |||
| Chthoniobacter flavus | NA | NA | Unfertilized | High ESN | 0.639 | 1.73 × 10−2 |
| Nitrosospira briensis | NA | NA | High ESN | Unfertilized | 3.256 | 2.58 × 10−10 |
| High ESN | Low ESN | 2.521 | 1.85 × 10−5 | |||
| High ESN | Low Urea | 2.325 | 5.64 × 10−4 | |||
| High Urea | Unfertilized | 2.498 | 9.11 × 10−6 | |||
| High Urea | Low ESN | 1.763 | 4.55 × 10−2 | |||
| Oligotropha carboxidovorans | NA | NA | High ESN | Unfertilized | 6.310 | 3.77 × 10−3 |
| Pusillimonas sp. ye3 | NA | NA | High Urea | Unfertilized | 7.030 | 2.51 × 10−2 |
| Soil | Treatment | Total | N Fixation | Nitrification | Denitrification | ||
|---|---|---|---|---|---|---|---|
| 16S rRNA | nifH | pmoA-amoA | hao | nirk | nosZ | ||
| Non-autoclaved | Unfertilized | 2.22 × 109 a | 1.23 × 108 a | 3.26 × 107 bcd | 3.12 × 107 bcd | 2.35 × 108 ab | 1.92 × 108 a |
| Low Urea | 2.33 × 109 a | 1.41 × 108 a | 5.04 × 107 bc | 4.89 × 107 bc | 2.71 × 108 ab | 2.12 × 108 a | |
| High Urea | 2.48 × 109 a | 1.50 × 108 a | 5.83 × 107 b | 5.62 × 107 ab | 3.09 × 108 a | 2.16 × 108 a | |
| Low ESN | 2.11 × 109 a | 1.10 × 108 ab | 3.86 × 107 bcd | 3.68 × 107 bcd | 2.23 × 108 ab | 1.76 × 108 a | |
| High ESN | 2.40 × 109 a | 1.36 × 108 a | 1.05 × 108 a | 9.31 × 107 a | 3.36 × 108 a | 2.02 × 108 a | |
| Autoclaved | Unfertilized | 1.13 × 109 b | 4.09 × 107 ac | 4.71 × 106 d | 4.64 × 106 d | 1.74 × 108 b | 1.31 × 108 b |
| Low Urea | 8.70 × 108 b | 3.24 × 107 c | 4.65 × 106 cd | 4.69 × 106 d | 1.41 × 108 b | 1.00 × 108 b | |
| High Urea | 1.01 × 109 b | 4.41 × 107 c | 1.70 × 107 bcd | 1.67 × 107 bc | 2.21 × 108 ab | 1.44 × 108 b | |
| Low ESN | 1.31 × 109 b | 6.74 × 107 bc | 1.46 × 107 bcd | 1.40 × 107 bc | 2.26 × 108 ab | 1.79 × 108 b | |
| High ESN | 1.17 × 109 b | 4.51 × 107 bc | 1.58 × 107 bcd | 1.47 × 107 bcd | 2.08 × 108 ab | 1.25 × 108 b | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rohrbaugh, C.R.; Dixon, M.M.; Delgado, J.A.; Manter, D.K.; Vivanco, J.M. Soil Bacteriome Resilience and Reduced Nitrogen Toxicity in Tomato by Controlled Release Nitrogen Fertilizer Compared to Urea. Appl. Microbiol. 2023, 3, 1262-1276. https://doi.org/10.3390/applmicrobiol3040087
Rohrbaugh CR, Dixon MM, Delgado JA, Manter DK, Vivanco JM. Soil Bacteriome Resilience and Reduced Nitrogen Toxicity in Tomato by Controlled Release Nitrogen Fertilizer Compared to Urea. Applied Microbiology. 2023; 3(4):1262-1276. https://doi.org/10.3390/applmicrobiol3040087
Chicago/Turabian StyleRohrbaugh, Carley R., Mary M. Dixon, Jorge A. Delgado, Daniel K. Manter, and Jorge M. Vivanco. 2023. "Soil Bacteriome Resilience and Reduced Nitrogen Toxicity in Tomato by Controlled Release Nitrogen Fertilizer Compared to Urea" Applied Microbiology 3, no. 4: 1262-1276. https://doi.org/10.3390/applmicrobiol3040087
APA StyleRohrbaugh, C. R., Dixon, M. M., Delgado, J. A., Manter, D. K., & Vivanco, J. M. (2023). Soil Bacteriome Resilience and Reduced Nitrogen Toxicity in Tomato by Controlled Release Nitrogen Fertilizer Compared to Urea. Applied Microbiology, 3(4), 1262-1276. https://doi.org/10.3390/applmicrobiol3040087






