Construction of an Escherichia coli Strain Capable of Utilizing Steamed Rice as the Sole Carbon and Energy Source by Extracellular Expression of Amylase and Its Use for the Production of Putrescine
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids and Oligonucleotides
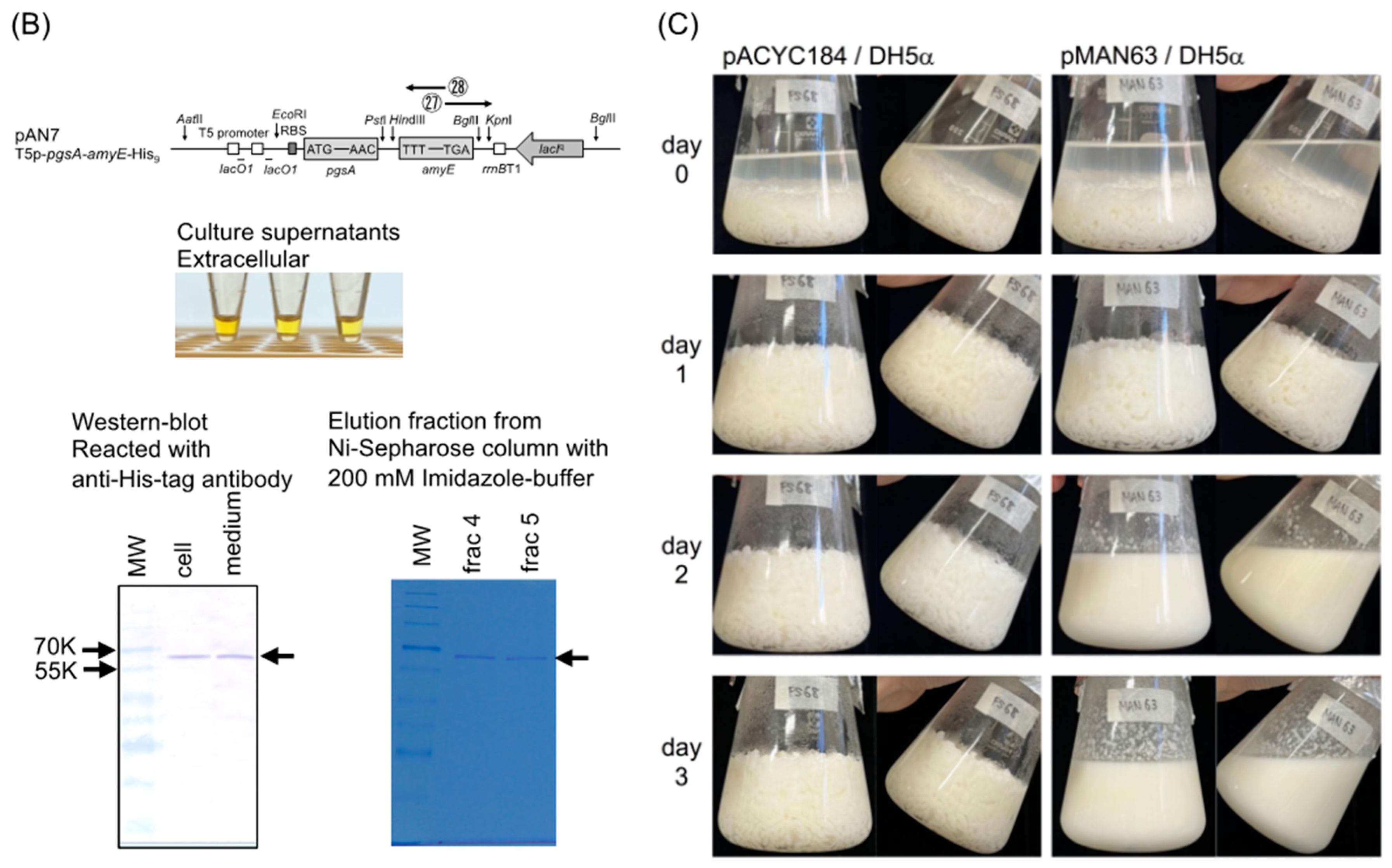
2.2. Construction of Plasmids Express α-Amylase in the Extracellular Space of E. coli
2.3. Comparison of Amylase Activity by the Iodine-Starch Test
2.4. Western Blot Analysis of α-Amylase Expressed by Strain AN7
2.5. Purification of α-Amylase from the Culture Medium of AN7 and Determination of its N-Terminal Amino Acid Sequence
2.6. Production of Putrescine in 1 × M9 Steamed Rice Medium
2.7. Measurement of Putrescine
3. Results and Discussion
3.1. Comparison of Amylase Activity Expressed from Various Plasmids

3.2. Comparison of Putrescine Production Expressed from Various Plasmids
3.3. Effect of Arginine Supplementation on Putrescine Production
3.4. Effect of IPTG Addition on Putrescine Production
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Kwak, D.; Lim, H.; Yang, J.; Seo, S.W.; Jung, G.Y. Synthetic redesign of Escherichia coli for cadaverine production from galactose. Biotechnol. Biofuels 2017, 10, 20. [Google Scholar] [CrossRef]
- Ma, W.; Chen, K.; Li, Y.; Hao, N.; Wang, X.; Ouyang, P. Advances in cadaverine bacterial production and its applications. Engineering 2017, 3, 308–317. [Google Scholar] [CrossRef]
- Meng, J.; Wang, B.; Liu, D.; Chen, T.; Wang, Z.; Zhao, X. High-yield anaerobic succinate production by strategically regulating multiple metabolic pathways based on stoichiometric maximum in Escherichia coli. Microb. Cell Fact. 2016, 15, 141. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Miyachi, M.; Tamaki, H.; Suzuki, H. The Escherichia coli CitT transporter can be used as a succinate exporter for succinate production. Biosci. Biotechnol. Biochem. 2021, 85, 981–988. [Google Scholar] [CrossRef]
- Atsumi, S.; Cann, A.F.; Connor, M.R.; Shen, C.R.; Smith, K.M.; Brynildsen, M.P.; Chou, K.J.Y.; Hanai, T.; Liao, J.C. Metabolic engineering of Escherichia coli for 1-butanol production. Metab. Eng. 2008, 10, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, S.; Pereira, R.; Wahl, S.A.; Rocha, I. Metabolic engineering strategies for butanol production in Escherichia coli. Biotech. Bioeng. 2020, 117, 2571–2587. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Khusnutdinova, S.; Chen, J.; Crisante, D.; Batyrova, K.; Raj, K.; Feigis, M.; Shirzadi, E.; Wang, X.; Dorakhan, R.; et al. Systems engineering of Escherichia coli for n-butane production. Metab. Eng. 2022, 74, 98–107. [Google Scholar] [CrossRef]
- Zhao, M.; Huang, D.; Zhang, X.; Koffas, M.A.G.; Zhou, J.; Deng, Y. Metabolic engineering of Escherichia coli for producing adipic acid through the reverse adipate-degradation pathway. Metab. Eng. 2018, 47, 254–262. [Google Scholar] [CrossRef]
- Ning, Y.; Liu, H.; Zhang, R.; Jin, Y.; Yu, Y.; Deng, L.; Wang, F. Research progress on the construction of artificial pathways for the biosynthesis of adipic acid by engineered microbes. Fermentation 2022, 8, 393. [Google Scholar] [CrossRef]
- Choi, Y.J.; Park, J.H.; Kim, T.Y.; Lee, S.Y. Metabolic engineering of Escherichia coli for the production of 1-propanol. Metab. Eng. 2012, 14, 477–486. [Google Scholar] [CrossRef]
- Jain, R.; Sun, X.; Yuan, Q.; Yan, Y. Systematically engineering Escherichia coli for enhanced production of 1,2-propanediol and 1-propanol. ACS Synth. Biol. 2015, 4, 746–756. [Google Scholar] [CrossRef] [PubMed]
- Ingram, L.O.; Conway, T.; Clark, D.P.; Sewell, G.W.; Preston, J.F. Genetic engineering of ethanol production in Escherichia coli. Appl. Environ. Microbiol. 1987, 53, 2420–2425. [Google Scholar] [CrossRef] [PubMed]
- Fithriani; Suryadarma, P.; Mangunwidjaja, D. Metabolic engineering of Escherichia coli cells for ethanol production under aerobic conditions. Procedia Chem. 2015, 16, 600–607. [Google Scholar] [CrossRef]
- Sierra-Ibarra, E.; Alcaraz-Cienfuegos, J.; Vargas-Tah, A.; Rosas-Aburto, A.; Valdivia-López, Á.; Hernández-Luna, M.G.; Vivaldo-Lima, E.; Martinez, A. Ethanol production by Escherichia coli from detoxified lignocellulosic teak wood hydrolysates with high concentration of phenolic compounds. J. Ind. Microbiol. Biotechnol. 2022, 49, kuab077. [Google Scholar] [CrossRef]
- Jayalakshmi, S.; Joseph, K.; Sukumaran, V. Methane production from kitchen waste using Escherichia coli. J. Environ. Sci. Eng. 2007, 49, 99–102. [Google Scholar]
- Han, M.J.; Lee, S.H. An efficient bacterial surface display system based on a novel outer membrane anchoring element from the Escherichia coli protein YiaT. FEMS Microbiol. Lett. 2015, 362, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Narita, J.; Okano, K.; Tateno, T.; Tanino, T.; Sewaki, T.; Sung, M.H.; Fukuda, H.; Kondo, A. Display of active enzymes on the cell surface of Escherichia coli using PgsA anchor protein and their application to bioconversion. Appl. Microbiol. Biotechnol. 2006, 70, 564–572. [Google Scholar] [CrossRef] [PubMed]
- Tateno, T.; Fukuda, H.; Kondo, A. Production of L-lysine from starch by Corynebacterium glutamicum displaying α-amylase on its cell surface. Appl. Microbiol. Biotechnol. 2007, 74, 1213–1220. [Google Scholar] [CrossRef]
- van Bloois, E.; Winter, R.T.; Kolmar, H.; Fraaije, M.W. Decorating microbes: Surface display of proteins on Escherichia coli. Trends Biotechnol. 2011, 29, 79–86. [Google Scholar] [CrossRef]
- Narita, J.; Okano, K.; Kitao, T.; Ishida, S.; Sewaki, T.; Sung, M.H.; Fukuda, H.; Kondo, A. Display of α-amylase on the surface of Lactobacillus casei cells by use of the PgsA anchor protein, and production of lactic acid from starch. Appl. Environ. Microbiol. 2006, 72, 269–275. [Google Scholar] [CrossRef]
- Suzuki, H.; Thongbhubate, K.; Muraoka, M.; Sasabu, S. Agmatine production by Escherichia coli cells expressing SpeA on the extracellular surface. Enzyme Microb. Technol. 2023, 162, 110139. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Sasabui, A. First example of the extracellular surface expression of intrinsically periplasmic Escherichia coli γ-glutamyltranspeptidase, a member of the N-terminal nucleophile hydrolase superfamily, and the use of cells as a catalyst for γ-glutamylvalylglycine production. J. Agric. Food Chem. 2023, 71, 1132–1138. [Google Scholar] [PubMed]
- Thongbhubate, K.; Irie, K.; Sakai, Y.; Itoh, A.; Suzuki, H. Improvement of putrescine production through the arginine decarboxylase pathway in Escherichia coli K-12. AMB Express 2021, 11, 168. [Google Scholar] [CrossRef]
- Chang, A.C.; Cohen, S.N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 1978, 134, 1141–1156. [Google Scholar] [CrossRef]
- Ashikaga, S.; Nanamiya, H.; Ohashi, Y.; Kawamura, F. Natural genetic competence in Bacillus subtilis natto OK2. J. Bacteriol. 2000, 182, 2411–2415. [Google Scholar] [CrossRef] [PubMed]
- Ashiuchi, M.; Soda, K.; Misono, H. A poly-γ-glutamate synthetic system of Bacillus subtilis IFO3336: Gene cloning and biochemical analysis of poly-γ-glutamate produced by Escherichia coli clone cells. Biochem. Biophys. Res. Commun. 1999, 263, 6–12. [Google Scholar] [CrossRef]
- Thongbhubate, K.; Nakafuji, Y.; Matsuoka, R.; Kakegawa, S.; Suzuki, H. Effect of spermidine on biofilm formation in Escherichia coli K-12. J. Bacteriol. 2021, 203, e00652-20. [Google Scholar] [CrossRef]
- Kurihara, S.; Oda, S.; Tsuboi, Y.; Kim, H.G.; Oshida, M.; Kumagai, H.; Suzuki, H. γ-Glutamylputrescine synthetase in the putrescine utilization pathway of Escherichia coli K-12. J. Biol. Chem. 2008, 283, 19981–19990. [Google Scholar] [CrossRef]
- Yamazaki, H.; Ohmura, K.; Nakayama, A.; Takeichi, Y.; Otozai, K.; Yamasaki, M.; Tamura, G.; Yamane, K. α-Amylase genes (amyR2 and amyE+) from an α-amylase-hyperproducing Bacillus subtilis strain: Molecular cloning and nucleotide sequences. J. Bacteriol. 1983, 156, 327–337. [Google Scholar] [CrossRef]
- Mäntsälä, P.; Zalkin, H. Membrane-bound and soluble extracellular alpha-amylase from Bacillus subtilis. J. Biol. Chem. 1979, 254, 8540–8547. [Google Scholar] [CrossRef]



| Strains and Plasmids | Genotype | Source and Reference |
|---|---|---|
| (Escherichia coli K-12) | ||
| AN7 | pAN7/DH5α | This study |
| AN14 | pMAN63 pSH1733/SH2204 | This study |
| DH5α | F− supE44 ΔlacU169(Φ80 lacZΔM15) | |
| hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | ||
| KT160 | pKN11/SH2204 | [23] |
| MG1655 | F− rph-1 | Carol A. Gross |
| SH2204 | F− rph-1 ΔargR::FRT ΔpatA::FRT | [23] |
| ΔpotE::FRT ΔspeD::FRT ΔspeG::FRT | ||
| ΔargA::FRT ΔpuuPA::FRT yifEQ100TAG | ||
| zie-296::Tn10 | ||
| SH2287 | pMAN63 pKN11/SH2204 | This study |
| (plasmids) | ||
| pACYC184 | p15A replicon cat+ tet+ | Stanley N. Cohen, [24] |
| pAN1 | ColE1 replicon bla+ lacIq | This study |
| T5p-pgsA+-amyE(Δsignal) | ||
| pAN2 | ColE1 replicon bla+ lacIq | This study |
| T5p-pgsA+-amyEΔsignalΔpropeptide | ||
| pAN5 | p15A replicon cat+ | This study |
| T5p-yiaTR232-amyEΔsignalΔpropeptideΔ12bp | ||
| pAN7 | p15A replicon cat+ | This study |
| tetS-T5p-pgsA+-amyE-His9-rrnBT1 lacIq | ||
| pFT06 | ColE1 replicon bla+ lacIq | This study |
| T5p-capA+-amyE+ | ||
| pKN11 | ColE1 replicon bla+ lacIq | [23] |
| T5p-speA+-speB+-argAATG Y19C | ||
| pMAN36 | ColE1 replicon bla+ lacIq T5p-pgsA+ | This study |
| pMAN39 | ColE1 replicon bla+ lacIq | This study |
| T5p-pgsA+-amyE+ | ||
| pMAN63 | p15A replicon cat+ | This study |
| tetS-T5p-pgsA+-amyE+-rrnBT1 lacIq | ||
| pMAN95 | ColE1 replicon bla+ lacIq | This study |
| T5p-yiaTR232-amyEΔsignalΔpropeptideΔ12bp | ||
| pMAN99 | ColE1 replicon bla+ lacIq | This study |
| T5p-capA+-amyEΔsignalΔpropeptideΔ12bp | ||
| pMAN100 | p15A replicon cat+ | This study |
| T5p-capA+-amyEΔsignalΔpropeptideΔ12bp | ||
| pQE-80L | ColE1 replicon bla+ lacIq T5p-(His)6 | Quiagen |
| pSH1733 | ColE1 replicon bla+ lacIq T5p-argAATG Y19C | [23] |
| (Bacillus subtilis) | ||
| 168 | trp | Pasteur Institute |
| subsp. natto OK2 | bio | Fujio Kawamura, [25] |
| No. | Name | DNA Sequence |
|---|---|---|
| 1 | pQE_EcoRI_ATG_pgsA | 5′-cccgaattcattaaagaggagaaattaactATGTTTAACTTA- |
| CCGAATAAAATCAC-3′ | ||
| 2 | PstI_natto_pgsA_Cterm | 5′-cccctgcagGTTAGATGTTTTTAACGCTTCC-3′ |
| 3 | Bsub168 alpha-amylase_C | 5′-attaagcttTCAATGGGGAAGAGAACCGC-3′ |
| 4 | Bsub 168 alpha-amylase Hind3_N | 5′-GCCAAGCTTATTTGCAAAACGATTCAAAAC-3′ |
| 5 | pgsA C-comp | 5′-GTTAGATGTTTTTAACGCTTCCCAG-3′ |
| 6 | amyE deltasignal_F | 5′-CCGGCGGCTGCGAGTGCTGAAACGG-3′ |
| 7 | amyE deltasignal_F2 | 5′-CTTACAGCACCGTCGATCAAAAGCG-3′ |
| 8 | pQE80L-yiaTR232_infusion-F | 5′-caatttcacacagaattattaaagaggagaaattaactATGTTA- |
| ATTAATCGCAATATTG-3′ | ||
| 9 | amyE(136bp)-F | 5′-TCGATCAAAAGCGGAACCATTCTTCATGCA- |
| TGG-3′ | ||
| 10 | yiaTR232-amyE(136bp)_infuR | 5′-GGTTCCGCTTTTGATCGAACGATCAATCATC- |
| GGGCTGTCG-3′ | ||
| 11 | pQE80L-amyE_infuR.new | 5′-gctaattaagcttggcTCAATGGGGAAGAGAACCGC- |
| TTAAGC-3′ | ||
| 12 | EcoRI_RBS_ATG-ywtBF | 5′-gaattcattaaagaggagaaattaactATGAAAAAAGAA- |
| CTGAGCTTTCATGAAAAGCTGC-3′ | ||
| 13 | RBS_EcoRI-T5pR | 5′-agttaatttctcctctttaatgaattctgtgtgaaattgttatccgctcac- |
| aattg-3′ | ||
| 14 | amyE(136bp)-ywtB_infuR | 5′-GGTTCCGCTTTTGATCGATTTAGATTTTAGTT- |
| TGTCACTATGATC-3′ | ||
| 15 | amyE(136bp)-F | 5′-TCGATCAAAAGCGGAACCATTCTTCATGCA- |
| TGG-3′ | ||
| 16 | infuF_EcoRI-RBS-ywtB(25bp) new | 5′-gataacaatttcacacagaattcattaaagaggagaaattaact- |
| ATGAAAAAAGAACTGAGCTTTCATG-3′ | ||
| 17 | infuR_amyE(15bp)-ywtB(25bp) | 5′-GAATCGTTTTGCAAATTTAGATTTTAGTTT- |
| GTCACTATGATC-3′ | ||
| 18 | amyE-F | 5′-TTTGCAAAACGATTCAAAACCTCTTTACTG- |
| CCG-3′ | ||
| 19 | pACYC184_T5p-F | 5′-gcgaccacacccgtcctgtggatcgacgtctaagaaaccattattat- |
| catgacattaacc-3′ | ||
| 20 | pACYC184_T5p-R | 5′-ttctcctctttaatgaattctgtgtgaaattgttatccgctcacaattg-3′ |
| 21 | EcoRI_RBS_ATG_iPCR-F | 5′-gaattcattaaagaggagaaattaactATG-3′ |
| 22 | rrnBT1terminator_lacI-R | 5′-cccgactggaaagcgggcagtgaggcggatttgtcctactcagga- |
| gagcg-3′ | ||
| 23 | lacI-F | 5′-tcactgcccgctttccagtcgggaaacctgtcg-3′ |
| 24 | pACYC184_lacIqp-R | 5′-ccgccgccgcaaggaatggtggacaccatcgaatggtgcaaaac- |
| ctttcgcgg-3′ | ||
| 25 | BamH1-tamakiN | 5′-cccggatccgaggccctttcgtcttcacctcgag-3′ |
| 26 | Hind3-amyE-C | 5′-cccaagcttggcTCAATGGGGAAGAGAACCGCTT- |
| AAGC-3′ | ||
| 27 | H5_TGA_BglII_KpnI | 5′-CATCACCACCATCATTGAagatctgagcatggtaccc- |
| ttgaggc-3′ | ||
| 28 | H5_amyE_comp | 5′-GTGATGATGATGATGGGGAAGAGAACCGC- |
| TTAAGCCCCAGTC-3′ | ||
| Plasmid Used to Transform KT160 | Yield of Putrescine (μM) | SD | 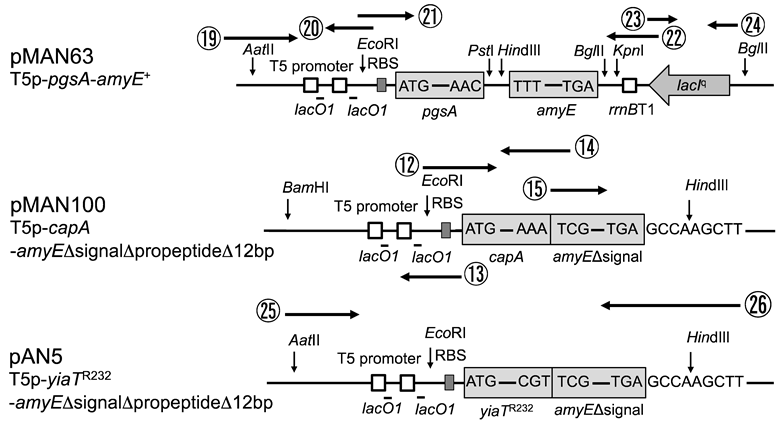 |
| pMAN63 | 292 | 3.4 | |
| pMAN100 | 229 | 17 | |
| pAN5 | 246 | 8.3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suzuki, H.; Iwamoto, N.; Nishimura, M. Construction of an Escherichia coli Strain Capable of Utilizing Steamed Rice as the Sole Carbon and Energy Source by Extracellular Expression of Amylase and Its Use for the Production of Putrescine. Appl. Microbiol. 2023, 3, 375-387. https://doi.org/10.3390/applmicrobiol3020026
Suzuki H, Iwamoto N, Nishimura M. Construction of an Escherichia coli Strain Capable of Utilizing Steamed Rice as the Sole Carbon and Energy Source by Extracellular Expression of Amylase and Its Use for the Production of Putrescine. Applied Microbiology. 2023; 3(2):375-387. https://doi.org/10.3390/applmicrobiol3020026
Chicago/Turabian StyleSuzuki, Hideyuki, Nana Iwamoto, and Manami Nishimura. 2023. "Construction of an Escherichia coli Strain Capable of Utilizing Steamed Rice as the Sole Carbon and Energy Source by Extracellular Expression of Amylase and Its Use for the Production of Putrescine" Applied Microbiology 3, no. 2: 375-387. https://doi.org/10.3390/applmicrobiol3020026
APA StyleSuzuki, H., Iwamoto, N., & Nishimura, M. (2023). Construction of an Escherichia coli Strain Capable of Utilizing Steamed Rice as the Sole Carbon and Energy Source by Extracellular Expression of Amylase and Its Use for the Production of Putrescine. Applied Microbiology, 3(2), 375-387. https://doi.org/10.3390/applmicrobiol3020026






