Abstract
Biochar, derived from the pyrolysis of plant materials has the potential to enhance plant growth in soilless media. Howevetar, little is known about the impact of biochar amendments to soilless growth media, microbial community composition, and fate of chemical constituents in the media. In this study, different concentrations of biochar were added to soilless media and microbial composition, and chemical constituents were analyzed using metagenomics and gamma spectroscopy techniques, respectively. Across treatments, carboxyl-C, phenolic-C, and aromatic-C were the main carbon sources that influenced microbial community composition. Flavobacterium (39.7%), was the predominantly bacteria genus, followed by Acidibacter (12.2%), Terrimonas (10.1%), Cytophaga (7.5%), Ferruginibacter (6.0%), Lacunisphaera (5.9%), Cellvibrio (5.8%), Opitutus (4.8%), Mucilaginibacter (4.0%) and Bryobacter (4.0%). Negative relationships were found between Cytophaga and 226Ra (r = −0.84, p = 0.0047), 40K (r = −0.82, p = 0.0069) and 137Cs (r = −0.93, p = 0.0002). Similarly, Mucilaginibacter was negatively correlated with 226Ra (r = −0.83, p = 0.0054) and 137Cs (r = −0.87, p = 0.0021). Overall, the data suggest that high % biochar amended samples have high radioactivity concentration levels. Some microorganisms have less presence in high radioactivity concentration levels.
1. Introduction
The use of soilless substrate as plant growth media over the past five and half decades has attracted high attention globally [1]. Organic (peat, compost, coir, bark, and wood fiber) and inorganic (Rockwool volcanic rock, tuff, expanded clay granules, vermiculite, zeolite, and pumice) materials have been utilized as soilless substrates, individually and in combination, and with additives such as fertilizers [2]. In the past, peat has been the major material used in container agriculture. But recent concerns about its economic cost and environmental implications [3], have led to renewable organic materials being used as substitutes. Some common renewable materials currently used as soilless substrates documented in the scientific literature are coconut (Cocos nucifera L.) husk fiber (coir), ground pine (Pinus taeda L.) bark and logs, rice (Oryza sativa L.) hulls, and switchgrass (Panicum virgatum L.) [1]. The physical and chemical properties of substrates can be affected by their inherent components [4], and these physicochemical properties can be manipulated with the use of additives such as biochar.
Biochar is a carbon (C) rich charred organic product derived from the pyrolysis of waste biomass in the absence of, or little oxygen [5]. Extensive studies and reviews have been conducted on biochar amendment to traditional or mineral soil [6]. The type of biomass, pyrolysis temperature, and storage conditions influence the physicochemical properties of biochar [7]. Biochar influences the physicochemical properties of soil by increasing soil pH to improve soil fertility, changing soil bulk density, and improving organic C and cation exchange capacity [8]. Additional benefits of biochar are soil water retention, improved nutrient retention to increase crop yield, and altered microbial populations and functions in soil [5]. The resistance of biochar to microbial degradation has been highlighted by [9]; this limits the release of C in the form of CO2 into the atmosphere to mitigate climate change.
Various environmental factors such as moisture content, temperature, and organic matter determine the abundance and activities of microorganisms in soil [10]. Soil microbial communities play a vital role in nutrient acquisition [11]. Microbial communities are considered important biological indicators of terrestrial ecosystem stress because of their sensitivity to abiotic changes, soil quality, and plant cover [10]. Understanding the composition and diversity of microbial communities in relation to environmental parameters is very important [12] and soil microbial community structure and activity, can be altered and enhanced to improve soil properties by biochar amendment [13].
Radionuclides can either be natural or anthropogenic [14]. The three major primordial radionuclides are 238U and 232Th, and 40K, and these occur naturally in minute concentrations depending on the geological, and geographical nature of the parent rock and soil [14,15]. Atmospheric nuclear weapons testing, nuclear accidents such as Chernobyl and Fukushima, and mining activities have been the primary sources of global anthropogenic radioactive contaminants in the environment for the past seven and half decades [16,17,18].
The release of radioactive materials into the environment leading to exposure to the population has stimulated intense public concern and has substantially led to research on the fate of major radionuclides in the environment [19,20]. Radionuclides in growth media for agriculture and horticulture crop production can potentially be a long-term source of radiation exposure to humans and the environment due to their accumulation by plants [21].
Literature has shown that the mobility and interaction of a range of radionuclides in both natural and engineered environments are influenced by microorganisms [20]. Microbial communities, since the origin of cellular life, have periodically been exposed to contaminated environments [22]. While there have been numerous studies of microbe-radionuclide interactions in agricultural soils [15,23,24,25], to our knowledge, there have not been any studies in biochar-amended soilless growth media on the relationships among microbial community diversity and the activity of radionuclides.
With fixed land area and a growing population worldwide, the causes and effects of the deterioration of agricultural land have been debated elsewhere [26]. Globally, anthropogenic activities are converting natural land cover into human-dominated ecosystems [27]. It has been reported that approximately 13 million hectares of natural land cover were converted to other land uses each year between 2000 and 2010 [27,28]. However, as a potential mitigation strategy for the deterioration of agricultural lands, soilless-biochar amendment systems for agriculture are gaining increasing attention globally [29,30].
However, studies involving relationships among C composition, microbial communities, and radionuclides in soilless growth media amended with biochar are in their infancy. For this reason, we examined C composition, bacteria composition, and radionuclide activities in soilless growth media amended with biochar samples collected from Florida Agriculture and Mechanical University Research and Extension Center (FAMU-REC) located in Quincy, Florida (FL). The aim of the present study was to determine the relationships among (i) C composition and thermal stability (R400) and microbial diversity, and (ii) microbial diversity and radionuclides activity levels, in growth media amended with biochar samples. The present study is the first to provide baseline data among these parameters in biochar-amended growth media, not only for FAMU-REC, but for Florida, the country, and the world.
2. Materials and Methods
2.1. Samples Collection Site Description
The study site where samples were collected was the FAMU-REC in Quincy, Gadsden County, FL in the USA. The site on the Florida-Georgia State line (30°67′ N and 84°61′ W) is approximately 30 miles from the main university campus in Tallahassee, FL. The FAMU-REC consists of more than 100 acres of farmland, pines forest, lakes and animal research facilities, and laboratories. It has annual high and low average temperatures of 26.1 °C (79.0° F) and 12.9 °C (55.3° F), respectively, with an annual average temperature of about 19.5 °C (67.2° F), annual precipitation of 59.67 inches, and humidity level of approximately 94.0%.
2.2. Media Composition
A soilless growth media consisting of a mixture of 60% coconut coir and 40% fine pine bark was used to prepare 8 biochar treatments containing 1%, 2%, 3%, 4%, 6%, 8%, 10%, and 12% biochar, respectively, plus a control, without biochar. Compressed coconut coir bricks of 8 × 4 × 2-inches were expanded and rehydrated by soaking in water, expanding volume to 5–7 times the original size. A cement mixer was used to mix the soilless media treatment and biochar amendments, which were dispensed into 3-gallon plastic containers to give 3 replicates per treatment. Triplicates of each treatment were collected for analysis and the means were reported.
2.3. Sample Analysis
2.3.1. pH Determination
1 g of sample was placed in 20 mL of deionized water (DI), shaken for 1.5 h, and then left for 5 min to equilibrate before measuring pH with a Fisher Scientific Accumet Basic AB15 pH meter [31,32,33].
2.3.2. Nuclear Magnetic Resonance Analysis (NMR)
Nuclear magnetic resonance technique was used to evaluate the carbon composition of the soilless media’s carbon functional groups. Finely milled powder samples of soilless media amended with various percentages of biochar by weight were analyzed using the magic angle spinning 13C solid-state nuclear magnetic resonance (MAS 13C SSNMR) technique, as previously employed by [31,34] using a Bruker 300 MHz DR NMR spectrometer equipped with a Bruker 4.0 mm double resonance NMR probe.
2.3.3. Multi-Elemental Scanning Thermal Analysis (MESTA)
Sample total C content was determined using the multi-elemental scanning thermal analysis (MESTA) technique previously employed by [31,35,36]. Carbon thermograms were created using C content. Due to the high C concentration in biochar [31], a dilution consisting of a 1:5 mixture of sample: talc by weight was applied before the MESTA analysis to improve thermogram resolution. The analyses were performed using Antek 9000HN Series Nitrogen Analyzer by SpectraLab Scientific Inc., Markham, ON Canada.
Low and high carbon stability were examined for C recovered at temperatures of < 400 °C and >400 °C, respectively [31]. R400 is the region identified below 400 °C normalization divided by the total surface area [37]. Using Equation (1) below, R400 was computed as the fraction of low thermal stability to total C of samples based on the data acquired from the low C thermal stability (<400 °C):
2.3.4. DNA Extraction, Quantification, and Purity, Metagenomics
Samples’ genomic DNA was extracted using DNeasy Powersoil Kit (QIAGEN Inc., Germantown, MD, USA) per the manufacturer’s instruction. A NanoDrop 1000 (NanoDrop Technologies, Wilmington, DE, USA) was used to quantify the total DNA of the extracted samples by measuring the concentrations (ng/µL) by absorbance at A260/280, and A260/230 ratios [38]. Sequence libraries were prepared using universal primers 345F (GTGCCAGCMGCCGCGGTAA) and 371R (CCGYCAATTYMTTTRAGTTT) to perform the amplification of the16S r RNA metagenomics. A mid-output kit with 2 × 150 paired-end sequencing was utilized using Illumina MiSeq437 equipment to do the sequencing [39].
PEAR was used to integrate forward and backward reads [40]. Based on a quality threshold of p = 0.01, combined readings were edited to remove ambiguous nucleotides and primer sequences. Any reads with sequences shorter than 300 bp or without a primer sequence were eliminated. Using the USEARCH algorithm and a comparison to the Silva v132 reference sequence database, chimeric sequences were found and eliminated [41,42].
To produce taxonomy summaries utilizing a sub-OTU resolution of the sequence collection, the conventional QIIME pipeline was modified [43,44]. The generated sequence files were then quickly combined with the sample data. The list of unique sequences was then produced by dereplicating each sequence. All sequences with at least 10 counts of abundance were referred to as seed sequences. The next step was to use USEARCH to locate the closest seed sequence for any non-seed sequence that met the 97% minimum identity requirement. If a non-seed sequence matched a seed sequence, its counts were combined with the counts from the seed sequence, and if it didn’t, it remained an independent sequence [41].
With a minimum similarity threshold of 90%, taxonomic annotations for seed and mismatched non-seed sequences were assigned using the USEARCH and Silva v132 reference [41,42]. The usual QIIME assignment algorithm was changed to only consider hits at each taxonomic level with an assigned name to increase the depth of annotation. When assigning the taxonomic kingdom through the family, a reference annotated as “k Bacteria; p Firmicutes; c Clostridia; o Clostridiales; f Ruminococcaceae; g; s_” would be considered, but it would not be used when assigning the genus or species. Additionally, to be considered for genus or species level designation, any hits in the reference database must have a minimum identity of 97% or 99%, respectively. Then, sequence abundance data and taxonomic annotations were combined into a single sequencing table.
2.3.5. Radionuclides Sample Preparation and Analysis
Samples of the soilless media with the various biochar amendments were dried at 110 °C for 48 h in an electric oven to remove moisture content [45]. The dried samples were transferred into 500 mL Marinelli beakers of the same geometry as the reference material, covered, sealed with parafilm to limit any possible escape of radon, and relabeled as above. The prepared samples were left for at least 30 days to reach secular equilibrium with radon and its daughters [45,46,47]. Each sample was handled carefully, and proper measures were taken to prevent cross-contamination.
Gamma spectrometry analyses were performed using high purity germanium (HPGe); detector by Canberra Industries/Merion Inc., Meriden, CT. The detector was shielded with a thick lead shield with Cu inner layer to reduce the detection of background radiation. The HPGe detector was coupled with a Canberra DSA-2000 data acquisition system and connected directly with a PC equipped with Canberra Genie 2000 software in which measured gamma spectra were stored and analyzed. The software internally calculates activity concentrations of radionuclides from all prominent gamma lines with background subtraction [48]. The instrument has an energy resolution of 0.5keV full width at half of the maximum (FWHM) for a 1332 keV channel (using Co-60) and a relative photopeak efficiency of 35%. The instrument was calibrated for energy and efficiency over the photon energy range of 2 to 2000 keV using a National Institute of Standards and Technology (NIST) traceable mixed gamma standard. Each sample was counted for a period of 86400s.
2.3.6. 235U, 226Ra, 232Th, 40K and 137Cs
The activity concentrations of the natural radionuclides 235U and 40K, and the anthropogenic radionuclide 137Cs were determined directly from their photopeak energies lines of 185.7 (54.0%) 1460.8 (10.7%), and 661.7 (85.1%) keV, respectively [45]. The weighted mean photopeak energy lines of 214Pb (295.2 and 351.9) and 214Bi (609.3 and 1120.3 keV) were used to estimate the activity concentration value of 226Ra. The weighted mean photopeak energies lines of 212Pb (238.6 keV), 212Bi (727.2 keV), and 228Ac (338.3, 911.6, and 969.1 keV) were used to determine the activity concentration value of 232Th [46,49]. Using the weighted mean photopeak procedure for multiple energy lines gives more accurate results with lower errors than using only one of the photopeak lines [50]. The measured and estimated activity concentration values of the radionuclides are reported in Bqkg−1.
2.3.7. Metagenomic Sequence Accession Numbers
The 16S metagenomic sequences obtained from this study are available from NCBI’s Sequence Read Archive under Bioproject accession # PRJNA773140.
2.3.8. Statistical Analysis
JMP software (version 13.2.1, SAS, Cary, NC 27513, USA) was used to conduct ANOVA and Pearson correlation analysis for this study. Data are reported as means and standard error of the mean (SEM). Analysis of variance using post-hoc Turkey HSD tests, where α = 0.05, was used to determine significant differences among treatment variables. Sigma plot (version 12.0) was used to plot correlations between variables.
3. Results
3.1. Samples’ Basic Physicochemical Characteristics
Physicochemical characteristics of growth media samples from control (0% biochar) and different percentages of biochar amendments are reported in (Table 1). pH increased with an increasing percentage of biochar amendments, with values ranging between 6.02–6.84. (Table 1). Similarly, TC, C composition; carboxyl-C, phenolic-C, and aromatic-C, increased with the increasing percentage of biochar amendments to growth media. TC, carboxyl-C, phenolic-C, and aromatic-C, concentration values ranged between 443.03 to 514.47, 14.58 to 38.29, 32.17 to 56.11, and 65.66–95.59 g kg−1, respectively (Table 1).

Table 1.
Samples’ physicochemical and C composition properties with TC, carboxyl-C, phenolic-C, and aromatic-C in g kg−1.
The R400 value for the growth media without added biochar was 0.61. with added biochar, the R400 values ranged from 0.50 to 0.70. However, the 1% biochar amended growth media recorded the highest R400 value: 0.7. The values then tend to decrease with an increasing percentage of biochar amendments (Table 1). Generally, R400 exhibited a declining trend with increasing biochar percentage amendments, decreasing from 0.7 for 1% biochar to 0.5 for 12% biochar (Table 1).
Cluster analysis revealed close similarity among M03, M04, M12, M08, and M10, representing 3%, 4%, 12%, 8%, and 10% biochar amendments, respectively (Figure 1). M0, M1, M2, and M6, representing biochar amendments of 0, 1, 2, and 6%, respectively, clustered independently. These findings indicate that biochar modifications had an impact on media qualities.
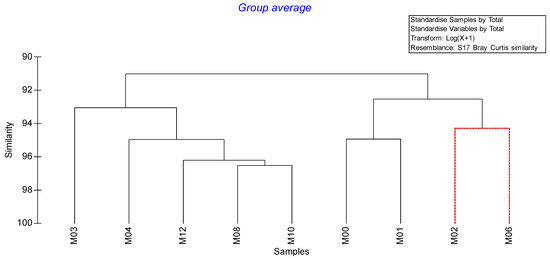
Figure 1.
Cluster analysis without considering biochar media amendments as a variable.
3.2. Media Microbial Composition
Sample microbial composition and diversity were evaluated by 16S rRNA gene sequencing-based metagenomics. Proteobacteria (41.6%), Bacteroidetes (27.0%), Verrucomicrobia (7.9%), Acidobacteria (5.9%), Planctomycetes (3.9%), Actinobacteria (2.9%), Chloroflexi (2.2%), Cyanobacteria (1.4%) and Patescibacteria (1.3%) were the 9 predominant bacteria phyla in all samples. The phyla level bacteria relative abundance is reported in Figure S1. At the genera level, Flavobacterium (39.7%) was the dominant bacteria identified in the samples, followed by Acidibacter (12.2%), Terrimonas (10.1%), Cytophaga (7.5%), Ferruginibacter (6.0%), Lacunisphaera (5.9%), Cellvibrio (5.8%), Opitutus (4.8%), Mucilaginibacter (4.0%) and Bryobacter (4.0%), with their relative abundance shown in Figure 2.
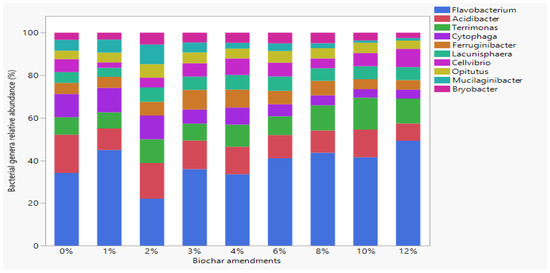
Figure 2.
Relative abundance of dominant bacteria genera in different % biochar amended soilless growth media.
3.3. Radioactivity Measurements
Radionuclide activities are summarized in Table 2. 235U, 226Ra, 232Th, and 40K were detected as natural radionuclides, as was anthropogenic 137Cs. The natural radionuclide with the highest recorded activity concentration (Bq kg−1) was 40K, followed by 226Ra, 232Th, and 235U in that order. Interestingly, even though 40K recorded the highest activity concentration in the analyzed samples, its presence was not detected in the control and 1% biochar amended samples. The activity concentrations for 235U, 226Ra, 232Th, 40K and 137Cs ranged between 0.81–0.99, 2.54–4.14, 0.57–1.05, 19.57–24.23 and 0.19–0.55 Bq kg−1, respectively (Table 2). Activity concentrations of 226Ra and 137Cs increased with an increasing percentage of biochar amendment to control media. Correlation analysis of radionuclides contents indicated that 226Ra and 40K concentrations correlated significantly (r = 0.67, p = 0.0497), suggesting 226Ra and 40K have dissolution similarities in media (Table 3). Similarly, 226Ra and 137Cs (r = 0.83, p = 0.0052), 40K and 137Cs (r = 0.74, p = 0.0231) concentrations significantly correlated (Table 3). However, 235U and 232Th were not significantly correlated with the other radionuclides (Table 3).

Table 2.
Activity Concentration of Radionuclides (Bq kg−1) from FAMU-RCE growth media.

Table 3.
Correlation matrix of detected radionuclides in analyzed samples.
3.4. Media Physicochemical Properties Relationship with Bacterial Composition and Radionuclides Activity Concentrations
Figure 3 shows that pH was related both positively and negatively to the percentage of various bacteria phyla in treatments. Specifically, media pH positively correlated with Chloroflexi (r = 0.73, p = 0.0268) and negatively correlated with Cyanobacteria (r = −0.70, p = 0.0368) (Figure 3a,b, respectively). At the bacterial genera level, there were significant negative correlations between pH and the genera Cytophaga (r = −0.91, p = 0.0007) and Mucilaginibacter (r = −0.90, p = 0.0008) (Figure 3c,d, respectively). Similarly, Figure 3e,f, respectively, show that pH was positively correlated with radionuclides 226Ra (r = 0.89, p = 0.0012) and 137Cs (r = 0.93, p = 0.0002).
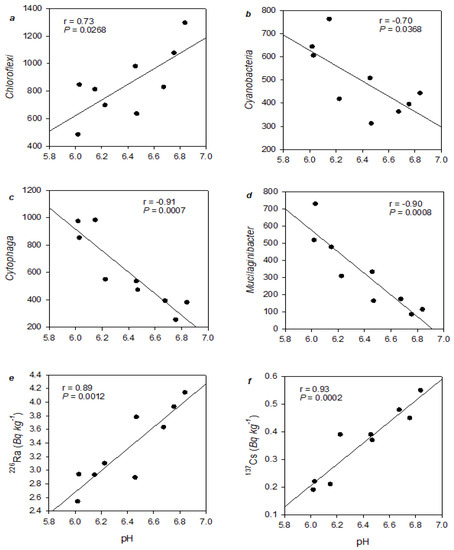
Figure 3.
Correlations of pH with number of genotypes detected in various bacterial phyla: (a) Chloroflexi and (b) Cyanobacteria, genera: (c) Cytophaga and (d) Mucilaginibacter, and correlations of pH with radionuclide activity concentrations: (e) 226Ra and (f) 137Cs.
Additionally, C composition showed relationships with bacterial composition. As reported in Figure S2, TC, phenolic-C, and aromatic-C and bacterial phyla Acidobacteria, Chloroflexi and Cyanobacteria exhibited positive and negative relationships. Acidobacteria exhibited a significant negative relationship with aromatic-C (r = −0.67, p = 0.0461) and phenolic-C (r = −0.68, p = 0.0436) (Figure S2a,b, respectively). In contrast to Acidobacteria, the bacteria phylum Chloroflexi was positively correlated with aromatic-C (r = 0.73, p = 0.0261), and TC (r = 0.94, p = 0.0225) (Figure S2c,d, respectively). Like Acidobacteria, there was a significant negative relationship between Cyanobacteria and phenolic-C (r = −0.70, p = 0.0369) (Figure S2e).
Furthermore, the genus Cytophaga was negatively correlated with carboxyl-C (r = −0.77, p = 0.0154) (Figure S3a), phenolic-C (r = −0.87, p = 0.0026) (Figure S3b) and aromatic-C (r = −0.86, p = 0.0027) (Figure S3c). Similarly, the genus Mucilaginibacter was negatively correlated with carboxyl-C (r = −0.79, p = 0.0114), phenolic-C (r = −0.84, p = 0.0045), and aromatic-C (r = −0.87, p = 0.0021) (Figure S3d–f, respectively). Aromatic-C and phenolic-C were correlated with the bacteria genera Terrimonas (r = 0.70, p = 0.0375) and Acidibacter (r = −0.71, p = 0.0318), respectively (Figure S4a,b).
Significant relationships among C composition and radionuclides were positive correlated between carboxyl-C and 226Ra (r = 0.78, p = 0.0129), phenolic-C and 226Ra (r = 0.81, p = 0.0077), aromatic-C and 226Ra (r = 0.91, p = 0.0006) (Figure S5a–c). Carboxyl-C and 137Cs (r = 0.90, p = 0.0009), phenolic-C and 137Cs (r = 0.90, p = 0.0009) and aromatic-C and 137Cs (r = 0.89, p = 0.0011) again correlated positively (Figure S5d–f). Similarly, TC was positively correlated with 226Ra (r = 0.91, p = 0.0008) and 137Cs (r = 0.89, p = 0.0012) (Figure S6a,b). The results from this study indicated that higher values of C composition (carboxyl-C, phenolic-C, and aromatic-C) are associated with higher activity concentration values of 226Ra and 137Cs, and vice versa (Table 1 and Table 2).
Relating to C thermal stability, which was translated to R400, bacterial phyla Chloroflexi (r = −0.92, p = 0.0004) and Patescibacteria (r = −0.71, p = 0.0335) both correlated negatively with R400, respectively (Figure S7a,b). Additionally, R400 was negatively correlated with the genera Terrimonas (r = −0.79, p = 0.0107) and Cellvibrio (r = −0.79, p = 0.0108) (Figure S7c,d). Similarly, R400 showed significant negative relationship with 226Ra (r = −0.74, p = 0.0227) and 137Cs (r = −0.72, p = 0.0292) (Figure S7e,f).
3.5. Samples Bacteria Composition Relationship with Radionuclides Contents
Bacterial phylum Cyanobacteria was negatively correlated with 226Ra (r = −0.73, p = 0.0270) (Figure 4a), 137Cs (r = −0.79, p = 0.0121) (Figure 4b) and 40K (r = −0.80, p = 0.0100) (Figure 4c). In contrast, Chloroflexi was positively correlated with 137Cs (r = 0.67, p = 0.0468) (Figure 4d). At the bacteria genera level, both Cytophaga and Mucilaginibacter negatively correlated with radionuclide activity concentrations. Cytophaga was positively correlated with 226Ra (r = −0.84, p = 0.0047), 40K (r = −0.82, p = 0.0069) and 137Cs (r = −0.93, p = 0.0002) (Figure 5a–c). Similarly, Mucilaginibacter was positively correlated only with 137Cs (r = 0.87, p = 0.0021) and 226Ra (r = −0.83, p = 0.0054) (Figure 5d,e). The negative correlations of the above bacteria phyla and genera with the radionuclides suggested that greater counts of these bacteria phyla and genera are associated with lower radionuclide content in the samples. However, there were no significant correlations for sample physicochemical parameters, and or for bacterial proportions with activity concentrations of 235U and 232Th.
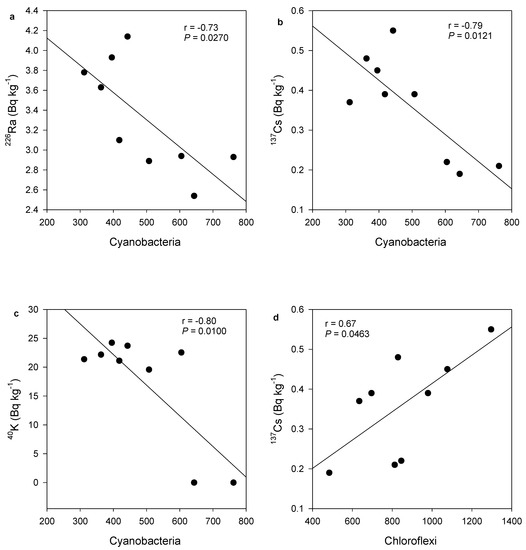
Figure 4.
Relationships between number of bacterial genotypes detected from various phyla and activity concentrations of radionuclides. Cyanobacteria correlations with (a) 226Ra, (b) 40K and (c) 137Cs. (d) Chloroflexi correlation with 137Cs.
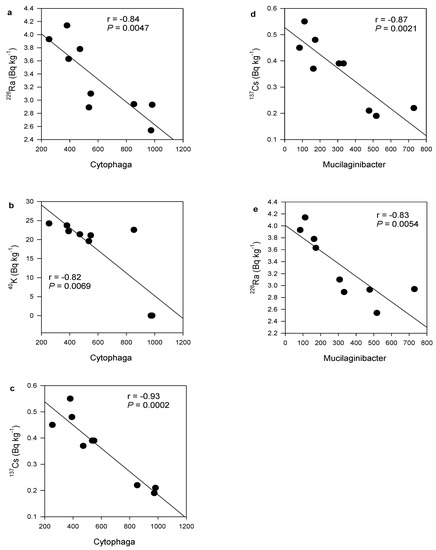
Figure 5.
Correlations between number of genotypes detected from the genus Cytophaga with activity concentrations of (a) 226Ra, (b) 40K, and (c) 137Cs, and from the genus Mucilaginibacter with (d) 137Cs and (e) 226Ra.
4. Discussion
The physiochemical factor pH strongly influences bacterial diversity in soils [51,52]. Microbial populations can be strongly influenced by media pH some bacteria can adapt to extreme pH, although many enzymes and proteins can become denatured and inactivated under high acidic (<4.0) and alkaline (>9.0) conditions, impairing many metabolic processes [51]. Ref. [51] reported that most bacteria favor circumneutral pH conditions (between 6.5 and 7.5). Amending biochar to media changed the media pH which, in turn, likely affected the bacterial community composition [53].
Despite the complex nature of environmental microbial communities, our results show patterns characterizing microbial abundance of Proteobacteria phylum in the biochar amended growth media. Literature has indicated the consistent abundance of Proteobacteria in many studied areas, and the relative abundance of Proteobacteria, regardless of the % biochar added to the growth media, was high compared to other bacterial phyla. Proteobacteria have been recognized as plant growth-promoting bacteria and facilitate nutrient acquisition and disease protection [54]. For that reason, a low relative abundance of Proteobacteria [54], can have a major effect on the plant productivity and media health in an agricultural environment. Proteobacteria are globally recognized for their important role in carbon cycling [55], consistent with the high abundance of Proteobacteria because of the high carbon concentration of biochar.
In agricultural systems, Bacteroidetes are abundant and noted for their ability to metabolize organic matter exploitation [56]. In the present study, the high abundance of Bacteroidetes, which was second only to Proteobacteria, regardless of the percentage of biochar added to growth media, agreed with previous findings [56]. According to [57,58], Bacteroidetes promote plant growth and plants’ resistance to environmental stress. Other authors reported that Bacteroidetes are enriched in environments with high C availability [59], and hence their high abundance in biochar amendment media in the present study was not a coincidence. Verrucomicrobia was third in abundance in all the samples analyzed, ranked behind Proteobacteria and Bacteroidetes Based on their lower growth rates and adaptations for growing on relatively recalcitrant forms of C, a major component of biochar, and a major constituent of the growth media in the present study, Verrucomicrobia is classified as an “oligotroph” [60].
Acidobacteria is also another diverse group of bacteria found in soil [61,62] and has been affiliated with the biogeochemical cycling of C, a major constituent of biochar [63]. Additionally, Acidobacteria has been associated with the degradation of recalcitrant polymer and is considered an important phylum ecologically in the turnover of soil organic matter [61,64]. Therefore, its presence in the biochar-amended media in this study can be attributed to its association with recalcitrant C compounds (the main constituent of biochar) [63]. Similarly, previous research suggested that Acidobacteria prefer an acidic environment [65]. This was consistent with the high abundance of Acidobacteria in lower pH samples in this study. Furthermore, Acidobacteria exhibits an oligotrophic lifestyle [66] consistent with our study because of their negative correlation with carbon composition as previously suggested [66].
In an agricultural setting, Actinobacteria enhance plant growth and improve plant nutrition. They provide direct plant growth-promoting mechanisms such as nitrogen fixation to improve plant nutrition [67]. Actinobacteria are found widely in different environments and play a critical role in organic matter decomposition [55,68]. This is in line with its negative relationship with R400 observed in this study. Also, Actinobacteria are known for the production of antibiotics and other secondary metabolites to suppress other bacteria [69].
An essential macronutrient limiting agricultural productivity, N, is the largest and most costly input in agriculture [70]. Atmospheric and dissolved dinitrogen (N2) are abundant in soil and water, yet unable for used by plants [70]. However, there is conclusive evidence that N is made available for plants and other organisms through N fixation by Cyanobacteria [71] Hence the presence of Cyanobacteria in this study was an important benefit in the biochar amended growth media for use in agriculture. In this study, Chloroflexi increased with increasing media parameters such as pH, carboxyl-C, phenolic-C, and aromatic-C, while Cyanobacteria decreased with increases in the same media parameters. This scenario agrees with an observation by [60] in agricultural systems of arid, continental, and temperate regions. It was reported that Chloroflexi metabolic flexibility can be a disadvantage in competition with Cyanobacteria for nutrient and physical space when they co-exist in the same environment [72].
Flavobacterium, the predominant bacterial genera found in samples collected from this study, has been shown to be widely distributed in nature and functions in different types of organic matter mineralization and rapid digestion of insoluble chitin [73,74]. The genus Cellvibrio has previously been reported to produce hydrolytic enzymes [73]. Media biochar amendment has been shown to stimulate the genera Flavobacterium and Cellvibrio. The genus Cellvibrio has been reported to promote plant growth and act as an inducer of plant systemic resistance [73]. This is in line with what resulted in this study where Cellvibrio was negatively correlated with R400 (p = 0.0108), translating into the decomposition of organic matter for available use in crops. The genus Cytophaga belongs to the phylum Bacteroidetes and is known for its ability to degrade cellulose [75] and other high molecular weight organic compounds [76].
In this study, increasing the percentage of biochar amendments increased pH, which was positively correlated with 226Ra and 137Cs activity concentrations. Therefore, it can be implied that pH has a great influence on radionuclides availability, a conclusion reported in the literature [22]. Additionally, it has been reported that media chemical parameters such as pH contributes to the various effect of radium mobility or adsorption [77,78], and we observed that 226Ra concentrations in samples varied with pH. The very significant correlations of the anthropogenic radionuclide 137Cs with C composition of the samples can be attributed to 137Cs fixation by organic C, in agreement with what has been previously reported [79]. The association of 40K with soil mineral fraction contributed to the lack of significant correlation of this radionuclide with samples’ C composition [79].
The relative abundance of Proteobacteria (41.9–47.2%), Bacteroidetes (25.1–31.8%), Acidobacteria (5.2–8.3%) and Actinobacteria (2.6–4.5%) in the biochar-amended growth media reported in this study reflects their frequent presence in radionuclide contaminated environments [39,80,81,82]. For example, Bryobacter identified in our samples is known to withstand extreme environments including uranium-contaminated samples [83,84]. However, the above predominant bacteria phyla and genera did not have a significant correlation with the detected radionuclides in the present study.
At the phyla level, Chloroflexi and Cyanobacteria were the two phyla that significantly correlated with some of the detected radionuclides. The presence of Chloroflexi has been reported in natural uranium ores [16,85] and uranium-contaminated sites [16,86]. Elsewhere, Chloroflexi has been proposed to have the ability to degrade plant polymers, lignocellulosic material, and tolerate uranium and its associated radioactive progenies [86]. However, in our study, Chloroflexi was not correlated with any of the uranium progenies detected in this study but was correlated with 137Cs. To our knowledge, this study is the first to report a significant negative correlation between Cyanobacteria and radionuclides 226Ra, 40K, and 137Cs. But with respect to heavy metals, Cyanobacteria can produce polyphosphate granules for Cu immobilization that could be adsorbed to transport heavy metals into cells of Cyanobacteria [87,88].
The identified bacteria genera, Cytophaga and Mucilaginibacter significantly correlated with the detected radionuclides in the present study. These two bacteria genera, Cytophaga and Mucilaginibacter can be suggested to resist 226Ra, 40K, and 137Cs, and 226Ra and 137Cs contaminants, respectively, due to their negative relationships. This is in line with previous reports where Mucilaginibacter was identified as a metal-resistant bacterium [87].
5. Conclusions and Future Research
This study investigated the relationships among the physiochemical properties, carbon composition, R400, and the indigenous microbial communities and radionuclide concentrations in biochar-amended growth media. We concluded that the samples’ pH significantly influences carbon composition, microbial composition, and the activity concentrations of the radionuclides. Simultaneously, Acidobacteria, Chloroflexi, and Cyanobacteria significantly correlated with carboxyl-C, phenolic-C, and aromatic-C. There were significantly negative relationships of Cyanobacteria with 226Ra, 40K, and 137Cs, which indicated resistance between Cyanobacteria and the radionuclides. Chloroflexi had a significant positive relationship with 137Cs, suggesting tolerance. Like the phylum Cyanobacteria, the genera Cytophaga and Mucilaginibacter had significant negative relationships with 226Ra, 40K, and 137Cs, and 226Ra and 137Cs.
The relationships among C composition, microbial diversity, and radionuclide distributions from biochar-amended growth media might provide a better understanding of the physiochemical properties of such media and the development of plant growth-promoting growth media in the future. Further analyses are required for deeper knowledge of other types of soilless media to ascertain their properties and behavior with respect to radionuclides.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/applmicrobiol2030051/s1, Figure S1: Relative abundance of dominant bacteria phyla as detected in different % biochar amended soilless growth media; Figure S2: Carbon composition relationships with bacteria phyla. a and b represent correlation between aromatic and phenolic-C, and Acidobacteria, c, and d is the relationships between aromatic-C and total carbon (TC) and Chloroflexi, while e shows the relationship between phenolic-C and Cyanobacteria; Figure S3 Carbon composition relationships with bacteria phyla. Cytophaga with (a) carboxyl-C, (b) phenolic-C, and (c) aromatic-C. and Mucilaginibacter with (a) carboxyl-C, (b) phenolic-C, and (c) aromatic-C; Figure S4: (a) Aromatic-C relationship with Terrimonas and (b) phenolic-C relationship with Acidibacter; Figure S5: Relationships of C composition with radionuclides 226Ra and 137Cs; Figure S6: Relationship between total C and (a) 226Ra and (b) 137Cs; Figure S7: R400 relationships with bacteria phyla: (a) Chloroflexi and (b) Patescibacteria, bacteria genera: (c) Terrimonas and (d) Cellvibrio, and radionuclides: (e) 226Ra and (f) 137Cs.
Author Contributions
Conceptualization, G.K.O., M.A., L.N. and A.B..; methodology, G.K.O., M.A., L.N. and A.C.; validation G.K.O., A.C. and C.J.; formal analysis, G.K.O.; writing—original draft preparation, G.K.O.; writing—review and editing, G.K.O., M.A., L.N., A.C., A.B., A.P. and C.J. All authors have read and agreed to the published version of the manuscript.
Funding
Funding numbers 1901377 and 2200615, as well as USDA award number, respectively. Financial support from FAMU’s title III program and the School of graduate studies is also acknowledged.
Data Availability Statement
Metagenomics data obtained from this study is available via National Center for Biotechnology Information’s (NCBI’s) Sequence Read Archive Bioproject accession # PRJNA773140.
Acknowledgments
Basic processing of the raw metagenomic data was performed by the University of Illinois at Chicago. The authors acknowledge faculty, staff, and students of the Nuclear Instrumentation Laboratory of Alcorn State University, Lorman, Mississippi for samples gamma spectroscopy analysis. We acknowledge the staff at FAMU-REC for growth media preparation. We also want to send our gratitude to Meenakshi Agarwal for the DNA extraction supervision and Djanan Nemours for MESTA analyses. Part of this study was made possible by support from the Title III Grant at Florida A&M University. A portion of this work was performed at the National High Magnetic Field Laboratory, which is supported by National Science Foundation Cooperative Agreement No. DMR-1644779* and the State of Florida.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Vaughn, S.F.; Kenar, J.A.; Thompson, A.R.; Peterson, S.C. Comparison of biochars derived from wood pellets and pelletized wheat straw as replacements for peat in potting substrates. Ind. Crop. Prod. 2013, 51, 437–443. [Google Scholar] [CrossRef]
- Gruda, N.S. Increasing Sustainability of Growing Media Constituents and Stand-Alone Substrates in Soilless Culture Systems. Agronomy 2019, 9, 298. [Google Scholar] [CrossRef]
- Barrett, G.E.; Alexander, P.D.; Robinson, J.S.; Bragg, N.C. Achieving environmentally sustainable growing media for soilless plant cultivation systems–A review. Sci. Hortic. 2016, 212, 220–234. [Google Scholar] [CrossRef]
- Jahromi, N.B.; Walker, F.; Fulcher, A.; Altland, J.; Wright, W.C. Growth response, mineral nutrition, and water utilization of con-tainer-grown woody ornamentals grown in biochar-amended pine bark. HortScience 2018, 53, 347–353. [Google Scholar]
- Graber, E.R.; Harel, Y.M.; Kolton, M.; Cytryn, E.; Silber, A.; David, D.R.; Tsechansky, L.; Borenshtein, M.; Elad, Y. Biochar impact on development and productivity of pepper and tomato grown in fertigated soilless media. Plant Soil 2010, 337, 481–496. [Google Scholar] [CrossRef]
- Ahmad, Z.; Mosa, A.; Zhan, L.; Gao, B. Biochar modulates mineral nitrogen dynamics in soil and terrestrial ecosystems: A critical review. Chemosphere 2021, 278, 130378. [Google Scholar] [CrossRef]
- Palansooriya, K.N.; Wong, J.T.F.; Hashimoto, Y.; Huang, L.; Rinklebe, J.; Chang, S.X.; Bolan, N.; Wang, H.; Ok, Y.S. Response of microbial communities to biochar-amended soils: A critical review. Biochar 2019, 1, 3–22. [Google Scholar] [CrossRef]
- Pérez-Guzmán, L.; Lower, B.H.; Dick, R.P. Corn and hardwood biochars affected soil microbial community and enzyme activities. Agrosystems Geosci. Environ. 2020, 3, e20082. [Google Scholar] [CrossRef]
- Sohi, S.P.; Krull, E.; Lopez-Capel, E.; Bol, R. A review of biochar and its use and function in soil. Adv. Agron. 2010, 105, 47–82. [Google Scholar] [CrossRef]
- Saul-Tcherkas, V.; Steinberger, Y. Substrate utilization patterns of desert soil microbial communities in response to xeric and mesic conditions. Soil Biol. Biochem. 2009, 41, 1882–1893. [Google Scholar] [CrossRef]
- Waghmode, T.R.; Chen, S.; Li, J.; Sun, R.; Liu, B.; Hu, C. Response of Nitrifier and Denitrifier Abundance and Microbial Community Structure to Experimental Warming in an Agricultural Ecosystem. Front. Microbiol. 2018, 9, 474. [Google Scholar] [CrossRef] [PubMed]
- Ligi, T.; Oopkaup, K.; Truu, M.; Preem, J.-K.; Nõlvak, H.; Mitsch, W.J.; Mander, Ü.; Truu, J. Characterization of bacterial communities in soil and sediment of a created riverine wetland complex using high-throughput 16S rRNA amplicon sequencing. Ecol. Eng. 2014, 72, 56–66. [Google Scholar] [CrossRef]
- Li, Y.; Li, Y.; Chang, S.X.; Yang, Y.; Fu, S.; Jiang, P.; Luo, Y.; Yang, M.; Chen, Z.; Hu, S.; et al. Biochar reduces soil heterotrophic respi-ration in a subtropical plantation through increasing soil organic carbon recalcitrancy and decreasing carbon-degrading mi-crobial activity. Soil Biol. Biochem. 2018, 122, 173–185. [Google Scholar]
- Khandaker, M.U.; Jojo, P.J.; Abu Kassim, H. Determination of Primordial Radionuclides in Natural Samples Using HPGe Gamma-Ray Spectrometry. APCBEE Procedia 2012, 1, 187–192. [Google Scholar] [CrossRef][Green Version]
- Shukla, A.; Parmar, P.; Saraf, M. Radiation, radionuclides and bacteria: An in-perspective review. J. Environ. Radioact. 2017, 180, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Theodorakopoulos, N.; Février, L.; Barakat, M.; Ortet, P.; Christen, R.; Piette, L.; Levchuk, S.; Beaugelin-Seiller, K.; Sergeant, C.; Berthomieu, C.; et al. Soil prokaryotic communities in Chernobyl waste disposal trench T22 are modulated by organic matter and radionuclide contamination. FEMS Microbiol. Ecol. 2017, 93. [Google Scholar] [CrossRef]
- Salbu, B.; Skipperud, L.; Lind, O.C. Sources contributing to radionuclides in the environment: With focus on radioactive particles. In Radionuclides in the Environment 2015; Springer: Cham, Switzerland, 2015; pp. 1–36. [Google Scholar]
- Hu, Q.-H.; Weng, J.-Q.; Wang, J.-S. Sources of anthropogenic radionuclides in the environment: A review. J. Environ. Radioact. 2010, 101, 426–437. [Google Scholar] [CrossRef]
- Correa, R.; Miranda, P.; Ortiz-Ramirez, P.; Wachter, J.; Camilla, S.; Mera, E.; Piñones, E. Activity concentration of NORM and 137Cs radionuclide in soil samples from the Andes Cordillera at latitude 33°56′ South. J. Physics Conf. Ser. 2018, 1043, 012028. [Google Scholar] [CrossRef]
- Lloyd, J.R.; Gadd, G.M. The geomicrobiology of radionuclides. Geomicrobiol. J. 2011, 28, 383–386. [Google Scholar]
- Belimov, A.A.; Kunakova, A.M.; Vasilyeva, N.D.; Kovatcheva, T.S.; Dritchko, V.F.; Kuzovatov, S.N.; Trushkina, I.R.; Alekseyev, Y.U. Accumulation of radionuclides by associative bacteria and the uptake of 134 Cs by the inoculated barley plants. In Nitrogen Fixation with Non-Legumes 1998; Springer: Dordrecht, The Netherlands, 1998; pp. 275–280. [Google Scholar]
- Yan, X.; Luo, X. Radionuclides distribution, properties, and microbial diversity of soils in uranium mill tailings from southeastern China. J. Environ. Radioact. 2015, 139, 85–90. [Google Scholar] [CrossRef]
- Newsome, L.; Morris, K.; Lloyd, J.R. The biogeochemistry and bioremediation of uranium and other priority radionuclides. Chem. Geol. 2013, 363, 164–184. [Google Scholar] [CrossRef]
- Merroun, M.L.; Selenska-Pobell, S. Bacterial interactions with uranium: An environmental perspective. J. Contam. Hydrol. 2008, 102, 285–295. [Google Scholar] [CrossRef]
- Cáliz, J.; Montserrat, G.; Martí, E.; Sierra, J.; Chung, A.P.; Morais, P.V.; Vila, X. Emerging resistant microbiota from an acidic soil exposed to toxicity of Cr, Cd and Pb is mainly influenced by the bioavailability of these metals. J. Soils Sediments 2012, 13, 413–428. [Google Scholar] [CrossRef]
- Wolman, M.G. Population, land use, and environment: A long history. In Population and Land Use in Developing Countries; The National Academies Press: Washington, DC, USA, 1993. [Google Scholar]
- Camill, P. Global change. Nat. Educ. Knowl. 2010, 3, 49. [Google Scholar]
- Assessment, G.F. Main Report; Food and Agriculture Organization of the United Nations: Rome, Italy, 2010. [Google Scholar]
- Huang, L.; Gu, M. Effects of Biochar on Container Substrate Properties and Growth of Plants—A Review. Horticulturae 2019, 5, 14. [Google Scholar] [CrossRef]
- Méndez, A.; Cárdenas-Aguiar, E.; Paz-Ferreiro, J.; Plaza, C.; Gascó, G. The effect of sewage sludge biochar on peat-based growing media. Biol. Agric. Hortic. 2016, 33, 40–51. [Google Scholar] [CrossRef]
- Ngatia, L.; Hsieh, Y.; Nemours, D.; Fu, R.; Taylor, R. Potential phosphorus eutrophication mitigation strategy: Biochar carbon composition, thermal stability and pH influence phosphorus sorption. Chemosphere 2017, 180, 201–211. [Google Scholar] [CrossRef]
- Rajkovich, S.; Enders, A.; Hanley, K.; Hyland, C.; Zimmerman, A.R.; Lehmann, J. Corn growth and nitrogen nutrition after additions of biochars with varying properties to a temperate soil. Biol. Fertil. Soils 2011, 48, 271–284. [Google Scholar] [CrossRef]
- Yao, Y.; Gao, B.; Zhang, M.; Inyang, M.; Zimmerman, A.R. Effect of biochar amendment on sorption and leaching of nitrate, ammonium, and phosphate in a sandy soil. Chemosphere 2012, 89, 1467–1471. [Google Scholar] [CrossRef]
- Knicker, H. Solid state CPMAS 13C and 15N NMR spectroscopy in organic geochemistry and how spin dynamics can either aggravate or improve spectra interpretation. Org. Geochem. 2011, 42, 867–890. [Google Scholar]
- Hsieh, Y.P.; Bugna, G.C. Analysis of black carbon in sediments and soils using multi-element scanning thermal analysis (MESTA). Org. Geochem. 2008, 39, 1562–1571. [Google Scholar] [CrossRef]
- Hsieh, Y.-P. A novel multielemental scanning thermal analysis (MESTA) method for the identification and characterization of solid substances. J. AOAC Int. 2007, 90. [Google Scholar]
- Disnar, J.R.; Jacob, J.; Morched-Issa, M.; Lottier, N.; Arnaud, F. Assessment of peat quality by molecular and bulk geochemical analysis: Application to the Holocene record of the Chautagne marsh (Haute Savoie, France). Chem. Geol. 2008, 254, 101–112. [Google Scholar] [CrossRef]
- Ning, J.; Liebich, J.; Kästner, M.; Zhou, J.; Schäffer, A.; Burauel, P. Different influences of DNA purity indices and quantity on PCR-based DGGE and functional gene microarray in soil microbial community study. Appl. Microbiol. Biotechnol. 2009, 82, 983–993. [Google Scholar] [CrossRef]
- Jaswal, R.; Pathak, A.; Chauhan, A. Metagenomic Evaluation of Bacterial and Fungal Assemblages Enriched within Diffusion Chambers and Microbial Traps Containing Uraniferous Soils. Microorganisms 2019, 7, 324. [Google Scholar] [CrossRef]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2013, 30, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Glöckner, F.O.; Yilmaz, P.; Quast, C.; Gerken, J.; Beccati, A.; Ciuprina, A.; Bruns, G.; Yarza, P.; Peplies, J.; Westram, R.; et al. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J. Biotechnol. 2017, 261, 169–176. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Gonzalez Peña, A.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Tikhonov, M.; Leach, R.W.; Wingreen, N.S. Interpreting 16S metagenomic data without clustering to achieve sub-OTU resolution. ISME J. 2014, 9, 68–80. [Google Scholar] [CrossRef]
- Jabbar, A.; Arshed, W.; Bhatti, A.S.; Ahmad, S.S.; Akhter, P.; Rehman, S.-U.; Anjum, M.I. Measurement of soil radioactivity levels and radiation hazard assessment in southern Rechna interfluvial region, Pakistan. Environ. Monit. Assess. 2009, 169, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Hilal, M.; Attallah, M.; Mohamed, G.Y.; Fayez-Hassan, M. Evaluation of radiation hazard potential of TENORM waste from oil and natural gas production. J. Environ. Radioact. 2014, 136, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Völgyesi, P.; Kis, Z.; Szabo, Z.; Szabó, C. Using the 186-keV peak for 226Ra activity concentration determination in Hungarian coal-slag samples by gamma-ray spectroscopy. J. Radioanal. Nucl. Chem. Artic. 2014, 302, 375–383. [Google Scholar] [CrossRef]
- Bikit, I.; Forkapic, S.; Nikolov, J.; Todorovic, N.; Mrdja, D. Radioactivity of the agricultural soil in northern province of Serbia, Vojvodina. Int. J. Environ. Ecol. Eng. 2011, 5, 232–237. [Google Scholar]
- Alnour, I.A.; Ibrahim, N.; Hossain, I. Concentrations of 214 Pb, 214 Bi in 238 U series and 208 Tl, 228 Ac in 232 Th series in granite rock in (Kadugli) Sudan. Available online: http://nopr.niscpr.res.in/bitstream/123456789/13996/1/IJPAP%2050%285%29%20285-288.pdf (accessed on 25 June 2022).
- Papp, Z.; Dezső, Z.; Daroczy, S. Measurement of the radioactivity of238U, 232Th, 226Ra, 137Cs and40K in soil using direct Ge (Li) γ-ray spectrometry. J. Radioanal. Nucl. Chem. 1997, 222, 171–176. [Google Scholar] [CrossRef]
- Thies, J.E.; Rillig, M.C.; Graber, E.R. Biochar effects on the abundance, activity and diversity of the soil biota. Biochar Environ. Manag. Sci. Technol. Implement. 2015, 2, 327–389. [Google Scholar]
- Husson, O. Redox potential (Eh) and pH as drivers of soil/plant/microorganism systems: A transdisciplinary overview pointing to integrative opportunities for agronomy. Plant Soil 2012, 362, 389–417. [Google Scholar] [CrossRef]
- Chen, J.; Liu, X.; Zheng, J.; Zhang, B.; Lu, H.; Chi, Z.; Pan, G.; Li, L.; Zheng, J.; Zhang, X.; et al. Biochar soil amendment increased bacterial but decreased fungal gene abundance with shifts in community structure in a slightly acid rice paddy from Southwest China. Appl. Soil Ecol. 2013, 71, 33–44. [Google Scholar] [CrossRef]
- Lugtenberg, B.; Kamilova, F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef]
- Rampelotto, P.H.; Ferreira, A.D.S.; Barboza, A.D.M.; Roesch, L.F.W. Changes in Diversity, Abundance, and Structure of Soil Bacterial Communities in Brazilian Savanna Under Different Land Use Systems. Microb. Ecol. 2013, 66, 593–607. [Google Scholar] [CrossRef]
- Acosta-Martínez, V.; Dowd, S.; Sun, Y.; Allen, V. Tag-encoded pyrosequencing analysis of bacterial diversity in a single soil type as affected by management and land use. Soil Biol. Biochem. 2008, 40, 2762–2770. [Google Scholar] [CrossRef]
- Naether, A.; Foesel, B.U.; Naegele, V.; Wüst, P.K.; Weinert, J.; Bonkowski, M.; Alt, F.; Oelmann, Y.; Polle, A.; Lohaus, G.; et al. Environmental factors affect acidobacterial communities below the subgroup level in grassland and forest soils. Appl. Environ. Microbiol. 2012, 78, 7398–7406. [Google Scholar] [CrossRef]
- Nacke, H.; Thürmer, A.; Wollherr, A.; Will, C.; HoDac, L.; Herold, N.; Schöning, I.; Schrumpf, M.; Daniel, R. Pyrosequencing-Based Assessment of Bacterial Community Structure Along Different Management Types in German Forest and Grassland Soils. PLoS ONE 2011, 6, e17000. [Google Scholar] [CrossRef]
- Zheng, J.; Chen, J.; Pan, G.; Liu, X.; Zhang, X.; Li, L.; Bian, R.; Cheng, K.; Jinwei, Z. Biochar decreased microbial metabolic quotient and shifted community composition four years after a single incorporation in a slightly acid rice paddy from southwest China. Sci. Total Environ. 2016, 571, 206–217. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.; Delgado-Baquerizo, M.; Anderson, I.C.; Singh, B.K. Response of Soil Properties and Microbial Communities to Agriculture: Implications for Primary Productivity and Soil Health Indicators. Front. Plant Sci. 2016, 7, 990. [Google Scholar] [CrossRef]
- Nielsen, S.; Minchin, T.; Kimber, S.; Van Zwieten, L.; Gilbert, J.; Munroe, P.; Joseph, S.; Thomas, T. Comparative analysis of the microbial communities in agricultural soil amended with enhanced biochars or traditional fertilisers. Agric. Ecosyst. Environ. 2014, 191, 73–82. [Google Scholar] [CrossRef]
- Lee, S.H.; Ka, J.O.; Cho, J.C. Members of the phylum Acidobacteria are dominant and metabolically active in rhizosphere soil. FEMS Microbiol. Lett. 2008, 285, 263–269. [Google Scholar] [CrossRef]
- Lehmann, J.; Rillig, M.C.; Thies, J.; Masiello, C.A.; Hockaday, W.C.; Crowley, D. Biochar effects on soil biota–a review. Soil Biol. Biochem. 2011, 43, 1812–1836. [Google Scholar] [CrossRef]
- Kirby, R. Actinomycetes and Lignin Degradation. Adv. Appl. Microbiol. 2005, 58, 125–168. [Google Scholar] [CrossRef]
- Xu, N.; Tan, G.; Wang, H.; Gai, X. Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur. J. Soil Biol. 2016, 74, 1–8. [Google Scholar] [CrossRef]
- Leff, J.W.; Jones, S.E.; Prober, S.M.; Barberán, A.; Borer, E.T.; Firn, J.L.; Harpole, W.S.; Hobbie, S.E.; Hofmockel, K.S.; Knops, J.M.H.; et al. Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proc. Natl. Acad. Sci. USA 2015, 112, 10967–10972. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.N.; Verma, P.; Kumar, S.; Kumar, V.; Kumar, M.; Sugitha, T.C.; Singh, B.P.; Saxena, A.K.; Dhaliwal, H.S. Actinobacteria from rhizosphere: Molecular diversity, distributions, and potential biotechnological applications. In New and Future Developments in Microbial Biotechnology and Bioengineering 2018 Jan 1; Elsevier: Amsterdam, The Netherlands, 2018; pp. 13–41. [Google Scholar]
- Ventura, M.; Canchaya, C.; Tauch, A.; Chandra, G.; Fitzgerald, G.F.; Chater, K.F.; van Sinderen, D. Genomics of Actinobacteria: Tracing the Evolutionary History of an Ancient Phylum. Microbiol. Mol. Biol. Rev. 2007, 71, 495–548. [Google Scholar] [CrossRef]
- Huang, L.N.; Zhu, S.; Zhou, H.; Qu, L.H. Molecular phylogenetic diversity of bacteria associated with the leachate of a closed municipal solid waste landfill. FEMS Microbiol. Lett. 2005, 242, 297–303. [Google Scholar] [PubMed]
- Singh, H.; Khattar, J.S.; Ahluwalia, A.S. Cyanobacteria and agricultural crops. Vegetos 2014, 27, 37. [Google Scholar] [CrossRef]
- Rana, A.; Joshi, M.; Prasanna, R.; Shivay, Y.S.; Nain, L. Biofortification of wheat through inoculation of plant growth promoting rhizobacteria and cyanobacteria. Eur. J. Soil Biol. 2012, 50, 118–126. [Google Scholar] [CrossRef]
- Strauss, G.; Fuchs, G. Enzymes of a novel autotrophic CO2 fixation pathway in the phototrophic bacterium Chloroflexus aurantiacus, the 3-hydroxypropionate cycle. Eur. J. Biochem. 1993, 215, 633–643. [Google Scholar]
- Kolton, M.; Harel, Y.M.; Pasternak, Z.; Graber, E.R.; Elad, Y.; Cytryn, E. Impact of Biochar Application to Soil on the Root-Associated Bacterial Community Structure of Fully Developed Greenhouse Pepper Plants. Appl. Environ. Microbiol. 2011, 77, 4924–4930. [Google Scholar] [CrossRef]
- McBride, M.J.; Xie, G.; Martens, E.C.; Lapidus, A.; Henrissat, B.; Rhodes, R.G.; Goltsman, E.; Wang, W.; Xu, J.; Hunnicutt, D.W.; et al. Novel Features of the Polysaccharide-Digesting Gliding Bacterium Flavobacterium johnsoniae as Revealed by Genome Sequence Analysis. Appl. Environ. Microbiol. 2009, 75, 6864–6875. [Google Scholar] [CrossRef]
- Pérez-Jaramillo, J.E.; De Hollander, M.; Ramírez, C.A.; Mendes, R.; Raaijmakers, J.M.; Carrión, V.J. Deciphering rhizosphere microbiome assembly of wild and modern common bean (Phaseolus vulgaris) in native and agricultural soils from Colombia. Microbiome 2019, 7, 114. [Google Scholar] [CrossRef]
- Jaiswal, A.K.; Elad, Y.; Paudel, I.; Graber, E.R.; Cytryn, E.; Frenkel, O. Linking the Belowground Microbial Composition, Diversity and Activity to Soilborne Disease Suppression and Growth Promotion of Tomato Amended with Biochar. Sci. Rep. 2017, 7, srep44382. [Google Scholar] [CrossRef]
- Vinson, D.S.; Vengosh, A.; Hirschfeld, D.; Dwyer, G.S. Relationships between radium and radon occurrence and hydrochemistry in fresh groundwater from fractured crystalline rocks, North Carolina (USA). Chem. Geol. 2008, 260, 159–171. [Google Scholar] [CrossRef]
- Szabo, Z.; DePaul, V.T.; Kraemer, T.F.; Parsa, B. Occurrence of Radium-224, Radium-226, and Radium-228 in Water of the Unconfined Kirkwood-Cohansey Aquifer System, Southern New Jersey; U. S. Geological Survey: Reston, VA, USA, 2005. [Google Scholar]
- Gaspar, L.; Lizaga, I.; Navas, A. Spatial distribution of fallout and lithogenic radionuclides controlled by soil carbon and water erosion in an agroforestry South-Pyrenean catchment. Geoderma 2021, 391, 114941. [Google Scholar] [CrossRef]
- Lopez-Fernandez, M.; Jroundi, F.; Ruiz-Fresneda, M.A.; Merroun, M.L. Microbial interaction with and tolerance of radionuclides: Underlying mechanisms and biotechnological applications. Microb. Biotechnol. 2020, 14, 810–828. [Google Scholar] [CrossRef]
- Kumar, V.; Chandra, R. Bacteria-assisted phytoremediation of industrial waste pollutants and ecorestoration. In Phytoremediation of Environmental Pollutants 2017 Dec 14; CRC Press: Boca Raton, FL, USA, 2017; pp. 159–200. [Google Scholar]
- Suriya, J.; Shekar, M.C.; Nathani, N.M.; Suganya, T.; Bharathiraja, S.; Krishnan, M. Assessment of bacterial community composition in response to uranium levels in sediment samples of sacred Cauvery River. Appl. Microbiol. Biotechnol. 2016, 101, 831–841. [Google Scholar] [CrossRef] [PubMed]
- Hoyos-Hernandez, C.; Courbert, C.; Simonucci, C.; David, S.; Vogel, T.M.; Larose, C. Community structure and functional genes in radionuclide contaminated soils in Chernobyl and Fukushima. FEMS Microbiol. Lett. 2019, 366. [Google Scholar] [CrossRef]
- Barns, S.M.; Cain, E.C.; Sommerville, L.; Kuske, C.R. Acidobacteria phylum sequences in uranium-contaminated subsurface sediments greatly expand the known diversity within the phylum. Appl. Environ. Microbiol. 2007, 73, 3113–3116. [Google Scholar]
- Mondani, L.; Benzerara, K.; Carrière, M.; Christen, R.; Mamindy-Pajany, Y.; Février, L.; Marmier, N.; Achouak, W.; Nardoux, P.; Berthomieu, C.; et al. Influence of Uranium on Bacterial Communities: A Comparison of Natural Uranium-Rich Soils with Controls. PLoS ONE 2011, 6, e25771. [Google Scholar] [CrossRef]
- Hug, L.A.; Castelle, C.J.; Wrighton, K.C.; Thomas, B.C.; Sharon, I.; Frischkorn, K.R.; Williams, K.H.; Tringe, S.G.; Banfield, J.F. Community genomic analyses constrain the distribution of metabolic traits across the Chloroflexi phylum and indicate roles in sediment carbon cycling. Microbiome 2013, 1, 22. [Google Scholar] [CrossRef]
- Banach, A.M.; Kuźniar, A.; Grządziel, J.; Wolińska, A. Azolla filiculoides L. as a source of metal-tolerant microorganisms. PLoS ONE 2020, 15, e0232699. [Google Scholar] [CrossRef]
- Shilpi, G.; Shilpi, S.; Sunita, S. Tolerance against heavy metal toxicity in cyanobacteria: Role of antioxidant defense system. Int. J. Pharm. Pharm. Sci. 2015, 7, 1–8. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).