Overview of Approaches to Improve Rhizoremediation of Petroleum Hydrocarbon-Contaminated Soils
Abstract
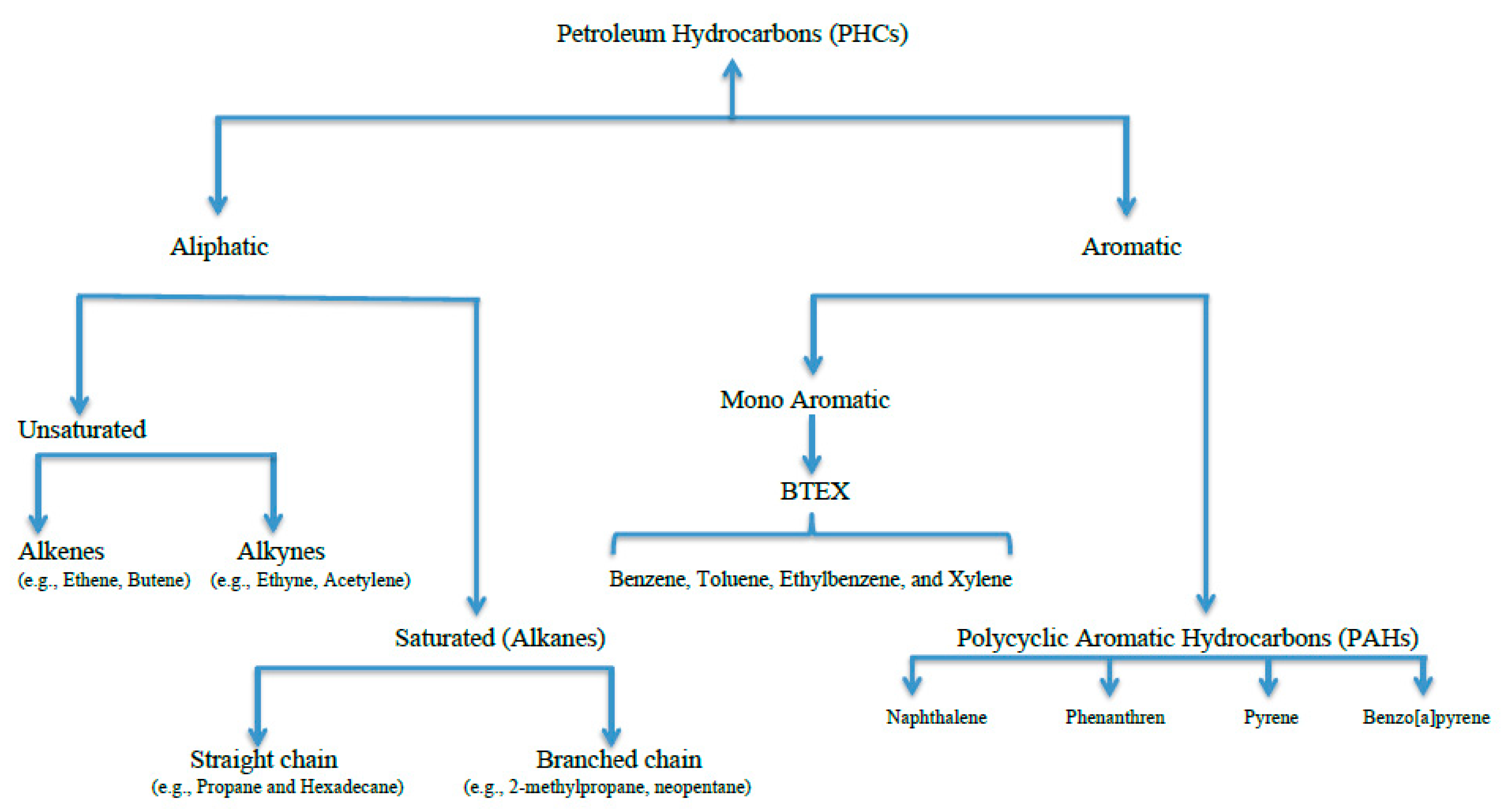
1. Introduction
2. Phytoremediation
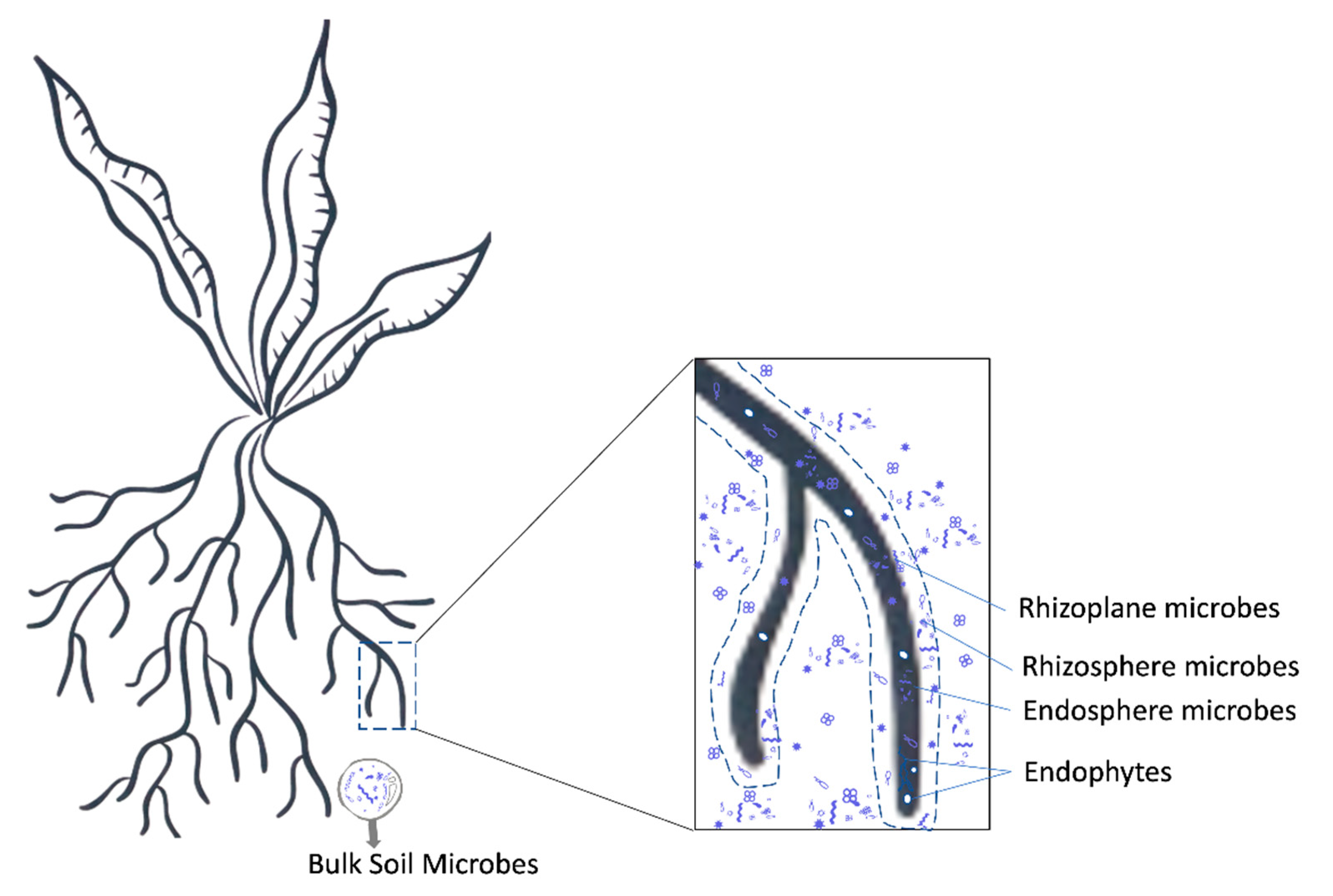
3. The Rhizosphere Microbiome
3.1. Plant Selection
3.2. Exploiting and Manipulating the Plant Microbiome through Inoculation
3.2.1. Plant Growth-Promoting Rhizobacteria (PGPR)
Enhanced Nutrient Acquisition (Biofertilizer)
Plant Growth Regulation (Phytostimulation)
3.2.2. Reduction of Plant Ethylene (Stress Alleviating)
Plant Growth-Promoting Rhizobacteria-Assisted Phytoremediation
3.2.3. Hydrocarbon-Degrading Bacteria
Ecology and Diversity of PHC-Degrading Bacteria
Alkane-Degrading Bacteria
Polycyclic Aromatic Hydrocarbon-Degrading Bacteria
4. Enhancing the Understanding of Mechanisms through Which Host Plants Assemble a Beneficial Microbiome, and How It Functions, under Pollutant Stress
4.1. High-Throughput Amplicon Sequencing
4.2. Metatranscriptomics
4.3. Genome Sequencing
| Bacterial Strain | Importance in Bioremediation | Isolation Source | PGPR Features | Genome Size | Reference |
|---|---|---|---|---|---|
| Pseudomonas veronii strain VI4T1 | degradation of aromatic and aliphatic hydrocarbons | long-term oil field-polluted soil | IAA, siderophore | 7.15 Mb | [184] |
| Pseudomonas sp. strain VI4.1 | degradation of aromatic and aliphatic hydrocarbons | long-term oil field-polluted soil | IAA, siderophore | 7.3 Mb | [184] |
| Halomonas sp. strain G11 | degradation of alkanes and polyaromatic hydrocarbons | hypersaline sediment | Salt-tolerance, biosurfactant production | 3.96 Mb | [185] |
| Pseudomonas aeruginosa strain DN1 | fluoranthene degradation | PHC-contaminated soil | N/D | 6.6 Mb | [186] |
| Alcaligenes aquatilis strain BU33N | degradation of n-alkanes and phenanthrene | PHC-polluted sediments | biosurfactant production; heavy metals resistance | 3.8 Mb | [187] |
| Gordonia paraffinivorans strain MTZ052 | degradation of n-hexadecane | composting pile | N/D | 4.8 Mb | [188] |
| Gordonia sihwensis strain MTZ096 | degradation of n-hexadecane | composting pile | N/D | 3.9 Mb | [188] |
| Klebsiella pneumoniae strain AWD5 | degradation of xenobiotic compounds | PAH-contaminated soil | siderophore production | 4.8 Mb | [189] |
| Bacillus licheniformis strain VSD4 | degradation of diesel fuel | leaves of Hedera helix plants growing at a high-traffic city center | IAA, siderophore | 4.19 Mb | [190] |
| Pseudomonas putida strain BS3701 | degradation of crude oil and PAHs | soil contaminated with coke by-product waste | N/D | 6.3 Mb | [191] |
5. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Brzeszcz, J.; Kaszycki, P. Aerobic bacteria degrading both n-alkanes and aromatic hydrocarbons: An undervalued strategy for metabolic diversity and flexibility. Biodegradation 2018, 29, 359–407. [Google Scholar] [CrossRef]
- Gkorezis, P.; Daghio, M.; Franzetti, A.; Van Hamme, J.; Sillen, W.; Vangronsveld, J. The Interaction between Plants and Bacteria in the Remediation of Petroleum Hydrocarbons: An Environmental Perspective. Front. Microbiol. 2016, 7, 1836. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Fingas, M.; Yang, C.; Christensen, J.H. Crude Oil and Refined Product Fingerprinting: Principles. In Environmental Forensics: Contaminant Specific Guide; Morrison, R.D., Murphy, B.L., Eds.; Academic Press: New York, NY, USA, 2006; pp. 339–407. [Google Scholar]
- Farrell-Jones, J. Petroleum Hydrocarbons and Polyaromatic Hydrocarbons; Blackwell Publishing CRC Press: New York, NY, USA, 2003. [Google Scholar]
- Tissot, B.P.; Welte, D.H. Petroleum Formation and Occurrence; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Pandey, P.; Pathak, H.; Dave, S. Microbial Ecology of Hydrocarbon Degradation in the Soil: A Review. Res. J. Environ. Toxicol. 2016, 10, 1–15. [Google Scholar] [CrossRef]
- Hoang, S.A.; Lamb, D.; Seshadri, B.; Sarkar, B.; Choppala, G.; Kirkham, M.; Bolan, N.S. Rhizoremediation as a green technology for the remediation of petroleum hydrocarbon-contaminated soils. J. Hazard. Mater. 2020, 401, 123282. [Google Scholar] [CrossRef] [PubMed]
- Secretariat. Federal Contaminated Sites Inventory. Available online: https://www.tbs-sct.gc.ca/fcsi-rscf/home-accueil-eng.aspx (accessed on 18 August 2020).
- Panagos, P.; Van Liedekerke, M.; Yigini, Y.; Montanarella, L. Contaminated Sites in Europe: Review of the Current Situation Based on Data Collected through a European Network. J. Environ. Public Health 2013, 2013, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Henner, P.; Schiavon, M.; Morel, J.-L.; Lichtfouse, E. Polycyclic aromatic hydrocarbon (PAH) occurrence and remediation methods. Analusis 1997, 25, M56–M59. [Google Scholar]
- Inoue, Y.; Katayama, A. Two-scale evaluation of remediation technologies for a contaminated site by applying economic input–output life cycle assessment: Risk–cost, risk–energy consumption and risk–CO2 emission. J. Hazard. Mater. 2011, 192, 1234–1242. [Google Scholar] [CrossRef] [PubMed]
- Yerushalmi, L.; Rocheleau, S.; Cimpoia, R.; Sarrazin, M.; Sunahara, G.; Peisajovich, A.; Leclair, G.; Guiot, S.R. Enhanced biodegradation of petroleum hydrocarbons in contaminated soil. J. Soil Contam. 1998, 7, 37–51. [Google Scholar] [CrossRef]
- Pilon-Smits, E. Phytoremediation. Annu. Rev. Plant Biol. 2005, 56, 15–39. [Google Scholar] [CrossRef]
- Salt, D.E.; Smith, R.D.; Raskin, I. Phytoremediation. Annu. Rev. Plant Biol. 1998, 49, 643–668. [Google Scholar] [CrossRef]
- Aken, B.V.; Doty, S.L. Transgenic plants and associated bacteria for phytoremediation of chlorinated compounds. Biotechnol. Genet. Eng. Rev. 2009, 26, 43–64. [Google Scholar] [CrossRef]
- Panz, K.; Miksch, K. Phytoremediation of explosives (TNT, RDX, HMX) by wild-type and transgenic plants. J. Environ. Manag. 2012, 113, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Leguizamo, M.A.O.; Gómez, W.D.F.; Sarmiento, M.C.G. Native herbaceous plant species with potential use in phytoremediation of heavy metals, spotlight on wetlands—A review. Chemosphere 2017, 168, 1230–1247. [Google Scholar] [CrossRef] [PubMed]
- Ch, J.A.J.; Romero, R.M. Evaluation of Cajanus cajan (pigeon pea) for phytoremediation of landfill leachate containing chromium and lead. Int. J. Phytoremediat. 2016, 18, 1122–1127. [Google Scholar] [CrossRef]
- Olette, R.; Couderchet, M.; Biagianti, S.; Eullaffroy, P. Toxicity and removal of pesticides by selected aquatic plants. Chemosphere 2008, 70, 1414–1421. [Google Scholar] [CrossRef]
- Newman, L.A.; Reynolds, C.M. Phytodegradation of organic compounds. Curr. Opin. Biotechnol. 2004, 15, 225–230. [Google Scholar] [CrossRef]
- Sharma, S.; Singh, B.; Manchanda, V.K. Phytoremediation: Role of terrestrial plants and aquatic macrophytes in the remediation of radionuclides and heavy metal contaminated soil and water. Environ. Sci. Pollut. Res. 2014, 22, 946–962. [Google Scholar] [CrossRef]
- Devi, S.; Nandwal, A.; Angrish, R.; Arya, S.; Kumar, N.; Sharma, S. Phytoremediation potential of some halophytic species for soil salinity. Int. J. Phytoremediat. 2015, 18, 693–696. [Google Scholar] [CrossRef] [PubMed]
- Transparency Market Research. BioremediationI Technology & Services Market to Reach Valuation gf ~US$ 20 BN by 2030. Available online: https://www.transparencymarketresearch.com/pressrelease/bioremediation-technology-services-market.htm (accessed on 18 September 2020).
- Mench, M.; Lepp, N.; Bert, V.; Schwitzguébel, J.-P.; Gawroński, S.; Schröder, P.; Vangronsveld, J. Successes and limitations of phytotechnologies at field scale: Outcomes, assessment and outlook from COST Action 859. J. Soils Sediments 2010, 10, 1039–1070. [Google Scholar] [CrossRef]
- Vangronsveld, J.; Herzig, R.; Weyens, N.; Boulet, J.; Adriaensen, K.; Ruttens, A.; Thewys, T.; Vassilev, A.; Meers, E.; Nehnevajova, E.; et al. Phytoremediation of contaminated soils and groundwater: Lessons from the field. Environ. Sci. Pollut. Res. 2009, 16, 765–794. [Google Scholar] [CrossRef] [PubMed]
- Arthur, E.L.; Rice, P.J.; Rice, P.J.; Anderson, T.A.; Baladi, S.M.; Henderson, K.L.; Coats, J.R. Phytoremediation—An overview. Crit. Rev. Plant Sci. 2005, 24, 109–122. [Google Scholar] [CrossRef]
- Wenzel, W.W. Rhizosphere processes and management in plant-assisted bioremediation (phytoremediation) of soils. Plant Soil 2008, 321, 385–408. [Google Scholar] [CrossRef]
- Kuiper, I.; Lagendijk, E.L.; Bloemberg, G.V.; Lugtenberg, B.J.J. Rhizoremediation: A Beneficial Plant-Microbe Interaction. Mol. Plant-Microbe Interact. 2004, 17, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.; Khan, E.; Sajad, M.A. Phytoremediation of heavy metals—Concepts and applications. Chemosphere 2013, 91, 869–881. [Google Scholar] [CrossRef]
- Behera, K.K. Phytoremediation, transgenic plants and microbes. In Sustainable Agriculture Reviews; Springer: Berlin/Heidelberg, Germany, 2014; pp. 65–85. [Google Scholar]
- Anderson, T.A.; Guthrie, E.A.; Walton, B.T. Bioremediation in the rhizosphere. Environ. Sci. Technol. 1993, 27, 2630–2636. [Google Scholar] [CrossRef]
- Martin, B.; George, S.J.; Price, C.A.; Ryan, M.; Tibbett, M. The role of root exuded low molecular weight organic anions in facilitating petroleum hydrocarbon degradation: Current knowledge and future directions. Sci. Total Environ. 2014, 472, 642–653. [Google Scholar] [CrossRef]
- Correa-García, S.; Pande, P.; Séguin, A.; St-Arnaud, M.; Yergeau, E. Rhizoremediation of petroleum hydrocarbons: A model system for plant microbiome manipulation. Microb. Biotechnol. 2018, 11, 819–832. [Google Scholar] [CrossRef]
- Ethijs, S.; Sillen, W.; Erineau, F.; Eweyens, N.; Vangronsveld, J. Towards an Enhanced Understanding of Plant–Microbiome Interactions to Improve Phytoremediation: Engineering the Metaorganism. Front. Microbiol. 2016, 7, 341. [Google Scholar] [CrossRef]
- Philippot, L.; Raaijmakers, J.; Lemanceau, P.; van der Putten, W. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Genet. 2013, 11, 789–799. [Google Scholar] [CrossRef]
- Raaijmakers, J.M.; Paulitz, T.; Steinberg, C.; Alabouvette, C.; Moënne-Loccoz, Y. The rhizosphere: A playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 2008, 321, 341–361. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.; Bakker, P.A. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Smalla, K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Bakker, P.A.H.M.; Berendsen, R.L.; Doornbos, R.F.; Wintermans, P.C.A.; Pieterse, C.M.J. The rhizosphere revisited: Root microbiomics. Front. Plant Sci. 2013, 4, 165. [Google Scholar] [CrossRef]
- Kuzyakov, Y.; Blagodatskaya, E. Microbial hotspots and hot moments in soil: Concept & review. Soil Biol. Biochem. 2015, 83, 184–199. [Google Scholar] [CrossRef]
- Smalla, K.; Wieland, G.; Buchner, A.; Zock, A.; Parzy, J.; Kaiser, S.; Roskot, N.; Heuer, H.; Berg, G. Bulk and Rhizosphere Soil Bacterial Communities Studied by Denaturing Gradient Gel Electrophoresis: Plant-Dependent Enrichment and Seasonal Shifts Revealed. Appl. Environ. Microbiol. 2001, 67, 4742–4751. [Google Scholar] [CrossRef]
- Loeppmann, S.; Blagodatskaya, E.; Pausch, J.; Kuzyakov, Y. Substrate quality affects kinetics and catalytic efficiency of exo-enzymes in rhizosphere and detritusphere. Soil Biol. Biochem. 2016, 92, 111–118. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Van Themaat, E.V.L.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nat. Cell Biol. 2012, 488, 91–95. [Google Scholar] [CrossRef]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; Del Rio, T.G.; et al. Defining the core Arabidopsis thaliana root microbiome. Nat. Cell Biol. 2012, 488, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Vieira, S.; Sikorski, J.; Dietz, S.; Herz, K.; Schrumpf, M.; Bruelheide, H.; Scheel, D.; Friedrich, M.W.; Overmann, J. Drivers of the composition of active rhizosphere bacterial communities in temperate grasslands. ISME J. 2019, 14, 463–475. [Google Scholar] [CrossRef]
- Vives-Peris, V.; de Ollas, C.; Gómez-Cadenas, A.; Pérez-Clemente, R.M. Root exudates: From plant to rhizosphere and beyond. Plant Cell Rep. 2019, 39, 3–17. [Google Scholar] [CrossRef]
- Rohrbacher, F.; St-Arnaud, M. Root Exudation: The Ecological Driver of Hydrocarbon Rhizoremediation. Agronomy 2016, 6, 19. [Google Scholar] [CrossRef]
- Jones, D.; Murphy, D. Microbial response time to sugar and amino acid additions to soil. Soil Biol. Biochem. 2007, 39, 2178–2182. [Google Scholar] [CrossRef]
- Gao, Y.; Yang, Y.; Ling, W.; Kong, H.; Zhu, X. Gradient Distribution of Root Exudates and Polycyclic Aromatic Hydrocarbons in Rhizosphere Soil. Soil Sci. Soc. Am. J. 2011, 75, 1694–1703. [Google Scholar] [CrossRef]
- Muratova, A.; Dubrovskaya, E.; Golubev, S.; Grinev, V.; Chernyshova, M.; Turkovskaya, O. The coupling of the plant and microbial catabolisms of phenanthrene in the rhizosphere of Medicago sativa. J. Plant Physiol. 2015, 188, 1–8. [Google Scholar] [CrossRef]
- Yergeau, E.; Sanschagrin, S.; Maynard, C.; St-Arnaud, M.; Greer, C.W. Microbial expression profiles in the rhizosphere of willows depend on soil contamination. ISME J. 2013, 8, 344–358. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Ren, L.; Ling, W.; Gong, S.; Sun, B.; Zhang, Y. Desorption of phenanthrene and pyrene in soils by root exudates. Bioresour. Technol. 2010, 101, 1159–1165. [Google Scholar] [CrossRef]
- Van Elsas, J.D.; Bailey, M.J. The ecology of transfer of mobile genetic elements. FEMS Microbiol. Ecol. 2002, 42, 187–197. [Google Scholar] [CrossRef]
- Top, E.M.; Springael, D. The role of mobile genetic elements in bacterial adaptation to xenobiotic organic compounds. Curr. Opin. Biotechnol. 2003, 14, 262–269. [Google Scholar] [CrossRef]
- Sentchilo, V.; Mayer, A.P.; Guy, L.; Miyazaki, R.; Tringe, S.; Barry, K.; Malfatti, S.; Goessmann, A.; Robinson-Rechavi, M.; van der Meer, J.R. Community-wide plasmid gene mobilization and selection. ISME J. 2013, 7, 1173–1186. [Google Scholar] [CrossRef]
- Khan, S.; Afzal, M.; Iqbal, S.; Khan, Q.M. Plant–bacteria partnerships for the remediation of hydrocarbon contaminated soils. Chemosphere 2013, 90, 1317–1332. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, Q.; Blom-Zandstra, M.; Gupta, S.K.; Joner, E.J. Utilising the Synergy between Plants and Rhizosphere Microorganisms to Enhance Breakdown of Organic Pollutants in the Environment (15 pp). Environ. Sci. Pollut. Res. 2004, 12, 34–48. [Google Scholar] [CrossRef] [PubMed]
- Aprill, W.; Sims, R.C. Evaluation of the use of prairie grasses for stimulating polycyclic aromatic hydrocarbon treatment in soil. Chemosphere 1990, 20, 253–265. [Google Scholar] [CrossRef]
- Gaskin, S.E.; Bentham, R.H. Rhizoremediation of hydrocarbon contaminated soil using Australian native grasses. Sci. Total Environ. 2010, 408, 3683–3688. [Google Scholar] [CrossRef]
- Hall, J.; Soole, K.; Bentham, R. Hydrocarbon Phytoremediation in the FamilyFabacea—A Review. Int. J. Phytoremediat. 2011, 13, 317–332. [Google Scholar] [CrossRef]
- Reynolds, C.M.; Wolf, D.C.; Gentry, T.J.; Perry, L.B.; Pidgeon, C.S.; Koenen, B.A.; Rogers, H.B.; Beyrouty, C.A. Plant enhancement of indigenous soil micro-organisms: A low-cost treatment of contaminated soils. Polar Rec. 1999, 35, 33–40. [Google Scholar] [CrossRef]
- Frick, C.; Germida, J.; Farrell, R. Assessment of phytoremediation as an in-situ technique for cleaning oil-contaminated sites. In Proceedings of the Technical Seminar on Chemical Spills, Caligary, AB, Canada, 31 May–1 June 1999; pp. 105a–124a. [Google Scholar]
- Lu, M.; Zhang, Z.; Sun, S.; Wei, X.; Wang, Q.; Su, Y. The Use of Goosegrass (Eleusine indica) to Remediate Soil Contaminated with Petroleum. Water Air Soil Pollut. 2009, 209, 181–189. [Google Scholar] [CrossRef]
- Kuzovkina, Y.A.; Volk, T.A. The characterization of willow (Salix L.) varieties for use in ecological engineering applications: Co-ordination of structure, function and autecology. Ecol. Eng. 2009, 35, 1178–1189. [Google Scholar] [CrossRef]
- Bell, T.; Hassan, S.E.-D.; Lauron-Moreau, A.; Al-Otaibi, F.; Hijri, M.; Yergeau, E.; St-Arnaud, M. Linkage between bacterial and fungal rhizosphere communities in hydrocarbon-contaminated soils is related to plant phylogeny. ISME J. 2013, 8, 331–343. [Google Scholar] [CrossRef] [PubMed]
- de Cárcer, D.A.; Martín, M.; Karlson, U.; Rivilla, R. Changes in Bacterial Populations and in Biphenyl Dioxygenase Gene Diversity in a Polychlorinated Biphenyl-Polluted Soil after Introduction of Willow Trees for Rhizoremediation. Appl. Environ. Microbiol. 2007, 73, 6224–6232. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yergeau, E.; Tremblay, J.; Joly, S.; Labrecque, M.; Maynard, C.; Pitre, F.E.; St-Arnaud, M.; Greer, C.W. Soil contamination alters the willow root and rhizosphere metatranscriptome and the root–rhizosphere interactome. ISME J. 2018, 12, 869–884. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Jaramillo, J.E.; Mendes, R.; Raaijmakers, J.M. Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol. Biol. 2015, 90, 635–644. [Google Scholar] [CrossRef]
- Timmis, K.N.; Pieper, D.H. Bacteria designed for bioremediation. Trends Biotechnol. 1999, 17, 201–204. [Google Scholar] [CrossRef]
- Brown, R.W. Revegetation of an Alpine Mine disturbance: Beartooth Plateau, Montana; US Department of Agriculture; Forest Service, Intermountain Forest & Range: Logan, Utah, 1976; Volume 206. [Google Scholar]
- Desjardins, D.; Nissim, W.G.; Pitre, F.E.; Naud, A.; Labrecque, M. Distribution patterns of spontaneous vegetation and pollution at a former decantation basin in southern Québec, Canada. Ecol. Eng. 2014, 64, 385–390. [Google Scholar] [CrossRef]
- Lee, S.-J.; Kong, M.; St-Arnaud, M.; Hijri, M. Arbuscular Mycorrhizal Fungal Communities of Native Plant Species under High Petroleum Hydrocarbon Contamination Highlights Rhizophagus as a Key Tolerant Genus. Microorganisms 2020, 8, 872. [Google Scholar] [CrossRef] [PubMed]
- Brundrett, M. Mycorrhizas in natural ecosystems. Adv. Ecol. Res. 1991, 21, 171–313. [Google Scholar]
- Bolduc, A.R.; Hijri, M. The Use of Mycorrhizae to Enhance Phosphorus Uptake: A Way Out the Phosphorus Crisis. J. Fertil. Pestic. 2011, 2, 1–5. [Google Scholar] [CrossRef]
- Miransari, M. Hyperaccumulators, arbuscular mycorrhizal fungi and stress of heavy metals. Biotechnol. Adv. 2011, 29, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Porcel, R.; Aroca, R.; Lozano, J.M.R. Salinity stress alleviation using arbuscular mycorrhizal fungi. A review. Agron. Sustain. Dev. 2011, 32, 181–200. [Google Scholar] [CrossRef]
- Smith, S.E.; Read, D.J. Mycorrhizal Symbiosis; Academic Press: New York, NY, USA, 2008. [Google Scholar]
- St-Arnaud, M.; Vujanovic, V. Effects of the Arbuscular Mycorrhizal Symbiosis on Plant Diseases and Pests. In Mycorrhizae in Crop Production; Hamel, C., Plenchette, C., Eds.; CRC Press: London, UK, 2007; pp. 67–123. [Google Scholar]
- Hassan, S.E.; Hijri, M.; St-Arnaud, M. Effect of arbuscular mycorrhizal fungi on trace metal uptake by sunflower plants grown on cadmium contaminated soil. New Biotechnol. 2013, 30, 780–787. [Google Scholar] [CrossRef]
- Hassan, S.E.-D.; Bell, T.H.; Stefani, F.O.P.; Denis, D.; Hijri, M.; St-Arnaud, M. Contrasting the Community Structure of Arbuscular Mycorrhizal Fungi from Hydrocarbon-Contaminated and Uncontaminated Soils following Willow (Salix spp. L.) Planting. PLoS ONE 2014, 9, e102838. [Google Scholar] [CrossRef]
- Reichenauer, T.G.; Germida, J. Phytoremediation of Organic Contaminants in Soil and Groundwater. ChemSusChem 2008, 1, 708–717. [Google Scholar] [CrossRef]
- Sandermann, H. Plant metabolism of xenobiotics. Trends Biochem. Sci. 1992, 17, 82–84. [Google Scholar] [CrossRef]
- Singer, A.C. Advances in development of transgenic plants for remediation of xenobiotic pollutants. In Phytoremediation and Rhizoremediation: Theoretical Background; Mackova, M., Dowling, D., Macek, T., Eds.; Springer: Dordrecht, UK, 2006; pp. 5–21. [Google Scholar]
- Glick, B.R.; Stearns, J.C. Making Phytoremediation Work Better: Maximizing a Plant’s Growth Potential in the Midst of Adversity. Int. J. Phytoremediat. 2011, 13, 4–16. [Google Scholar] [CrossRef]
- Hardoim, P.; van Overbeek, L.S.; van Elsas, J.D. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 2008, 16, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Rughöft, S.; Herrmann, M.; Lazar, C.; Cesarz, S.; Levick, S.; Trumbore, S.E.; Küsel, K. Community Composition and Abundance of Bacterial, Archaeal and Nitrifying Populations in Savanna Soils on Contrasting Bedrock Material in Kruger National Park, South Africa. Front. Microbiol. 2016, 7, 1638. [Google Scholar] [CrossRef]
- Richardson, A.E.; Barea, J.M.; McNeill, A.M.; Prigent-Combaret, C. Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil 2009, 321, 305–339. [Google Scholar] [CrossRef]
- Schlaeppi, K.; Bulgarelli, D. The Plant Microbiome at Work. Mol. Plant-Microbe Interact. 2015, 28, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Vessey, J.K. Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 2003, 255, 571–586. [Google Scholar] [CrossRef]
- Bloemberg, G.V.; Lugtenberg, B.J. Molecular basis of plant growth promotion and biocontrol by rhizobacteria. Curr. Opin. Plant Biol. 2001, 4, 343–350. [Google Scholar] [CrossRef]
- Haas, D.; Défago, G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Genet. 2005, 3, 307–319. [Google Scholar] [CrossRef]
- Lugtenberg, B.; Kamilova, F. Plant-Growth-Promoting Rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef]
- Stevens, C.J. Nitrogen in the environment. Science 2019, 363, 578–580. [Google Scholar] [CrossRef] [PubMed]
- Boddey, R.M.; De Oliveira, O.; Urquiaga, S.; Reis, V.; De Olivares, F.; Baldani, V.; Döbereiner, J. Biological nitrogen fixation associated with sugar cane and rice: Contributions and prospects for improvement. In Management of Biological Nitrogen Fixation for the Development of More Productive and Sustainable Agricultural Systems; Springer: Berlin/Heidelberg, Germany, 1995; pp. 195–209. [Google Scholar]
- Dobbelaere, S.; Vanderleyden, J.; Okon, Y. Plant Growth-Promoting Effects of Diazotrophs in the Rhizosphere. Crit. Rev. Plant Sci. 2003, 22, 107–149. [Google Scholar] [CrossRef]
- Sessitsch, A.; Howieson, J.; Perret, X.; Antoun, H.; Martinez-Romero, E. Advances in Rhizobium research. Crit. Rev. Plant Sci. 2002, 21, 323–378. [Google Scholar] [CrossRef]
- Alori, E.T.; Glick, B.R.; Babalola, O.O. Microbial Phosphorus Solubilization and Its Potential for Use in Sustainable Agriculture. Front. Microbiol. 2017, 8, 971. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, F.J.; Cole, M.A. Cycles of Soils: Carbon, Nitrogen, Phosphorus, Sulfur, Micronutrients, 2nd ed.; John Wiley & Sons: New York, NY, USA, 1999. [Google Scholar]
- Glass, A.D. Plant Mineral Nutrition. An Introduction to Current Concepts; Jones and Bartlett Publishers, Inc.: Burlington, MA, USA, 1989. [Google Scholar]
- Kim, K.Y.; Jordan, D.; McDonald, G.A. Effect of phosphate-solubilizing bacteria and vesicular-arbuscular mycorrhizae on tomato growth and soil microbial activity. Biol. Fertil. Soils 1997, 26, 79–87. [Google Scholar] [CrossRef]
- Rodríguez, H.; Fraga, R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 1999, 17, 319–339. [Google Scholar] [CrossRef]
- Sharma, S.B.; Sayyed, R.; Trivedi, M.H.; Gobi, T.A. Phosphate solubilizing microbes: Sustainable approach for managing phosphorus deficiency in agricultural soils. SpringerPlus 2013, 2, 1–14. [Google Scholar] [CrossRef]
- Jorquera, M.A.; Crowley, D.E.; Marschner, P.; Greiner, R.; Fernández, M.T.; Romero, D.; Blackburn, D.; Mora, M.L. Identification of β-propeller phytase-encoding genes in culturable Paenibacillus and Bacillus spp. from the rhizosphere of pasture plants on volcanic soils. FEMS Microbiol. Ecol. 2010, 75, 163–172. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, D.; Sun, W.; Wang, T. The Adaptive Mechanism of Plants to Iron Deficiency via Iron Uptake, Transport, and Homeostasis. Int. J. Mol. Sci. 2019, 20, 2424. [Google Scholar] [CrossRef]
- Crowley, D.E.; Kraemer, S.M. Function of siderophores in the plant rhizosphere. In The Rhizosphere, Biochemistry and Organic Substances at the Soil–Plant Interface; CRC Press: Boca Raton, FL, USA, 2007; pp. 73–109. [Google Scholar]
- Rajkumar, M.; Ae, N.; Prasad, M.N.V.; Freitas, H. Potential of siderophore-producing bacteria for improving heavy metal phytoextraction. Trends Biotechnol. 2010, 28, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Sessitsch, A.; Kuffner, M.; Kidd, P.; Vangronsveld, J.; Wenzel, W.W.; Fallmann, K.; Puschenreiter, M. The role of plant-associated bacteria in the mobilization and phytoextraction of trace elements in contaminated soils. Soil Biol. Biochem. 2013, 60, 182–194. [Google Scholar] [CrossRef] [PubMed]
- Gururani, M.; Upadhyaya, C.P.; Baskar, V.; Venkatesh, J.; Nookaraju, A.; Park, S.W. Plant Growth-Promoting Rhizobacteria Enhance Abiotic Stress Tolerance in Solanum tuberosum Through Inducing Changes in the Expression of ROS-Scavenging Enzymes and Improved Photosynthetic Performance. J. Plant Growth Regul. 2012, 32, 245–258. [Google Scholar] [CrossRef]
- Sharma, A.; Johri, B.; Glick, B. Plant growth-promoting bacterium Pseudomonas sp. strain GRP3 influences iron acquisition in mung bean (Vigna radiata L. Wilzeck). Soil Biol. Biochem. 2003, 35, 887–894. [Google Scholar] [CrossRef]
- Dos Santos, R.M.; Diaz, P.A.E.; Lobo, L.L.B.; Rigobelo, E.C. Use of Plant Growth-Promoting Rhizobacteria in Maize and Sugarcane: Characteristics and Applications. Front. Sustain. Food Syst. 2020, 4, 136. [Google Scholar] [CrossRef]
- Mitter, E.K.; Tosi, M.; Obregón, D.; Dunfield, K.E.; Germida, J.J. Rethinking Crop Nutrition in Times of Modern Microbiology: Innovative Biofertilizer Technologies. Front. Sustain. Food Syst. 2021, 5, 29. [Google Scholar] [CrossRef]
- McSteen, P. Auxin and Monocot Development. Cold Spring Harb. Perspect. Biol. 2010, 2, a001479. [Google Scholar] [CrossRef]
- Salisbury, F.B. The role of plant hormones. In Plant—Environment Interactions; Wilkinson, R.E., Ed.; Marcel Dekker: New York, NY, USA, 1994; pp. 39–81. [Google Scholar]
- Martínez, C.; Espinosa-Ruiz, A.; Prat, S. Gibberellins and plant vegetative growth. Annu. Plant Rev. Online 2018, 49, 285–322. [Google Scholar]
- Shu, K.; Zhou, W.; Chen, F.; Luo, X.; Yang, W. Abscisic Acid and Gibberellins Antagonistically Mediate Plant Development and Abiotic Stress Responses. Front. Plant Sci. 2018, 9, 416. [Google Scholar] [CrossRef]
- Abts, W.; Vandenbussche, B.; De Proft, M.P.; Van De Poel, B. The Role of Auxin-Ethylene Crosstalk in Orchestrating Primary Root Elongation in Sugar Beet. Front. Plant Sci. 2017, 8, 444. [Google Scholar] [CrossRef]
- Weyens, N.; van der Lelie, D.; Taghavi, S.; Newman, L.; Vangronsveld, J. Exploiting plant–microbe partnerships to improve biomass production and remediation. Trends Biotechnol. 2009, 27, 591–598. [Google Scholar] [CrossRef]
- Patten, C.L.; Glick, B.R. Bacterial biosynthesis of indole-3-acetic acid. Can. J. Microbiol. 1996, 42, 207–220. [Google Scholar] [CrossRef]
- Vacheron, J.; Desbrosses, G.; Bouffaud, M.-L.; Touraine, B.; Moënne-Loccoz, Y.; Muller, D.; Legendre, L.; Wisniewski-Dyé, F.; Prigent-Combaret, C. Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 2013, 4, 356. [Google Scholar] [CrossRef]
- Li, J.; Sun, J.; Yang, Y.; Guo, S.; Glick, B.R. Identification of hypoxic-responsive proteins in cucumber roots using a proteomic approach. Plant Physiol. Biochem. 2012, 51, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Saleh-Lakha, S.; Glick, B.R. The effect of native and ACC deaminase-containing Azospirillum brasilense Cd1843 on the rooting of carnation cuttings. Can. J. Microbiol. 2005, 51, 511–514. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. Modulation of plant ethylene levels by the bacterial enzyme ACC deaminase. FEMS Microbiol. Lett. 2005, 251, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Sandhya, V.; Ali, S.Z.; Grover, M.; Reddy, G.; Venkateswarlu, B. Effect of plant growth promoting Pseudomonas spp. on compatible solutes, antioxidant status and plant growth of maize under drought stress. Plant Growth Regul. 2010, 62, 21–30. [Google Scholar] [CrossRef]
- Ali, S.; Kim, W.-C. Plant Growth Promotion Under Water: Decrease of Waterlogging-Induced ACC and Ethylene Levels by ACC Deaminase-Producing Bacteria. Front. Microbiol. 2018, 9, 1096. [Google Scholar] [CrossRef] [PubMed]
- Mayak, S.; Tirosh, T.; Glick, B.R. Plant growth-promoting bacteria confer resistance in tomato plants to salt stress. Plant Physiol. Biochem. 2004, 42, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Belimov, A.A.; Safronova, V.I.; Sergeyeva, T.A.; Egorova, T.N.; Matveyeva, V.A.; Tsyganov, V.E.; Borisov, A.Y.; Tikhonovich, I.A.; Kluge, C.; Preisfeld, A.; et al. Characterization of plant growth promoting rhizobacteria isolated from polluted soils and containing 1-aminocyclopropane-1-carboxylate deaminase. Can. J. Microbiol. 2001, 47, 642–652. [Google Scholar] [CrossRef] [PubMed]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant Growth-Promoting Rhizobacteria: Context, Mechanisms of Action, and Roadmap to Commercialization of Biostimulants for Sustainable Agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef] [PubMed]
- Bashan, Y.; De-Bashan, L.E.; Prabhu, S.R.; Hernandez, J. Advances in plant growth-promoting bacterial inoculant technology: Formulations and practical perspectives (1998–2013). Plant Soil 2013, 378, 1–33. [Google Scholar] [CrossRef]
- Owen, D.; Williams, A.P.; Griffith, G.; Withers, P.J.A. Use of commercial bio-inoculants to increase agricultural production through improved phosphrous acquisition. Appl. Soil Ecol. 2015, 86, 41–54. [Google Scholar] [CrossRef]
- Khan, M.S.; Zaidi, A.; Wani, P.A.; Oves, M. Role of plant growth promoting rhizobacteria in the remediation of metal contaminated soils. Environ. Chem. Lett. 2008, 7, 1–19. [Google Scholar] [CrossRef]
- Oleńska, E.; Małek, W.; Wójcik, M.; Swiecicka, I.; Thijs, S.; Vangronsveld, J. Beneficial features of plant growth-promoting rhizobacteria for improving plant growth and health in challenging conditions: A methodical review. Sci. Total Environ. 2020, 743, 140682. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Prasad, M.; Rajkumar, M.; Freitas, H. Plant growth promoting rhizobacteria and endophytes accelerate phytoremediation of metalliferous soils. Biotechnol. Adv. 2011, 29, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Arslan, M.; Afzal, M.; Amin, I.; Iqbal, S.; Khan, Q.M. Nutrients Can Enhance the Abundance and Expression of Alkane Hydroxylase CYP153 Gene in the Rhizosphere of Ryegrass Planted in Hydrocarbon-Polluted Soil. PLoS ONE 2014, 9, e111208. [Google Scholar] [CrossRef]
- Balseiro-Romero, M.; Gkorezis, P.; Kidd, P.; Van Hamme, J.; Weyens, N.; Monterroso, C.; Vangronsveld, J. Use of plant growth promoting bacterial strains to improve Cytisus striatus and Lupinus luteus development for potential application in phytoremediation. Sci. Total Environ. 2017, 581-582, 676–688. [Google Scholar] [CrossRef]
- Pizarro-Tobias, P.; Niqui, J.L.; Roca, A.; Solano, J.; Fernández, M.; Bastida, F.; García, C.; Ramos, J.L. Field trial on removal of petroleum-hydrocarbon pollutants using a microbial consortium for bioremediation and rhizoremediation. Environ. Microbiol. Rep. 2014, 7, 85–94. [Google Scholar] [CrossRef]
- Hou, J.; Liu, W.; Wang, B.; Wang, Q.; Luo, Y.; Franks, A. PGPR enhanced phytoremediation of petroleum contaminated soil and rhizosphere microbial community response. Chemosphere 2015, 138, 592–598. [Google Scholar] [CrossRef] [PubMed]
- Ptaszek, N.; Pacwa-Płociniczak, M.; Noszczyńska, M.; Płociniczak, T. Comparative Study on Multiway Enhanced Bio- and Phytoremediation of Aged Petroleum-Contaminated Soil. Agronomy 2020, 10, 19. [Google Scholar] [CrossRef]
- Płociniczak, T.; Fic, E.; Pacwa-Płociniczak, M.; Pawlik, M.; Piotrowska-Seget, Z. Improvement of phytoremediation of an aged petroleum hydrocarbon-contaminated soil by Rhodococcus erythropolis CD 106 strain. Int. J. Phytoremediat. 2017, 19, 614–620. [Google Scholar] [CrossRef]
- Whyte, L.G.; Schultz, A.; Beilen, J.B.; Luz, A.P.; Pellizari, V.; Labbã, D.; Greer, C.W.; Labbé, D. Prevalence of alkane monooxygenase genes in Arctic and Antarctic hydrocarbon-contaminated and pristine soils1. FEMS Microbiol. Ecol. 2002, 41, 141–150. [Google Scholar] [CrossRef][Green Version]
- Yakimov, M.; Timmis, K.N.; Golyshin, P. Obligate oil-degrading marine bacteria. Curr. Opin. Biotechnol. 2007, 18, 257–266. [Google Scholar] [CrossRef]
- Afzal, M.; Yousaf, S.; Reichenauer, T.G.; Sessitsch, A. Ecology of Alkane-Degrading Bacteria and Their Interaction with the Plant. Mol. Microb. Ecol. Rhizosphere 2013, 2, 975–989. [Google Scholar] [CrossRef]
- Johnsen, A.R.; Karlson, U. PAH Degradation Capacity of Soil Microbial Communities—Does It Depend on PAH Exposure? Microb. Ecol. 2005, 50, 488–495. [Google Scholar] [CrossRef]
- Tremblay, J.; Yergeau, E.; Fortin, N.; Cobanli, S.; Elias, M.; King, T.L.; Lee, K.; Greer, C.W. Chemical dispersants enhance the activity of oil- and gas condensate-degrading marine bacteria. ISME J. 2017, 11, 2793–2808. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Liu, W.; Tian, S.; Wang, W.; Qi, Q.; Jiang, P.; Gao, X.; Li, F.; Li, H.; Yu, H. Petroleum Hydrocarbon-Degrading Bacteria for the Remediation of Oil Pollution Under Aerobic Conditions: A Perspective Analysis. Front. Microbiol. 2018, 9, 2885. [Google Scholar] [CrossRef]
- Varjani, S.J. Microbial degradation of petroleum hydrocarbons. Bioresour. Technol. 2017, 223, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-B.; Chi, C.-Q.; Nie, Y.; Tang, Y.-Q.; Tan, Y.; Wu, G.; Wu, X.-L. Degradation of petroleum hydrocarbons (C6–C40) and crude oil by a novel Dietzia strain. Bioresour. Technol. 2011, 102, 7755–7761. [Google Scholar] [CrossRef]
- Sarkar, P.; Roy, A.; Pal, S.; Mohapatra, B.; Kazy, S.K.; Maiti, M.K.; Sar, P. Enrichment and characterization of hydrocarbon-degrading bacteria from petroleum refinery waste as potent bioaugmentation agent for in situ bioremediation. Bioresour. Technol. 2017, 242, 15–27. [Google Scholar] [CrossRef]
- Rojo, F. Degradation of alkanes by bacteria. Environ. Microbiol. 2009, 11, 2477–2490. [Google Scholar] [CrossRef]
- Post-Beittenmiller, D. Biochemistry and molecular biology of wax production in plants. Annu. Rev. Plant Biol. 1996, 47, 405–430. [Google Scholar] [CrossRef]
- Schirmer, A.; Rude, M.A.; Li, X.; Popova, E.; del Cardayre, S.B. Microbial Biosynthesis of Alkanes. Science 2010, 329, 559–562. [Google Scholar] [CrossRef]
- Ji, Y.; Mao, G.; Wang, Y.; Bartlam, M. Structural insights into diversity and n-alkane biodegradation mechanisms of alkane hydroxylases. Front. Microbiol. 2013, 4, 58. [Google Scholar] [CrossRef]
- Nie, Y.; Chi, C.-Q.; Fang, H.; Liang, J.-L.; Lu, S.-L.; Lai, G.-L.; Tang, Y.-Q.; Wu, X.-L. Diverse alkane hydroxylase genes in microorganisms and environments. Sci. Rep. 2014, 4, 4968. [Google Scholar] [CrossRef] [PubMed]
- Van Beilen, J.B.; Funhoff, E.G. Alkane hydroxylases involved in microbial alkane degradation. Appl. Microbiol. Biotechnol. 2007, 74, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Shao, Z. Enzymes and genes involved in aerobic alkane degradation. Front. Microbiol. 2013, 4, 116. [Google Scholar] [CrossRef] [PubMed]
- Bihari, Z.; Szvetnik, A.; Szabó, Z.; Blastyák, A.; Zombori, Z.; Balázs, M.; Kiss, I. Functional analysis of long-chain n-alkane degradation by Dietzia spp. FEMS Microbiol. Lett. 2011, 316, 100–107. [Google Scholar] [CrossRef]
- Piccolo, L.L.; DE Pasquale, C.; Fodale, R.; Puglia, A.M.; Quatrini, P. Involvement of an Alkane Hydroxylase System of Gordonia sp. Strain SoCg in Degradation of Solid n -Alkanes. Appl. Environ. Microbiol. 2011, 77, 1204–1213. [Google Scholar] [CrossRef]
- van Beilen, J.B.; Funhoff, E.G.; van Loon, A.; Just, A.; Kaysser, L.; Bouza, M.; Holtackers, R.; Röthlisberger, M.; Li, Z.; Witholt, B. Cytochrome P450 Alkane Hydroxylases of the CYP153 Family Are Common in Alkane-Degrading Eubacteria Lacking Integral Membrane Alkane Hydroxylases. Appl. Environ. Microbiol. 2006, 72, 59–65. [Google Scholar] [CrossRef]
- Nie, Y.; Liang, J.-L.; Fang, H.; Tang, Y.-Q.; Wu, X.-L. Characterization of a CYP153 alkane hydroxylase gene in a Gram-positive Dietzia sp. DQ12-45-1b and its “team role” with alkW1 in alkane degradation. Appl. Microbiol. Biotechnol. 2013, 98, 163–173. [Google Scholar] [CrossRef]
- Whyte, L.G.; Smits, T.H.M.; Labbé, D.; Witholt, B.; Greer, C.W.; van Beilen, J.B. Gene Cloning and Characterization of Multiple Alkane Hydroxylase Systems in Rhodococcus Strains Q15 and NRRL B-16531. Appl. Environ. Microbiol. 2002, 68, 5933–5942. [Google Scholar] [CrossRef]
- Wang, L.; Wang, W.; Lai, Q.; Shao, Z. Gene diversity of CYP153A and AlkB alkane hydroxylases in oil-degrading bacteria isolated from the Atlantic Ocean. Environ. Microbiol. 2010, 12, 1230–1242. [Google Scholar] [CrossRef] [PubMed]
- Long, H.; Wang, Y.; Chang, S.; Liu, G.; Chen, T.; Huo, G.; Zhang, W.; Wu, X.; Tai, X.; Sun, L.; et al. Diversity of crude oil-degrading bacteria and alkane hydroxylase (alkB) genes from the Qinghai-Tibet Plateau. Environ. Monit. Assess. 2017, 189, 116. [Google Scholar] [CrossRef]
- Varjani, S.J.; Gnansounou, E.; Pandey, A. Comprehensive review on toxicity of persistent organic pollutants from petroleum refinery waste and their degradation by microorganisms. Chemosphere 2017, 188, 280–291. [Google Scholar] [CrossRef]
- Perez-Pantoja, D.; Gonza’lez, B.; Pieper, D.H. Aerobic Degradation of Aromatic Hydrocarbons. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 800–837. [Google Scholar]
- Kotoky, R.; Rajkumari, J.; Pandey, P. The rhizosphere microbiome: Significance in rhizoremediation of polyaromatic hydrocarbon contaminated soil. J. Environ. Manag. 2018, 217, 858–870. [Google Scholar] [CrossRef]
- Cerniglia, C.E. Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation 1992, 3, 351–368. [Google Scholar] [CrossRef]
- Peng, R.-H.; Xiong, A.-S.; Xue, Y.; Fu, X.-Y.; Gao, F.; Zhao, W.; Tian, Y.-S.; Yao, Q.-H. Microbial biodegradation of polyaromatic hydrocarbons. FEMS Microbiol. Rev. 2008, 32, 927–955. [Google Scholar] [CrossRef] [PubMed]
- Mallick, S.; Chakraborty, J.; Dutta, T.K. Role of oxygenases in guiding diverse metabolic pathways in the bacterial degradation of low-molecular-weight polycyclic aromatic hydrocarbons: A review. Crit. Rev. Microbiol. 2010, 37, 64–90. [Google Scholar] [CrossRef] [PubMed]
- Ghosal, D.; Ghosh, S.; Dutta, T.K.; Ahn, Y. Current State of Knowledge in Microbial Degradation of Polycyclic Aromatic Hydrocarbons (PAHs): A Review. Front. Microbiol. 2016, 7, 1369. [Google Scholar] [CrossRef]
- Kweon, O.; Kim, S.-J.; Cerniglia, C.E. An Update on the Genomic View of Mycobacterial High-Molecular-Weight Polycyclic Aromatic Hydrocarbon Degradation. In Aerobic Utilization of Hydrocarbons, Oils and Lipids, Handbook of Hydrocarbon and Lipid Microbiology; Rojo, F., Ed.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 1–16. [Google Scholar]
- Wick, A.D.M.L.; De Munain, A.R.; Springael, D.; Harms, H. Responses of Mycobacterium sp. LB501T to the low bioavailability of solid anthracene. Appl. Microbiol. Biotechnol. 2002, 58, 378–385. [Google Scholar] [CrossRef]
- Simon, M.J.; Osslund, T.D.; Saunders, R.; Ensley, B.D.; Suggs, S.; Harcourt, A.; Wen-Chen, S.; Cruder, D.L.; Gibson, D.T.; Zylstra, G.J. Sequences of genes encoding naphthalene dioxygenase in Pseudomonas putida strains G7 and NCIB 9816-4. Gene 1993, 127, 31–37. [Google Scholar] [CrossRef]
- Laurie, A.D.; Lloyd-Jones, G. The phn Genes of Burkholderia sp. Strain RP007 Constitute a Divergent Gene Cluster for Polycyclic Aromatic Hydrocarbon Catabolism. J. Bacteriol. 1999, 181, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.A.; Wang, R.-F.; Cao, W.-W.; Doerge, D.R.; Wennerstrom, D.; Cerniglia, C.E. Molecular Cloning, Nucleotide Sequence, and Expression of Genes Encoding a Polycyclic Aromatic Ring Dioxygenase from Mycobacterium sp. Strain PYR-1. Appl. Environ. Microbiol. 2001, 67, 3577–3585. [Google Scholar] [CrossRef] [PubMed]
- Pagnout, C.; Frache, G.; Poupin, P.; Maunit, B.; Muller, J.-F.; Férard, J.-F. Isolation and characterization of a gene cluster involved in PAH degradation in Mycobacterium sp. strain SNP11: Expression in Mycobacterium smegmatis mc2155. Res. Microbiol. 2007, 158, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Kivisaar, M.; Kasak, L.; Nurk, A. Sequence of the plasmid-encoded catechol 1,2-dioxygenase-expressing gene, pheB, of phenol-degrading Pseudomonas sp. strain EST1001. Gene 1991, 98, 15–20. [Google Scholar] [CrossRef]
- Hiraoka, S.; Yang, C.-C.; Iwasaki, W. Metagenomics and Bioinformatics in Microbial Ecology: Current Status and Beyond. Microbes Environ. 2016, 31, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Equiza, L.; St-Arnaud, M.; Eyergeau, E. Harnessing phytomicrobiome signaling for rhizosphere microbiome engineering. Front. Plant Sci. 2015, 6, 507. [Google Scholar] [CrossRef]
- Tardif, S.; Yergeau, E.; Tremblay, J.; Legendre, P.; Whyte, L.G.; Greer, C.W. The Willow Microbiome Is Influenced by Soil Petroleum-Hydrocarbon Concentration with Plant Compartment-Specific Effects. Front. Microbiol. 2016, 7, 1363. [Google Scholar] [CrossRef] [PubMed]
- Mitter, E.K.; De Freitas, J.R.; Germida, J.J. Bacterial Root Microbiome of Plants Growing in Oil Sands Reclamation Covers. Front. Microbiol. 2017, 8, 849. [Google Scholar] [CrossRef]
- Ofek-Lalzar, M.; Sela, N.; Goldman-Voronov, M.; Green, S.; Hadar, Y.; Minz, D. Niche and host-associated functional signatures of the root surface microbiome. Nat. Commun. 2014, 5, 4950. [Google Scholar] [CrossRef] [PubMed]
- Bell, T.H.; Joly, S.; Pitre, F.E.; Yergeau, E. Increasing phytoremediation efficiency and reliability using novel omics approaches. Trends Biotechnol. 2014, 32, 271–280. [Google Scholar] [CrossRef]
- Pagé, A.P.; Yergeau, E.; Greer, C.W. Salix purpurea Stimulates the Expression of Specific Bacterial Xenobiotic Degradation Genes in a Soil Contaminated with Hydrocarbons. PLoS ONE 2015, 10, e0132062. [Google Scholar] [CrossRef]
- Zhao, Q.; Hu, H.; Wang, W.; Peng, H.; Zhang, X. Genome Sequence of Sphingobium yanoikuyae B1, a Polycyclic Aromatic Hydrocarbon-Degrading Strain. Genome Announc. 2015, 3, e01522-14. [Google Scholar] [CrossRef]
- Imperato, V.; Portillo-Estrada, M.; McAmmond, B.; Douwen, Y.; Van Hamme, J.D.; Gawronski, S.W.; Vangronsveld, J.; Thijs, S. Genomic Diversity of Two Hydrocarbon-Degrading and Plant Growth-Promoting Pseudomonas Species Isolated from the Oil Field of Bóbrka (Poland). Genes 2019, 10, 443. [Google Scholar] [CrossRef]
- Neifar, M.; Chouchane, H.; Najjari, A.; El Hidri, D.; Mahjoubi, M.; Ghedira, K.; Naili, F.; Soufi, L.; Raddadi, N.; Sghaier, H.; et al. Genome analysis provides insights into crude oil degradation and biosurfactant production by extremely halotolerant Halomonas desertis G11 isolated from Chott El-Djerid salt-lake in Tunisian desert. Genomics 2018, 111, 1802–1814. [Google Scholar] [CrossRef]
- He, C.; Li, Y.; Huang, C.; Chen, F.; Ma, Y. Genome Sequence and Metabolic Analysis of a Fluoranthene-Degrading Strain Pseudomonas aeruginosa DN1. Front. Microbiol. 2018, 9, 2595. [Google Scholar] [CrossRef] [PubMed]
- Mahjoubi, M.; Aliyu, H.; Cappello, S.; Naifer, M.; Souissi, Y.; Cowan, D.; Cherif, A. The genome of Alcaligenes aquatilis strain BU33N: Insights into hydrocarbon degradation capacity. PLoS ONE 2019, 14, e0221574. [Google Scholar] [CrossRef]
- Silva, N.M.; De Oliveira, A.M.S.A.; Pegorin, S.; Giusti, C.E.; Ferrari, V.; Barbosa, D.; Martins, L.F.; Morais, C.; Setubal, J.C.; Vasconcellos, S.P.; et al. Characterization of novel hydrocarbon-degrading Gordonia paraffinivorans and Gordonia sihwensis strains isolated from composting. PLoS ONE 2019, 14, e0215396. [Google Scholar] [CrossRef]
- Rajkumari, J.; Singha, L.P.; Pandey, P. Draft Genome Sequence of Klebsiella pneumoniae AWD5. Genome Announc. 2017, 5, e01531-16. [Google Scholar] [CrossRef]
- Stevens, V.; Thijs, S.; McAmmond, B.; Langill, T.; Van Hamme, J.; Weyens, N.; Vangronsveld, J. Draft Genome Sequence of Bacillus licheniformis VSD4, a Diesel Fuel–Degrading and Plant Growth–Promoting Phyllospheric Bacterium. Genome Announc. 2017, 5, e00027-17. [Google Scholar] [CrossRef]
- Filonov, A.; Delegan, Y.; Puntus, I.; Valentovich, L.; Akhremchuk, A.; Evdokimova, O.; Funtikova, T.; Zakharova, M.; Akhmetov, L.; Vetrova, A.; et al. Complete Genome Sequence of Pseudomonas putida BS3701, a Promising Polycyclic Aromatic Hydrocarbon-Degrading Strain for Bioremediation Technologies. Microbiol. Resour. Announc. 2020, 9, e00892-20. [Google Scholar] [CrossRef]
| Category | Mechanism | Target Pollutants | Region of Activity | Reference |
|---|---|---|---|---|
| Phytoextraction | Uptake and concentrate contaminants | Metals (e.g., Cd,Ni), radionuclides (e.g., Pu) | Shoot tissue | [21,29] |
| Phytostabilization | Immobilization and sequestration of contaminants | Primarily metals (e.g., Cu, Zn, Pb) | Root tissue | [30] |
| Phytotransformation | Enzymatic actions | Chlorinated solvents, ammunition waste, herbicides, monoaromatic hydrocarbons | Plant tissue | [15,30] |
| Phytovolatilization | Uptake and evapotranspiration | Volatile organics (e.g., TCE, toluene, MTBE), | Shoot tissue | [14] |
| Rhizoremediation | Breakdown of organic pollutants by using plants and root-associated microbiomes | PHC (e.g., diesel), pesticides (e.g., dimethomorph) | Root | [19,28] |
| Plant | Contaminants | Conditions | Bacteria | Role of PGPR | Reference |
|---|---|---|---|---|---|
| Lolium perenne | Diesel | Greenhouse | Pantoea sp. BTRH79 | ACCD | [133] |
| Cytisus striatus | Diesel | Greenhouse | Bradyrhizobium sp. ER33 | IAA, organic acids | [134] |
| Lupinus luteus | Diesel | Greenhouse | Streptomyces sp. RP92 | IAA, siderophore, organic acid | [134] |
| Trifolium repens | Oil refinery sludge | Field trial | Psudomonas putida BIRD-1 | P-solublization, IAA, siderophore | [135] |
| Festuca arundinacea | Aliphatic hydrocarbons | Field trail | PGPR consortia | N/A | [136] |
| Lolium perenne | Aged PHCs | Greenhouse | Rhodococcus erythropolis CDEL254 | Several PGP traits | [137] |
| Lolium perenne | Aged PHCs | Greenhouse | Rhodococcus erythropolis CD 106 | Several PGP traits | [138] |
| Enzyme Class | Substrate Range | Gene | Bacterial Species |
|---|---|---|---|
|
Soluble methane Monooxygenase | C1–C8 | mmoX | Gordonia, Methylococcus, Methylosinus, Methylocystis, Methylomonas, Methylocella. |
| Particulate methane Monooxygenase | C1–C5 | pmoC | Methylococcus, Methylosinus, Methylocystis, Methylobacter, Methylomonas, Methylomicrobium, Nocardioides. |
| Alkane 1-monooxygenase | C10–C20 | alkB | Acinetobacter, Alcanivorax, Burkholderia, Mycobacterium, Pseudomonas, Rhodococcus |
| Soluble cytochrome P450 | C5–C16 | CYP153 | Acinetobacter, Alcanivorax, Caulobacter, Mycobacterium, Rhodococcus, Sphingomonas. |
| Flavin-binding monooxygenase | C20–C36 | Alma | Alcanivorax, Marinobacter, Acinetobacter. |
| Thermophilic flavin-dependent monooxygenase | C10–C36 | LadA | Geobacillus thermodenitrificans NG80-2 |
| Enzyme | Gene | Bacterial Source | Reference |
|---|---|---|---|
| Naphthalene dioxygenase | Nah | Pseudomonas putida strain G7 | [171] |
| Phenanthrene dioxygenase | phnAc | Burkholderia sp. strain RP007 | [172] |
| Pyrene dioxygenase | nidA | Mycobacterium sp. strain PYR-1 | [173] |
| Extradiol dioxygenase | phdF | Mycobacterium sp. strain SNP11 | [174] |
| Catechol 1,2-dioxygenase | C12O | Pseudomonas sp. strain EST1001 | [175] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alotaibi, F.; Hijri, M.; St-Arnaud, M. Overview of Approaches to Improve Rhizoremediation of Petroleum Hydrocarbon-Contaminated Soils. Appl. Microbiol. 2021, 1, 329-351. https://doi.org/10.3390/applmicrobiol1020023
Alotaibi F, Hijri M, St-Arnaud M. Overview of Approaches to Improve Rhizoremediation of Petroleum Hydrocarbon-Contaminated Soils. Applied Microbiology. 2021; 1(2):329-351. https://doi.org/10.3390/applmicrobiol1020023
Chicago/Turabian StyleAlotaibi, Fahad, Mohamed Hijri, and Marc St-Arnaud. 2021. "Overview of Approaches to Improve Rhizoremediation of Petroleum Hydrocarbon-Contaminated Soils" Applied Microbiology 1, no. 2: 329-351. https://doi.org/10.3390/applmicrobiol1020023
APA StyleAlotaibi, F., Hijri, M., & St-Arnaud, M. (2021). Overview of Approaches to Improve Rhizoremediation of Petroleum Hydrocarbon-Contaminated Soils. Applied Microbiology, 1(2), 329-351. https://doi.org/10.3390/applmicrobiol1020023







