Abstract
Exuded plant metabolites play an important role in fostering beneficial interactions with the surrounding soil microbiota, thereby helping plants to better adjust to changing environmental conditions. These metabolites act as signals to attract or enhance the colonization of plant roots with specific groups of beneficial microbes and they modulate the dynamics of plant–microbe interactions in fulfilling plant niche-based requirements, directly and/or indirectly. This review emphasizes the expression, levels, modes of action, and net effects of the signaling metabolites that help food crop plants to become colonized by microbes that promote plant growth and development under periods of biotic stress.
1. Introduction
The world’s population is currently estimated to be around eight billion people and is growing at the rate of approximately 72 million people per year (https://www.worldometers.info/; accessed on 27 December 2023). At the same time, it is estimated that there are nearly 900 million undernourished people in the world with ~14,000 people dying of hunger every day. With the prospect of a significantly increased global population in the next 50 years, it is essential that the worldwide availability of food crops increases dramatically.
Food availability can be significantly increased in several different ways. In the first instance, it has been estimated that “over a third of all food produced (~2.5 billion tons) is lost or wasted each year” [1]. Food wastage is not limited to richer and more developed countries; it is also a huge problem in poorer and less developed countries. Second, the genetic engineering of food crops, including genome editing using CRISPR/CAS9 technology, promises to significantly increase both the quality and the quantity of food that is currently available worldwide [2,3]. Third, the increased use of PGPB in agriculture, a technology that is currently in its early stages of development but is growing rapidly, promises to both increase crop productivity and quality as well as to decrease the use of potentially harmful chemicals in agricultural practice. Unfortunately, there is no one simple solution to the problem of insufficient global food availability. Thus, it is likely that dramatically increasing the food supply within the next 50 years will require all three of the abovementioned approaches.
Soil bacteria have interacted closely with plants for millions of years. These bacteria dramatically impact plant growth and development including increasing the ability of the plant to function in the presence of a wide range of environmental stresses, both abiotic and biotic [4].
The soil contains an enormous number, typically many thousands, of different types of microbes. Moreover, soil microbes are generally found in concentrations in the millions to hundreds of millions of organisms per gram of soil. These microbes, including various bacteria, fungi, cyanobacteria, archaea, and viruses, can impact the growth and development of plants positively (plant growth-promoting microbes), negatively (phytopathogenic microbes) or not at all (commensal microbes) [5,6]. Bacteria are by far the most predominant of all soil microbes and their interactions with plants have been studied to the greatest extent [4].
Soil bacteria may facilitate plant growth either directly (where bacteria and their metabolites interact directly with plants to promote their growth and development) or indirectly (where bacteria prevent various phytopathogens from inhibiting plant growth). Direct mechanisms include (i) facilitating the solubilization and uptake of minerals from the soil environment, including iron, potassium, and phosphorus; (ii) providing plants with fixed nitrogen; (iii) synthesizing the plant hormones cytokinin, gibberellin, and auxin, all of which stimulate different aspects of plant growth [7,8]; and (iv) decreasing plant ethylene using the enzyme ACC deaminase [4,9,10,11,12,13].
Indirect mechanisms decrease the damage or growth inhibition to the plant with various phytopathogens; these mechanisms include (i) the production of antibiotics and hydrogen cyanide which inhibit the growth of some bacterial and fungal phytopathogens; (ii) the solubilization and subsequent sequestration of iron by bacterially synthesized siderophores, thereby limiting pathogens access to iron and preventing their proliferation; (iii) the synthesis and secretion of fungal pathogen cell-wall-degrading enzymes; (iv) outcompeting phytopathogens by (a) direct competition of nutrients and (b) establishing symbiosis with beneficial microorganism (e.g., arbuscular mycorrhizal fungi) [14], and as a result preventing the phytopathogens from binding extensively to plant roots; (v) the synthesis of many different volatile organic compounds that can inhibit pathogen proliferation; (vi) lowering the amount of potentially inhibitory plant ethylene concentrations by the enzyme ACC deaminase, thereby decreasing plant stress levels; and (vii) the induction of the plant’s systemic resistance system, turning on the plant’s defenses against various pathogens [4,11,15,16,17,18].
While the indirect mechanisms of plant growth promotion with PGPB appear to be applicable to the vast majority of plants, the bulk of the scientific literature deals with the protection afforded with PGPB to agricultural food crops [19,20]. Moreover, as of mid-2023, several dozen companies were marketing specific strains of PGPB which were purported to reduce the losses of agricultural food crops from both abiotic and biotic environmental stresses [20].
The largest numbers of bacteria are generally found in the plant rhizosphere, i.e., the portion of the soil that is present in the immediate vicinity of plant roots [5]. The reason for the high level of bacteria around plant roots is because of the exudation of a significant fraction (often 5–40%) of the carbon that is fixed by the plant through photosynthesis [21,22,23,24]. In addition to their presence in the rhizosphere, many plant growth-promoting bacteria (PGPB) are also able to colonize a plant’s interior tissues (i.e., they are endophytic bacteria and are typically found in the spaces between plant cells). In addition, some bacteria form highly specific nodules on plant roots (they are symbiotic bacteria, typically including different types of Rhizobia). In the recent 10–15 years, many studies of how PGPB promote plant growth and development have been undertaken; they have focused on how groups (consortia) of bacteria, rather than individual strains, are able to act together/synergistically to positively impact plants [18]. These studies include an understanding of how the microbiomes (i.e., the groups of microbes that together influence plant behavior) of different plants attract different portions of the soil bacterial population. Each plant exudes (secretes) a unique mixture of small molecules through its roots that attracts a specific fraction of the soil bacterial community. As a result of this interaction, various plants producing different compositions of root exudate metabolites attract unique subsets of bacteria from the soil to the plant roots. Thus, the specific chemical composition of each plant’s root exudates is involved in attracting from the soil the bacteria that make up a plant’s root microbiome [25,26].
2. How Plants Recruit Beneficial Microbes and Exclude Pathogens
The rhizosphere is a metabolically dynamic space that contains a wide range of primary and secondary metabolites that originate from both plants and microbes. Chemicals present within the rhizosphere at any given time can act as signals that connect the surrounding organisms (food crops and microbes in this case) and thereby facilitate communication. Plants program their communication according to their needs, mainly based on the environmental changes and the developmental stage of the crop by exuding these signal molecules through their root system (Figure 1). Being in a nutritionally scarce soil hub, microbes are attracted towards this valuable source of nutrition and energy [27]. The biochemical nature of root secretions ultimately determines the microbial function and diversity that host crops utilize under certain environmental conditions [28,29,30,31].
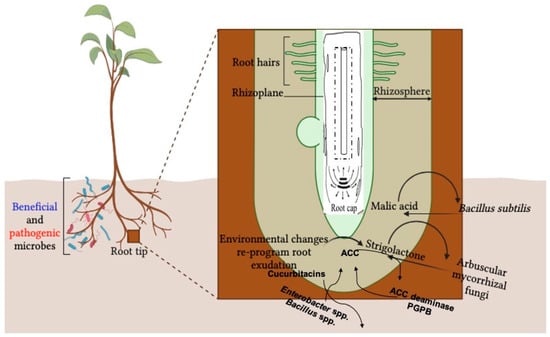
Figure 1.
Schematic representation of how biotic stress stimulates cellular and transcriptomic levels of plants’ secondary metabolites in root exudates resulting in an altered rhizomicrobiome.
Under a nitrogen-deficient environment, legume crops secrete flavonoids as a component of their root exudates [32] that attract nitrogen-fixing microbes from the surrounding soil, regulate nod gene expression, and help food crop proliferation through the establishment of nodulation [33,34]. Similarly, malic acid and strigolactones, which are plant signal molecules, help to recruit PGPB [35] and foster plant–mycorrhiza interactions [36] in food crops.
Under drought stress conditions, A. thaliana roots recruit selective bacteria (Pseudomonas chlororaphis) with stress remediation functions, i.e., decreased expression of ethylene and abscisic acid and increased synthesis of salicylic acid, jasmonic acid, and reactive oxygen species defense enzymes [37]. Studies by Creus et al. [38] and Molina-Favero et al. [39] have demonstrated that tomato plants can selectively recruit Azospirillum brasilense by secreting nitric oxide into their rhizosphere; this bacterium is a PGPB that mediates tomato root architecture by supplying IAA to its host plant. Similarly, a complex array of groundnut root exudates that contains threonine, glyoxylic oxime acid, serine, pentanoic acid, glucopyranoside, tartaric acid, and 2-pyrrolidinone enhances the selective interactions of groundnut with two PGPB, Pseudomonas aeruginosa RP2 and Bacillus sonorensis RS4 [40]. Moreover, Arabinogalactan proteins found in the root exudates of legume and non-legume food crops [41], sugars and strigolactones in non-legume food crops [42] and canavanine from legumes [43], help host crops to establish beneficial interactions with PGPB, Rhizobia, and mycorrhizal fungi, respectively.
Root exudates not only attract plant-beneficial microbes in facilitating mutualistic symbiotic relationships from a huge variety of microbial taxa [44], but also act as a biocontrol agent of unwanted opportunistic microbes. For example, maize exudes benzoxazinoids, which are antimicrobial in nature and clear the rhizosphere of actinobacteria, proteobacteria, and all pathogenic microbial populations [45]. Similarly, coumarins, plant-based secondary metabolites, have shown antimicrobial activity and helped the model plant A. thaliana to shape its microbiome [46]. In addition, Badri et al. [47] demonstrated that there are at least 25 genes directly involved in A. thaliana root exudation and that a single gene mutation can cause a significant change to the root exudate pattern, a change that may negatively impact the plant’s interaction with soil microbes. Plant root exudates may, however, cause harmful effects on plant growth and development [48]. Thus, inefficient cross-talk between plant root systems, the rhizospheric environment, and the surrounding microbial community can sometimes lead to an outbreak of soil-borne diseases, imbalance in soil physiochemical properties, disproportion in the soil microbiome [49,50], root cell death, and accumulation of phytopathogens in the root zone [47,51].
In order to inhabit a plant’s rhizosphere or colonize a plant’s interior, a microbe must be able to live on the carbon sources that the host plant offers and bypass the host plant’s defense mechanisms. Similarly, plants must have the ability to decide whether to allow a microbe to be the part of its microbiome and have it involved in mutualistic interactions or combat it through its defense systems [52]. Food crops, like most other plants, present multiple layers of scrutiny checkpoints for a foreign microbe (not native to the plant) to pass through and become resident on or in the plant. For example, the host–microbe interactions are highly dependent on the genetics and the growth stage of the participants, which is largely triggered by environmental biotic and abiotic factors [53]. Plants’ cell membranes are equipped with hundreds of pattern recognition receptors (PRRs) and Nod-like receptors (NLRs) [52,54,55]. These receptors recognize a variety of conserved microbe-associated molecular patterns—MAMPs (e.g., chitin, flagellin, peptidoglycan, etc.)—and distinguish between a potential symbiont and a pathogen, the latter of which is eventually dealt with by the activation of pattern-triggered immunity (PTI) responses. NLRs are believed to be involved in fostering beneficial plant–microbe interactions such as nitrogen-fixing bacteria with legumes. Nod factors secreted by nitrogen-fixing bacteria are recognized by a variety of LysM receptors (LysM is a small widespread globular protein domain involved in binding bacterial peptidoglycan moieties) situated on the plasma membranes of legume crops [56] and establish a symbiotic relationship with host plants.
Since many microorganisms (both pathogenic and non-pathogenic) share the conserved core of MAMPs, and only a weak immune response is generated when a PRR recognizes a microbe-associated molecular pattern; therefore, the recognition of microbial antigenic entities by multiple receptors is required to implement an intense immune response [57]. Perception of a microbe-associated molecular pattern at the same time by multiple receptors on the membrane can be validated by the simultaneous recognition of an effector [57] and damage-associated molecular pattern [58,59], thereby leading to a significantly more rigorous immune response via the increased expression of genes involved in the plant host defense mechanism [52,58]. Unfortunately, not all of the members of the rhizospheric microbiota exhibit and follow clear signaling patterns with the host plant. Ultimately, it is the choice that a host plant makes to allow a particular microbe to be a part of its microbiome, depending on the potentially host beneficial activities that the microbe offers. Nevertheless, being sessile in nature, plants have to make endless critical decisions throughout their life regarding what to keep and what to fight, considering the resources that they possess and the environmental challenges that they face at any given time. The exact mechanism(s) regarding how plants chose one or the other still remains to be uncovered [52].
3. Plant Root Exudates
Root exudates are a chemically diverse class of substances that plants, throughout their lifetime, secrete into their surrounding environment (rhizosphere) via their root system. It is estimated that plant roots secrete about 5–40% of their photosynthetic assimilated carbon as root exudates [21,24]. Plant root exudates are comprised of high- and low-molecular-weight compounds belonging to organic acids, sugars, phenols, amino acids, hormones, flavonoids, and growth regulators [60].
Root exudates affect soil microflora differentially; they exhibit both stimulatory and inhibitory activities toward a variety of soil microorganisms [60]. There is a large body of literature indicating that plants utilize their root exudates to shape their microbiome [60,61,62]. Root exudates play important roles in plant growth and development by regulating micro-niches within the rhizosphere, recruiting and harboring selective microbiota, and communicating to soil microbes under various environmental conditions [62].
Root exudates are dynamic in nature; the composition, concentration, and balance between these two variables changes at different time points. For example, the levels of sugars and secondary metabolites in the root exudates of the model plant Arabidopsis thaliana, which were collected at various developmental stages over time, were found to be significantly different from each other and attracted different consortia of microbes that contained a diverse range of functions, and were able to help the plant respond to a range of environmental changes [63,64,65].
Many factors influence the composition of root exudates and shifts in root exudate patterns, with changes in climatic conditions being the most pivotal. Other factors that have been reported to change root exudation patterns include soil pH, soil type, soil temperature, soil oxygen content (aeration), soil physio-chemical properties, plant developmental stage, and plant genotype [24,64,66,67,68]. Intercropping has also been reported to alter the pattern of root exudates and to help food crops (e.g., tomato) to recruit beneficial microbiota to the rhizosphere [69]. Many mechanisms are employed to secrete root exudates into the surrounding soil medium; that is, low-molecular-weight compounds are secreted by passive transport (diffusion, ion channels, vesicular transport), whereas high-molecular-weight compounds are transported via active transport (membrane transport of proteins at the expense of energy driven by ATP hydrolysis) [70]. The level of root exudates is highest at the tip and the lateral branches of roots, and it becomes diluted as the root system proliferates [21]. The root system architecture is the first point at the plant–soil interface that becomes affected by climatic change due to any stress stimuli [24]. The nature and amount of root exudates are therefore primarily determined by the variations that occur in the root system under changing environments [71,72]. Moreover, it has also been shown that different structures serve as specialized locations to secrete specific root chemistries. For example, asparagine and threonine are exudated from the root meristematic and elongation zones; amino acids like glutamic acid, valine, leucine, and phenylalanine are secreted from the root hairs; and aspartic acid can be secreted from any part of the whole root system [24,73].
4. Overview of How Food Crop Root Exudates Facilitate Plant Biocontrol
Biocontrol in food crops begins at the plant’s root–soil interface. It is neither energy efficient nor genetically possible for a plant to synthesize tailored antimicrobial weapons for each pathogen they might encounter during their life span. Rather, they have evolved ways to attract and recruit a microbial army with weapons (biocidal functions) to fight for them at the expense of a small portion of their atmospherically fixed carbon [10,20,25,74,75,76,77,78,79]. Thus, many PGPB have evolved together with their hosts to continue to offer them the functions necessary for the survival of the host plant in challenging environmental conditions.
Available knowledge of the molecular mechanisms and changes that occur within plants during biocontrol by soil microbes at transcriptomic and metabolomic levels are somewhat limited, although this research area is still growing. Fortunately, a few reports have discussed the role of some known biomolecules that have been identified in root exudates of food crops and have been suggested to shape the microbiome of a host plant’s rhizosphere and help it to better cope with various sorts of biotic stresses (phytopathogens). Table 1 presents such information and includes the food crop, the chemical signal found in root exudates, the changes to the microbiome as a result of adding this chemical signal, and the effects of such interactions on the biocontrol of known phytopathogens.


Table 1.
Effect of root exudate metabolite on plant microbiome overcoming biotic stress.
Table 1.
Effect of root exudate metabolite on plant microbiome overcoming biotic stress.
| Food Crop | Root Exudate | Affected Microbiome (↑ * or ↓ *) | Biocontrol/Outcome | Reference |
|---|---|---|---|---|
| Arabidopsis # | Malic acid | Bacillus subtilis FB17 ↑ | Pseudomonas syringae pv tomato | [80,81,82] |
| Arabidopsis # | Amino acids and long-chain fatty acids | Pseudomonas sp. ↑ | Pseudomonas syringae pv tomato | [83] |
| Barley | Phenolic compound | Pseudomonas fluorescens ↑ | Pythium ultimum | [84] |
| Barley | Phenolic compounds | Beneficial rhizosphere community | Fusarium graminearum | [85,86] |
| Bayberry | Humic acid | Mycobacterium and Crossiella ↑ Acidothermus, Bryobacter, Acidibacter Geminibasidium, and Mycena ↓ | Plant growth promotion and enhance disease resistance | [87] |
| Carex | Volatile organic compound (Monoterpene (Z)-limonene-oxide) | Janthinobacterium, Collimonas, and Paenibacillus ↑ | Fusarium culmorum | [88] |
| Pepper | L-Malic acid | B. subtilis GB03 ↑ | Aphids and Ralstonia solanacearum SL1931 control | [72,89] |
| Cucumber | Fumaric acid and citric acid | Bacillus amyloliquefaciens ↑ | Fusarium oxysporum f. sp. Cucumerinum | [82] |
| Ginseng | Organic acids, sugars, and amino acids | PGPB ↑ | Alternaria panax | [90] |
| Maize | Benzoxazinoids | Flavobacteriaceae and Comamonadacea ↓ | Increased plant growth and disease resistance | [91] |
| Maize | Sesquiterpene (E)-b-caryophyllene (Ebc) | Soil-borne entomopathogenic nematode ↑ | Diabrotica virgifera larvae | [92,93] |
| Maize | Phenolic compounds | Acinetobacter calcoaceticus ↑ | Larvae of western corn rootworms | [94] |
| Melon and watermelon | Cucurbitacins | Enterobacter and Bacillus ↑ | Increased resistance to fungal pathogens | [95] |
| Canola | Trehalose, indolyl, glucosinolates, and sulfur | Gammaproteobacteria, Firmicutes, Bacillus, Paenibacillus, Pseudomonas, and Stenotrophomonas ↑ | Biocontrol of cabbage root fly (Delia radicum) | [96] |
| Peanut | Ethylene | Actinobaterial species ↑ | Cassava (Manihot esculenta) | [97] |
| Potato | Volatile compounds | Phytophthora infestans ↓ | Biotic and abiotic stress reduction | [98] |
| Sorghum | Strigolactone | AMF colonization ↑ | Striga hermonthica | [99] |
| Sweet wormwood | Artemisinin | Sphingomonas and Sphingobium ↑ | Enhanced plant environmental fitness | [100] |
| Tomato | β-Aldehyde | Lysobacter sp. ↑ | Plant growth and defense enhancement | [101] |
| Tomato | β-Caryophyllene | B. amyloliquefaciens ↑ and Pseudomonas aeruginosa ↓ | Disease reduction and plant growth-promotion | [102] |
| Tomato | Strigolactone | AMF colonization ↑ | Meloidogyne incognita | [103] |
| Tomato | Volatile organic compound | Bacillus sp. ↑ | Fusarium oxysporum | [104] |
| Watermelon | Trans-chlorogenic acid, caffeic acid, and trans-cinnamic acid | PGPB ↑ | Fusarium oxysporum | [105] |
* ↑ increased abundance; ↓ decreased abundance; # a model plant but not a food crop.
5. Bioengineering of Rhizospheric Soils for Disease Suppression
Production of food crops mainly relies on the soil’s overall health and productivity. Yield loss due to biotic stress varies depending on the extent, spread, time of disease incidence, and nature of the pathogen(s) in the field; unfortunately, complete crop losses have been reported for many economically important food crops due to pathogen attacks [106,107]. Disease incidence is not a normal physiological state, and it prevails when the symbiotic homeostasis between a plant and its ecological niche is disturbed. Plants continuously send chemical signals into the surrounding soils, via root exudates, as communication measures. These messages keep changing in response to changes in the environment and are primarily aimed at facilitating a quick and smooth plant adaptation to the new environment.
Under biotic stress conditions, plants undergo a complex array of physiological events, including altered gene expression and excessive secretion of root exudates; the increased root exudation attracts and recruits more beneficial bacteria from the soil, a situation termed as a “cry for help” [108]. If the newly recruited microbial army works as needed, plants overcome the stress, the microbiome is shifted, and a new symbiotic homeostasis is obtained. However, if the stress stimuli overcome the plant and its microbiome combined defense mechanisms, disease incidence and severity increase, and the entire homeostasis is challenged by the intruder phytopathogens, eventually leading to plant death. The rhizosphere soil microbiome of a plant works in harmony with the host defense system and provides valuable services to the ecological niches by enhancing disease resilience [64]. A large number of studies have shown the potential capabilities of microorganisms to mitigate biotic stress in planta. Table 1 provides research-based evidence of hosts’ needs-based microbial help to ameliorate biotic stress, i.e., how plants through the use of root exudates call for help and gather the required functions in close proximity to stave off phytopathogens. Considering the interactions of plants with their microbiomes and the dynamics of such interactions, a plant’s rhizosphere can be engineered to achieve an optimized balance in chemical signals, the introduction of microbes that could respond and utilize such signals, and the optimization of the microbial-pesticidal functions to overcome present or future disease incidences. When it comes to the decision of which microbe(s) should be part of the synthetic community, there are certain parameters to be considered: bio-compatibility of microbial members of the community within and with their plant host, a stable pool of desired functions (pesticides in this case), and an overall congruence of functions to be fit into various environmental conditions. One study showed that species-rich synthetic communities of similar genera seem to be more efficient and highly stable compared to species-poor (genera-rich) communities in controlling Rhizoctonia solanacearum densities in the rhizosphere of tomato plants [109]. Moreover, a synthetic community of distantly related bacterial and fungal genera could be useful in obtaining fitness over various plant genotypes and/or diverse environmental conditions [27]. Several attempts at microbial synthetic community formulations based on plants’ “cry for help” have been documented [110,111]. A machine learning computational technique was used to compose a synthetic community using root exudates from plants growing in phosphate-deficient soil. A pool of bacterial strains was grown in root exudates collected from A. thaliana growing under Pi-deficient conditions and monitored individually for their effects on the phosphate contents in plant shoots, which helped to design a synthetic community for a consistent predictable plant phenotype [110]. Similarly, under conditions of a downy mildew pathogen attack, A. thaliana plants selected bacterial consortia that worked synergistically with plant defense systems, resulting in phenotypically healthier plants [111].
Moreover, such knowledge can be used to create functionally reliable synthetic microbial communities that improve plant fitness. The right plant, the right time, and most importantly, the right microbe in the right formulation is what will make these bio-based-control systems a success story. Taking the complexity of plant–microbe interactions into consideration, agricultural biologicals do not appear to be stand-alone solutions [111,112]; plants need to be fostered and the desired microbial populations need to be promulgated. Therefore, it is suggested to combine a synthetic beneficial microbial community with the traces of chemicals previously proven to establish specific microbial populations in the rhizosphere. Such combinations could enhance the survival chances of the target microbes and offer them a competitive advantage for colonizing plants over their unwanted and potentially deleterious counterparts.
Bioengineering of rhizospheric soil for disease suppression and to promote plant fitness is an emerging field, offering great promise towards achieving agricultural sustainability, but also requires extreme care in selecting what is working under controlled conditions with the understanding that things may behave differently in the natural environment. Microbial consortia have often been demonstrated to be better options compared to single microbial inoculants in protecting plants from environmental stresses [79] but care should be taken in increasing complexities in the design of functionally reliable synthetic communities, as this may decrease the overall feasibility of the large-scale production and application of such biological tools.
6. Conclusions and Future Perspectives
One of the biggest challenges that the agriculture sector is facing is to feed the ever-increasing population of the world. As many strategies have been discussed earlier, the use of PGPB has shown promise in many facets. While individual strains of PGPB are highly effective in laboratory conditions, they are often found to be much less effective in the field. However, one way to make PGPB more effective, especially in addressing crop plant biotic stresses, is to first develop an understanding of the elements of crop plant root exudates which attract the microbes that select for a beneficial root microbiome. This information might then be used to develop microbial consortia with which to beneficially treat crop plant seeds and seedlings, thereby increasing the reliability of PGPB use in the field.
Funding
This research received no external funding.
Data Availability Statement
The data presented in this study are openly available in publicly accessible repositories and all the references have been provided in the reference section.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Safdie, S. Global Food Waste in 2024. Greenly, 22 August 2023. Available online: https://greenly.earth/en-us/blog/ecology-news/global-food-waste-in-2022 (accessed on 28 December 2023).
- Aziz, M.A.; Brini, F.; Rouached, H.; Masmoudi, K. Genetically engineered crops for sustainably enhanced food production systems. Front. Plant Sci. 2022, 13, 1027828. [Google Scholar] [CrossRef]
- Sworder, C. Genetic Engineering for Crops and Food: Climate Adaptation, Resilience, and Food Security through Innovation. In Agriculture & Food, Cleantech Insights; Cleantech Group: San Francisco, CA, USA, 2021. [Google Scholar]
- Glick, B.R. Beneficial Plant-Bacterial Interactions; Springer Science and Business Media LLC.: Berlin/Heidelberg, Germany, 2020; p. 383. [Google Scholar] [CrossRef]
- Lynch, M.J. Development and Interaction Between Microbial Communities on the Root Surface. Dev. Soil Sci. 1989, 18, 5–12. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; van Themaat, E.V.L.; Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef]
- Santner, A.; Calderon-Villalobos, L.I.A.; Estelle, M. Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 2009, 5, 301–307. [Google Scholar] [CrossRef]
- Fahad, S.; Hussain, S.; Bano, A.; Saud, S.; Hassan, S.; Shan, D.; Khan, F.A.; Khan, F.; Chen, Y.; Wu, C.; et al. Potential role of phytohormones and plant growth-promoting rhizobacteria in abiotic stresses: Consequences for changing environment. Environ. Sci. Pollut. Res. 2015, 22, 4907–4921. [Google Scholar] [CrossRef]
- Glick, B.R.; Penrose, D.M.; Li, J. A model for the lowering of plant ethylene concentrations by plant growth-promoting bacteria. J. Theor. Biol. 1998, 190, 63–68. [Google Scholar] [CrossRef]
- Gamalero, E.; Glick, B.R. The use of plant growth-promoting bacteria to prevent nematode damage to plants. Biology 2020, 9, 381. [Google Scholar] [CrossRef]
- Glick, B.R. Plant Growth-Promoting Bacteria: Mechanisms and Applications. Scientifica 2012, 2012, 963401. [Google Scholar] [CrossRef]
- Ali, S.; Charles, T.C.; Glick, B.R. Delay of flower senescence by bacterial endophytes expressing 1-aminocyclopropane-1-carboxylate deaminase. J. Appl. Microbiol. 2012, 113, 1139–1144. [Google Scholar] [CrossRef]
- Ali, S.; Charles, T.C.; Glick, B.R. Amelioration of high salinity stress damage by plant growth-promoting bacterial endophytes that contain ACC deaminase. Plant Physiol. Biochem. 2014, 80, 160–167. [Google Scholar] [CrossRef]
- Frew, A.; Öpik, M.; Oja, J.; Vahter, T.; Hiiesalu, I.; Aguilar-Trigueros, C.A. Herbivory-driven shifts in arbuscular mycorrhizal fungal community assembly: Increased fungal competition and plant phosphorus benefits. New Phytol. 2023, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Zhao, J.; Wen, T.; Zhao, M.; Li, R.; Goossens, P.; Huang, Q.; Bai, Y.; Vivanco, J.M.; Kowalchuk, G.A.; et al. Root exudates drive the soil-borne legacy of aboveground pathogen infection. Microbiome 2018, 6, 156. [Google Scholar] [CrossRef]
- D’amelio, R.; Berta, G.; Gamalero, E.; Massa, N.; Avidano, L.; Cantamessa, S.; D’agostino, G.; Bosco, D.; Marzachì, C. Increased plant tolerance against chrysanthemum yellows phytoplasma (‘Candidatus Phytoplasma asteris’) following double inoculation with Glomus mosseae BEG12 and Pseudomonas putida S1Pf1Rif. Plant Pathol. 2011, 60, 1014–1022. [Google Scholar] [CrossRef]
- Ali, S.; Glick, B.R. The biochemistry and molecular biology of the enzyme ACC deaminase. In Microbes: The Foundation Stone of the Biosphere; Hurst, C., Ed.; Springer: Cham, Switzerland, 2021; pp. 365–390. [Google Scholar]
- Glick, B.R.; Gamalero, E. Recent developments in the study of plant microbiomes. Microorganisms 2021, 9, 1533. [Google Scholar] [CrossRef]
- Lucy, M.; Reed, E.; Glick, B.R. Applications of free living plant growth-promoting rhizobacteria. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2004, 86, 1–25. [Google Scholar] [CrossRef]
- Reed, L.; Glick, B.R. The Recent Use of Plant-Growth-Promoting Bacteria to Promote the Growth of Agricultural Food Crops. Agriculture 2023, 13, 1089. [Google Scholar] [CrossRef]
- Upadhyay, S.K.; Srivastava, A.K.; Rajput, V.D.; Chauhan, P.K.; Bhojiya, A.A.; Jain, D.; Chaubey, G.; Dwivedi, P.; Sharma, B.; Minkina, T. Root Exudates: Mechanistic Insight of Plant Growth Promoting Rhizobacteria for Sustainable Crop Production. Front. Microbiol. 2022, 13, 916488. [Google Scholar] [CrossRef]
- Walker, T.S.; Bais, H.P.; Grotewold, E.; Vivanco, J.M. Root exudation and rhizosphere biology. Plant Physiol. 2003, 132, 44–51. [Google Scholar] [CrossRef]
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef] [PubMed]
- Sharma, I.; Kashyap, S.; Agarwala, N. Biotic stress-induced changes in root exudation confer plant stress tolerance by altering rhizospheric microbial community. Front. Plant Sci. 2023, 14, 1132824. [Google Scholar] [CrossRef]
- Zhang, J.; Cook, J.; Nearing, J.T.; Zhang, J.; Raudonis, R.; Glick, B.R.; Langille, M.G.I.; Cheng, Z. Harnessing the plant microbiome to promote the growth of agricultural crops. Microbiol. Res. 2021, 245, 126690. [Google Scholar] [CrossRef]
- Hartmann, A.; Schmid, M.; van Tuinen, D.; Berg, G. Plant-driven selection of microbes. Plant Soil 2009, 321, 235–257. [Google Scholar] [CrossRef]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef]
- Singh, R.K.; Singh, P.; Sharma, A.; Guo, D.-J.; Upadhyay, S.K.; Song, Q.-Q.; Verma, K.K.; Li, D.-P.; Malviya, M.K.; Song, X.-P.; et al. Unraveling Nitrogen Fixing Potential of Endophytic Diazotrophs of Different Saccharum Species for Sustainable Sugarcane Growth. Int. J. Mol. Sci. 2022, 23, 6242. [Google Scholar] [CrossRef] [PubMed]
- Korenblum, E.; Dong, Y.; Szymanski, J.; Panda, S.; Jozwiak, A.; Massalha, H.; Meir, S.; Rogachev, I.; Aharoni, A. Rhizosphere microbiome mediates systemic root metabolite exudation by root-to-root signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 3874–3883. [Google Scholar] [CrossRef] [PubMed]
- Bowya, T.; Balachandar, D. Rhizosphere engineering through exogenous growth-regulating small molecules improves the colonizing efficiency of a plant growth-promoting rhizobacterium in rice. 3 Biotech 2020, 10, 277. [Google Scholar] [CrossRef] [PubMed]
- Chamam, A.; Sanguin, H.; Bellvert, F.; Meiffren, G.; Comte, G.; Wisniewski-Dyé, F.; Bertrand, C.; Prigent-Combaret, C. Plant secondary metabolite profiling evidences strain-dependent effect in the Azospirillum–Oryza sativa association. Phytochemistry 2013, 87, 65–77. [Google Scholar] [CrossRef]
- Coronado, C.; Zuanazzi, J.A.S.; Sallaud, C.; Quirion, J.C.; Esnault, R.; Husson, H.P.; Kondorosi, A.; Ratet, P. Alfalfa root flavonoid production is nitrogen regulated. Plant Physiol. 1995, 108, 533–542. [Google Scholar] [CrossRef]
- Abdel-Lateif, K.; Bogusz, D.; Hocher, V. The role of flavonoids in the establishment of plant roots endosymbioses with arbuscular mycorrhiza fungi, rhizobia and Frankia bacteria. Plant Signal. Behav. 2012, 7, 636–641. [Google Scholar] [CrossRef] [PubMed]
- Szoboszlay, M.; White-Monsant, A.; Moe, L.A. The effect of root exudate 7,4′-dihydroxyflavone and naringenin on soil bacterial community structure. PLoS ONE 2016, 11, e0146555. [Google Scholar] [CrossRef] [PubMed]
- Rudrappa, T.; Czymmek, K.J.; Paré, P.W.; Bais, H.P. Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol. 2008, 148, 1547–1556. [Google Scholar] [CrossRef]
- Akiyama, K.; Matsuzaki, K.-I.; Hayashi, H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 2005, 435, 824–827. [Google Scholar] [CrossRef]
- Cho, S.-M.; Kim, Y.H.; Anderson, A.J.; Kim, Y.C. Nitric oxide and hydrogen peroxide production are involved in systemic drought tolerance induced by 2R,3R-butanediol in Arabidopsis thaliana. Plant Pathol. J. 2013, 29, 427–434. [Google Scholar] [CrossRef]
- Creus, C.M.; Graziano, M.; Casanovas, E.M.; Pereyra, M.A.; Simontacchi, M.; Puntarulo, S.; Barassi, C.A.; Lamattina, L. Nitric oxide is involved in the Azospirillum brasilense-induced lateral root formation in tomato. Planta 2005, 221, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Molina-Favero, C.; Creus, C.M.; Simontacchi, M.; Puntarulo, S.; Lamattina, L. Aerobic nitric oxide production by Azospirillum brasilense Sp245 and its influence on root architecture in tomato. Mol. Plant-Microbe Interact. 2008, 21, 1001–1009. [Google Scholar] [CrossRef] [PubMed]
- Ankati, S.; Podile, A.R. Metabolites in the root exudates of groundnut change during interaction with plant growth promoting rhizobacteria in a strain-specific manner. J. Plant Physiol. 2019, 243, 153057. [Google Scholar] [CrossRef] [PubMed]
- Nguema-Ona, E.; Vicré-Gibouin, M.; Cannesan, M.-A.; Driouich, A. Arabinogalactan proteins in root–microbe interactions. Trends Plant Sci. 2013, 18, 440–449. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Leger, R.J.S. Mrt, a gene unique to fungi, encodes an oligosaccharide transporter and facilitates rhizosphere competency in Metarhizium robertsii. Plant Physiol. 2010, 154, 1549–1557. [Google Scholar] [CrossRef] [PubMed]
- Cai, T.; Cai, W.; Zhang, J.; Zheng, H.; Tsou, A.M.; Xiao, L.; Zhong, Z.; Zhu, J. Host legume-exuded antimetabolites optimize the symbiotic rhizosphere. Mol. Microbiol. 2009, 73, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Müller, D.B.; Vogel, C.; Bai, Y.; Vorholt, J.A. The Plant Microbiota: Systems-Level Insights and Perspectives. Annu. Rev. Genet. 2016, 50, 211–234. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Robert, C.A.M.; Cadot, S.; Zhang, X.; Ye, M.; Li, B.; Manzo, D.; Chervet, N.; Steinger, T.; Van Der Heijden, M.G.A.; et al. Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 2018, 9, 2738. [Google Scholar] [CrossRef] [PubMed]
- Voges, M.J.E.E.E.; Bai, Y.; Schulze-Lefert, P.; Sattely, E.S. Plant-derived coumarins shape the composition of an Arabidopsis synthetic root microbiome. Proc. Natl. Acad. Sci. USA 2019, 116, 12558–12565. [Google Scholar] [CrossRef]
- Badri, D.V.; Chaparro, J.M.; Zhang, R.; Shen, Q.; Vivanco, J.M. Application of natural blends of phytochemicals derived from the root exudates of arabidopsis to the soil reveal that phenolic-related compounds predominantly modulate the soil microbiome. J. Biol. Chem. 2013, 288, 4502–4512. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; He, Y.; Zhang, W.; Chen, L.; Zhang, J.; Zhang, X.; Dawson, W.; Ding, J. Greater chemical signaling in root exudates enhances soil mutualistic associations in invasive plants compared to natives. New Phytol. 2022, 236, 1140–1153. [Google Scholar] [CrossRef] [PubMed]
- Tong, A.-Z.; Liu, W.; Liu, Q.; Xia, G.-Q.; Zhu, J.-Y. Diversity and composition of the Panax ginseng rhizosphere microbiome in various cultivation modesand ages. BMC Microbiol. 2021, 21, 18. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Dai, S.; Wang, B.; Jiang, Y.; Ma, Y.; Pan, L.; Wu, K.; Huang, X.; Zhang, J.; Cai, Z.; et al. Autotoxic ginsenoside disrupts soil fungal microbiomes by stimulating potentially pathogenic microbes. Appl. Environ. Microbiol. 2020, 86, e00130-20. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Yang, M.; Yin, R.; Wang, L.; Luo, L.; Zi, B.; Liu, H.; Huang, H.; Liu, Y.; He, X.; et al. Autotoxin Rg 1 Induces Degradation of Root Cell Walls and Aggravates Root Rot by Modifying the Rhizospheric Microbiome. Microbiol. Spectr. 2021, 9, e0167921. [Google Scholar] [CrossRef]
- Thoms, D.; Liang, Y.; Haney, C.H. Maintaining symbiotic homeostasis: How do plants engage with beneficial microorganisms while at the same time restricting pathogens? Mol. Plant-Microbe Interact. 2021, 34, 462–469. [Google Scholar] [CrossRef]
- Ali, S.; Glick, B.R. Plant-bacterial interactions in management of plant growth under abiotic stresses. In New and Future Developments in Microbial Biotechnology and Bioengineering: Microbes in Soil, Crop and Environmental Sustainability; Elsevier: Amsterdam, The Netherlands, 2019; pp. 21–45. [Google Scholar] [CrossRef]
- Monteiro, F.; Nishimura, M.T. Structural, functional, and genomic diversity of plant NLR proteins: An evolved resource for rational engineering of plant immunity. Annu. Rev. Phytopathol. 2018, 56, 243–267. [Google Scholar] [CrossRef]
- Zipfel, C. Plant pattern-recognition receptors. Trends Immunol. 2014, 35, 345–351. [Google Scholar] [CrossRef]
- Gough, C.; Cullimore, J. Lipo-chitooligosaccharide signaling in endosymbiotic plant-microbe interactions. Mol. Plant-Microbe Interact. 2011, 24, 867–878. [Google Scholar] [CrossRef]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; Wang, Y.; Cai, B.; Zhou, J.-M.; He, S.Y.; Xin, X.-F. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 2021, 592, 105–109. [Google Scholar] [CrossRef]
- Zhou, F.; Emonet, A.; Tendon, V.D.; Marhavy, P.; Wu, D.; Lahaye, T.; Geldner, N. Co-incidence of Damage and Microbial Patterns Controls Localized Immune Responses in Roots. Cell 2020, 180, 440–453.e18. [Google Scholar] [CrossRef]
- Sun, J.; Yang, J.; Zhao, S.; Yu, Q.; Weng, L.; Xiao, C. Root exudates influence rhizosphere fungi and thereby synergistically regulate Panax ginseng yield and quality. Front. Microbiol. 2023, 14, 1194224. [Google Scholar] [CrossRef]
- Reinhold-Hurek, B.; Bünger, W.; Burbano, C.S.; Sabale, M.; Hurek, T. Roots Shaping Their Microbiome: Global Hotspots for Microbial Activity. Annu. Rev. Phytopathol. 2015, 53, 403–424. [Google Scholar] [CrossRef] [PubMed]
- Salem, M.A.; Wang, J.Y.; Al-Babili, S. Metabolomics of plant root exudates: From sample preparation to data analysis. Front. Plant Sci. 2022, 13, 1062982. [Google Scholar] [CrossRef] [PubMed]
- Chaparro, J.M.; Badri, D.V.; Vivanco, J.M. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014, 8, 790–803. [Google Scholar] [CrossRef] [PubMed]
- Pascale, A.; Proietti, S.; Pantelides, I.S.; Stringlis, I.A. Modulation of the Root Microbiome by Plant Molecules: The Basis for Targeted Disease Suppression and Plant Growth Promotion. Front. Plant Sci. 2020, 10, 1741. [Google Scholar] [CrossRef] [PubMed]
- Chaparro, J.M.; Badri, D.V.; Bakker, M.G.; Sugiyama, A.; Manter, D.K.; Vivanco, J.M. Root Exudation of Phytochemicals in Arabidopsis Follows Specific Patterns That Are Developmentally Programmed and Correlate with Soil Microbial Functions. PLoS ONE 2013, 8, e55731. [Google Scholar] [CrossRef]
- Inceoǧlu, O.; Salles, J.F.; van Elsas, J.D. Soil and Cultivar Type Shape the Bacterial Community in the Potato Rhizosphere. Microb. Ecol. 2012, 63, 460–470. [Google Scholar] [CrossRef]
- Ling, N.; Wang, T.; Kuzyakov, Y. Rhizosphere bacteriome structure and functions. Nat. Commun. 2022, 13, 836. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, N.B.; Shaikh, N.; Rochlani, A.; Dalwani, A.; Sharma, S.; Saraf, S.M. Rhizobacteria that Promote Plant Growth and their Impact on Root System Architecture, Root Development, and Function. Acta Sci. Microbiol. 2022, 5, 53–62. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, J.; Rahman, M.K.U.; Gao, D.; Wei, Z.; Wu, F.; Dini-Andreote, F. Interspecific plant interaction via root exudates structures the disease suppressiveness of rhizosphere microbiomes. Mol. Plant 2023, 16, 849–864. [Google Scholar] [CrossRef]
- Rohrbacher, F.; St-Arnaud, M. Root exudation: The ecological driver of hydrocarbon rhizoremediation. Agronomy 2016, 6, 19. [Google Scholar] [CrossRef]
- Canarini, A.; Kaiser, C.; Merchant, A.; Richter, A.; Wanek, W. Root exudation of primary metabolites: Mechanisms and their roles in plant responses to environmental stimuli. Front. Plant Sci. 2019, 10, 157. [Google Scholar] [CrossRef]
- Badri, D.V.; Weir, T.L.; van der Lelie, D.; Vivanco, J.M. Rhizosphere chemical dialogues: Plant–microbe interactions. Curr. Opin. Biotechnol. 2009, 20, 642–650. [Google Scholar] [CrossRef]
- Haldar, S.; Sengupta, S. Plant-microbe cross-talk in the rhizosphere: Insight and biotechnological potential. Open Microbiol. J. 2015, 9, 1–7. [Google Scholar] [CrossRef]
- Etesami, H.; Jeong, B.R.; Glick, B.R. Biocontrol of plant diseases by Bacillus spp. Physiol. Mol. Plant Pathol. 2023, 126, 102048. [Google Scholar] [CrossRef]
- Narayanan, Z.; Glick, B.R. Secondary Metabolites Produced by Plant Growth-Promoting Bacterial Endophytes. Microorganisms 2022, 10, 2008. [Google Scholar] [CrossRef]
- Orozco-Mosqueda, M.D.C.; Fadiji, A.E.; Babalola, O.O.; Glick, B.R.; Santoyo, G. Rhizobiome engineering: Unveiling complex rhizosphere interactions to enhance plant growth and health. Microbiol. Res. 2022, 263, 127137. [Google Scholar] [CrossRef]
- Sehrawat, A.; Sindhu, S.S.; Glick, B.R. Hydrogen cyanide production by soil bacteria: Biological control of pests and promotion of plant growth in sustainable agriculture. Pedosphere 2022, 32, 15–38. [Google Scholar] [CrossRef]
- Akanmu, A.O.; Babalola, O.O.; Venturi, V.; Ayilara, M.S.; Adeleke, B.S.; Amoo, A.E.; Sobowale, A.A.; Fadiji, A.E.; Glick, B.R. Plant Disease Management: Leveraging on the Plant-Microbe-Soil Interface in the Biorational Use of Organic Amendments. Front. Plant Sci. 2021, 12, 700507. [Google Scholar] [CrossRef]
- Santoyo, G.; Guzmán-Guzmán, P.; Parra-Cota, F.I.; de los Santos-Villalobos, S.; Orozco-Mosqueda, M.C.; Glick, B.R. Plant growth stimulation by microbial consortia. Agronomy 2021, 11, 219. [Google Scholar] [CrossRef]
- Lakshmanan, V.; Kitto, S.L.; Caplan, J.L.; Hsueh, Y.-H.; Kearns, D.B.; Wu, Y.-S.; Bais, H.P. Microbe-associated molecular patterns-triggered root responses mediate beneficial Rhizobacterial recruitment in Arabidopsis. Plant Physiol. 2012, 160, 1642–1661. [Google Scholar] [CrossRef]
- Chen, Y.; Yan, F.; Chai, Y.; Liu, H.; Kolter, R.; Losick, R.; Guo, J.-H. Biocontrol of tomato wilt disease by Bacillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation. Environ. Microbiol. 2013, 15, 848–864. [Google Scholar] [CrossRef] [PubMed]
- Dastogeer, K.M.G.; Tumpa, F.H.; Sultana, A.; Akter, M.A.; Chakraborty, A. Plant microbiome—An account of the factors that shape community composition and diversity. Curr. Plant Biol. 2020, 23, 100161. [Google Scholar] [CrossRef]
- Wen, T.; Zhao, M.; Yuan, J.; Kowalchuk, G.A.; Shen, Q. Root exudates mediate plant defense against foliar pathogens by recruiting beneficial microbes. Soil Ecol. Lett. 2021, 3, 42–51. [Google Scholar] [CrossRef]
- Jousset, A.; Rochat, L.; Lanoue, A.; Bonkowski, M.; Keel, C.; Scheu, S.; Lombardi, N.; Vitale, S.; Turrà, D.; Reverberi, M.; et al. Plants respond to pathogen infection by enhancing the antifungal gene expression of root-associated bacteria. Mol. Plant-Microbe Interact. 2011, 24, 352–358. [Google Scholar] [CrossRef]
- Afridi, M.S.; Fakhar, A.; Kumar, A.; Ali, S.; Medeiros, F.H.V.; Muneer, M.A.; Ali, H.; Saleem, M. Harnessing microbial multitrophic interactions for rhizosphere microbiome engineering. Microbiol. Res. 2022, 265, 127199. [Google Scholar] [CrossRef]
- Phour, M.; Sehrawat, A.; Sindhu, S.S.; Glick, B.R. Interkingdom signaling in plant-rhizomicrobiome interactions for sustainable agriculture. Microbiol. Res. 2020, 241, 126589. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Islam, M.S.; Wang, H.; Guo, H.; Wang, Z.; Qi, X.; Zhang, S.; Guo, J.; Wang, Q.; Li, B. Effect of Humic Acid on Soil Physical and Chemical Properties, Microbial Community Structure, and Metabolites of Decline Diseased Bayberry. Int. J. Mol. Sci. 2022, 23, 14707. [Google Scholar] [CrossRef]
- Schulz-Bohm, K.; Gerards, S.; Hundscheid, M.; Melenhorst, J.; De Boer, W.; Garbeva, P. Calling from distance: Attraction of soil bacteria by plant root volatiles. ISME J. 2018, 12, 1252–1262. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Lee, S.; Ryu, C.-M. Foliar aphid feeding recruits rhizosphere bacteria and primes plant immunity against pathogenic and non-pathogenic bacteria in pepper. Ann. Bot. 2012, 110, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Xu, H.; Chu, C.; He, F.; Fang, S. High Temperature can Change Root System Architecture and Intensify Root Interactions of Plant Seedlings. Front. Plant Sci. 2020, 11, 160. [Google Scholar] [CrossRef]
- Cadot, S.; Guan, H.; Bigalke, M.; Walser, J.-C.; Jander, G.; Erb, M.; van der Heijden, M.G.A.; Schlaeppi, K. Specific and conserved patterns of microbiota-structuring by maize benzoxazinoids in the field. Microbiome 2021, 9, 103. [Google Scholar] [CrossRef]
- Degenhardt, J.; Hiltpold, I.; Köllner, T.G.; Frey, M.; Gierl, A.; Gershenzon, J.; Hibbard, B.E.; Ellersieck, M.R.; Turlings, T.C.J. Restoring a maize root signal that attracts insect-killing nematodes to control a major pest. Proc. Natl. Acad. Sci. USA 2009, 106, 13213–13218. [Google Scholar] [CrossRef]
- Rasmann, S.; Köllner, T.G.; Degenhardt, J.; Hiltpold, I.; Toepfer, S.; Kuhlmann, U.; Gershenzon, J.; Turlings, T.C.J. Recruitment of entomopathogenic nematodes by insect-damaged maize roots. Nature 2005, 434, 732–737. [Google Scholar] [CrossRef]
- Dematheis, F.; Zimmerling, U.; Flocco, C.; Kurtz, B.; Vidal, S.; Kropf, S.; Smalla, K. Multitrophic interaction in the rhizosphere of maize: Root feeding of western corn rootworm larvae alters the microbial community composition. PLoS ONE 2012, 7, e37288. [Google Scholar] [CrossRef]
- Zhong, Y.; Xun, W.; Wang, X.; Tian, S.; Zhang, Y.; Li, D.; Zhou, Y.; Qin, Y.; Zhang, B.; Zhao, G.; et al. Root-secreted bitter triterpene modulates the rhizosphere microbiota to improve plant fitness. Nat. Plants 2022, 8, 887–896. [Google Scholar] [CrossRef]
- Ourry, M.; Lebreton, L.; Chaminade, V.; Guillerm-Erckelboudt, A.-Y.; Hervé, M.; Linglin, J.; Marnet, N.; Ourry, A.; Paty, C.; Poinsot, D.; et al. Influence of belowground herbivory on the dynamics of root and rhizosphere microbial communities. Front. Ecol. Evol. 2018, 6, 91. [Google Scholar] [CrossRef]
- Chen, Y.; Bonkowski, M.; Shen, Y.; Griffiths, B.S.; Jiang, Y.; Wang, X.; Sun, B. Root ethylene mediates rhizosphere microbial community reconstruction when chemically detecting cyanide produced by neighbouring plants. Microbiome 2020, 8, 4. [Google Scholar] [CrossRef]
- Mhlongo, M.I.; Piater, L.A.; Dubery, I.A. Profiling of Volatile Organic Compounds from Four Plant Growth-Promoting Rhizobacteria by SPME–GC–MS: A Metabolomics Study. Metabolites 2022, 12, 763. [Google Scholar] [CrossRef]
- Lendzemo, V.W.; Kuyper, T.W.; Matusova, R.; Bouwmeester, H.J.; Van Ast, A. Colonization by arbuscular mycorrhizal fungi of sorghum leads to reduced germination and subsequent attachment and emergence of Striga hermonthica. Plant Signal. Behav. 2007, 2, 58–62. [Google Scholar] [CrossRef]
- Shi, Y.; Pan, Y.; Xiang, L.; Zhu, Z.; Fu, W.; Hao, G.; Geng, Z.; Chen, S.; Li, Y.; Han, D. Assembly of rhizosphere microbial communities in Artemisia annua: Recruitment of plant growth-promoting microorganisms and inter-kingdom interactions between bacteria and fungi. Plant Soil 2022, 470, 127–139. [Google Scholar] [CrossRef]
- Fu, R.; Feng, H.; Dini-Andreote, F.; Wang, Z.; Bo, C.; Cao, W.; Yang, K.; Liu, M.; Yang, T.; Shen, Q.; et al. Modulation of the tomato rhizosphere microbiome via changes in root exudation mediated by the ethylene receptor nr. Microorganisms 2021, 9, 2456. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.G.; Song, G.C.; Sim, H.-J.; Ryu, C.-M. Achieving similar root microbiota composition in neighbouring plants through airborne signalling. ISME J. 2021, 15, 397–408. [Google Scholar] [CrossRef]
- Xu, X.; Fang, P.; Zhang, H.; Chi, C.; Song, L.; Xia, X.; Shi, K.; Zhou, Y.; Zhou, J.; Yu, J. Strigolactones positively regulate defense against root-knot nematodes in tomato. J. Exp. Bot. 2019, 70, 1325–1337. [Google Scholar] [CrossRef] [PubMed]
- Gulati, S.; Ballhausen, M.-B.; Kulkarni, P.; Grosch, R.; Garbeva, P. A non-invasive soil-based setup to study tomato root volatiles released by healthy and infected roots. Sci. Rep. 2020, 10, 12704. [Google Scholar] [CrossRef]
- Ling, N.; Zhang, W.; Wang, D.; Mao, J.; Huang, Q.; Guo, S.; Shen, Q. Root Exudates from Grafted-Root Watermelon Showed a Certain Contribution in Inhibiting Fusarium oxysporum f. sp. niveum. PLoS ONE 2013, 8, e63383. [Google Scholar] [CrossRef] [PubMed]
- Mwangi, R.W.; Mustafa, M.; Charles, K.; Wagara, I.W.; Kappel, N. Selected emerging and reemerging plant pathogens affecting the food basket: A threat to food security. J. Agric. Food Res. 2023, 14, 100827. [Google Scholar] [CrossRef]
- Barnwal, M.K.; Kotasthane, A.; Magculia, N.; Mukherjee, P.K.; Savary, S.; Sharma, A.K.; Singh, H.B.; Singh, U.S.; Sparks, A.H.; Variar, M.; et al. A review on crop losses, epidemiology and disease management of rice brown spot to identify research priorities and knowledge gaps. Eur. J. Plant Pathol. 2013, 136, 443–457. [Google Scholar] [CrossRef]
- Bakker, P.A.H.M.; Pieterse, C.M.J.; de Jonge, R.; Berendsen, R.L. The Soil-Borne Legacy. Cell 2018, 172, 1178–1180. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wei, Z.; Friman, V.-P.; Gu, S.-H.; Wang, X.-F.; Eisenhauer, N.; Yang, T.-J.; Ma, J.; Shen, Q.-R.; Xu, Y.-C.; et al. Probiotic diversity enhances rhizosphere microbiome function and plant disease suppression. mBio 2016, 7, e01790-16. [Google Scholar] [CrossRef] [PubMed]
- Paredes, S.H.; Gao, T.; Law, T.F.; Finkel, O.M.; Mucyn, T.; Teixeira, P.J.P.L.; González, I.S.; Feltcher, M.E.; Powers, M.J.; Shank, E.A.; et al. Design of synthetic bacterial communities for predictable plant phenotypes. PLoS Biol. 2018, 16, e2003962. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Vismans, G.; Yu, K.; Song, Y.; De Jonge, R.; Burgman, W.P.; Burmølle, M.; Herschend, J.; Bakker, P.A.H.M.; Pieterse, C.M.J. Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 2018, 12, 1496–1507. [Google Scholar] [CrossRef]
- Tsolakidou, M.-D.; Stringlis, I.A.; Fanega-Sleziak, N.; Papageorgiou, S.; Tsalakou, A.; Pantelides, I.S. Rhizosphere-enriched microbes as a pool to design synthetic communities for reproducible beneficial outputs. FEMS Microbiol. Ecol. 2019, 95, fiz138. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).