High-Precision, Automatic, and Fast Segmentation Method of Hepatic Vessels and Liver Tumors from CT Images Using a Fusion Decision-Based Stacking Deep Learning Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Cohort and Data Collection
2.1.1. External Dataset
2.1.2. Internal Dataset
2.2. Acquisition, Processing, and Segmentation of Ground-Truth CT Images
2.2.1. CT Image Acquisition
2.2.2. CT Image Processing
2.2.3. Ground Truth Region of Interest Segmentation
2.3. Segmentation Based on Precision Fusion, Model Stacking, and Deep Transfer Learning
2.4. Decision Fusion
2.5. Workflow of the Proposed Deep Transfer Stacking Precision Diffusion Segmentation Model
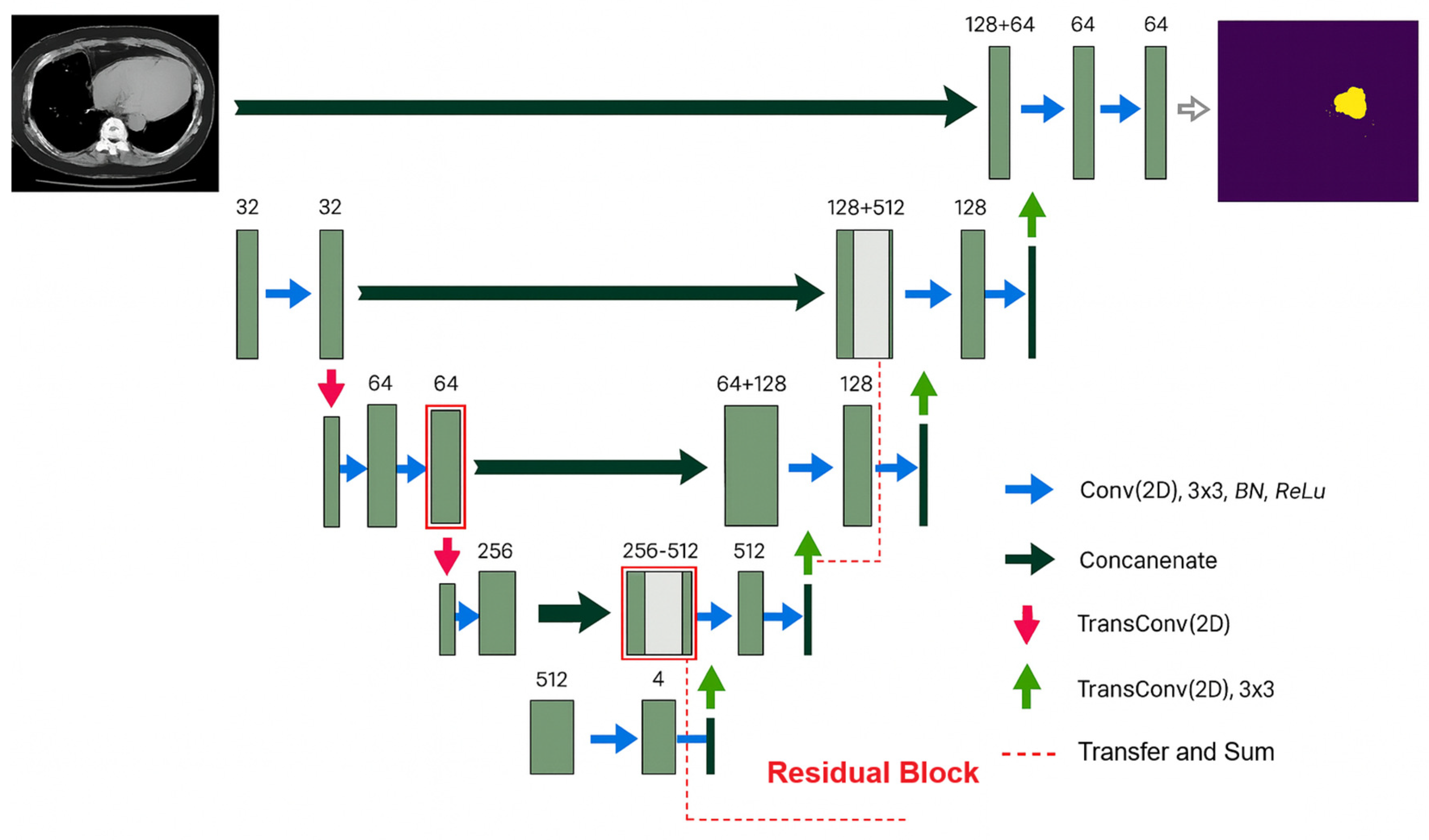
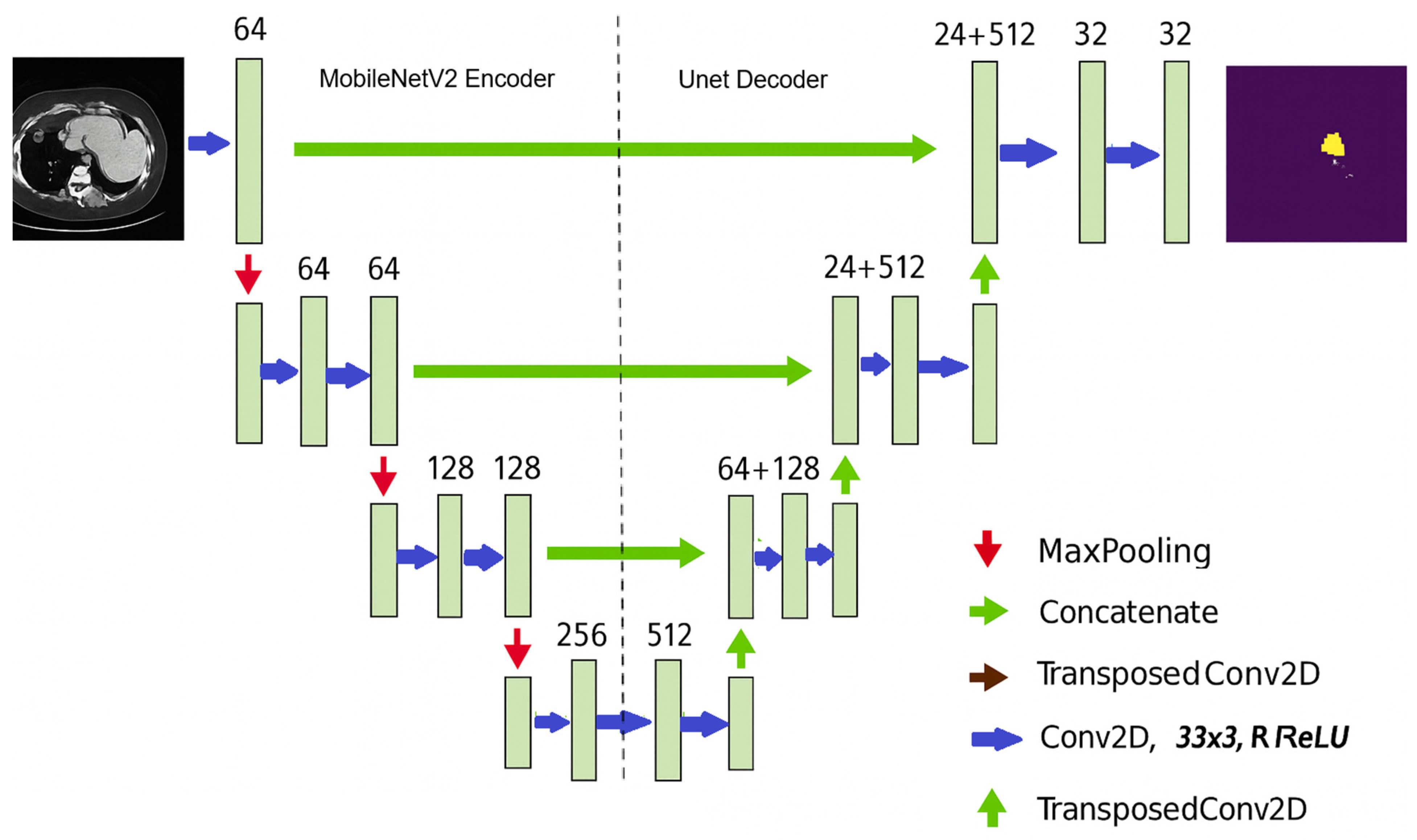
2.6. Stacking Model Architecture
- VGG16/19U-Net: We replaced the classical U-Net encoder with the convolutional blocks of VGG16 and VGG19, which were pretrained on ImageNet. This allowed for the extraction of detailed vascular features while preserving tumor boundaries.
- ResU-Net: We integrated residual blocks into the contracting path to improve gradient flow and mitigate vanishing gradients. This enhancement improved the detection of vessel connectivity.
- MobileNetV2U-Net: We utilized depthwise separable convolutions to reduce computational costs and accelerate inference while maintaining segmentation accuracy. By fine-tuning the pretrained MobileNetV2 encoder on our dataset, we made the model suitable for real-time clinical applications.
- Transfer Learning: For all variants, we initialized encoder layers with pretrained ImageNet weights and fine-tuned them on hepatic CT images. This approach significantly reduced training time and improved convergence.
2.6.1. U-Net Architecture Description
2.6.2. VGGU-Net Architecture
2.6.3. ResU-Net Architecture
2.6.4. MobileNetU-Net Architecture
2.7. Evaluating Model Performance
3. Results
3.1. Training and Testing Data
3.2. Training Process
3.2.1. Simulation Parameters
3.2.2. Loss Function
3.2.3. Segmentation Performance
3.3. Testing Process Performances
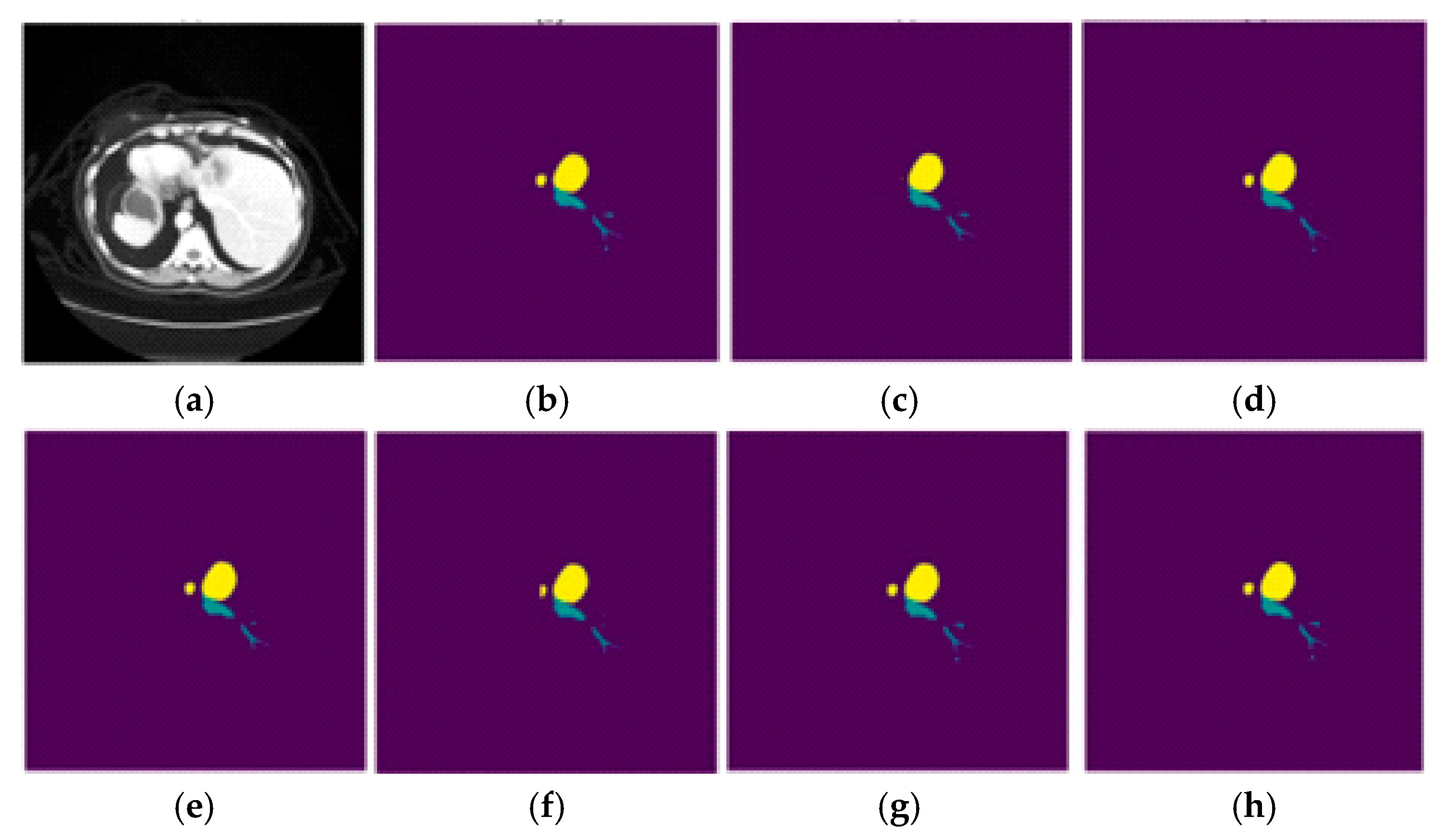
3.4. Visual Representation of the Segmentation
3.5. Benchmarking Vessel and Tumor Liver Segmentation
4. Discussion
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CT | Computed Tomography |
| DSC | Dice Similarity Coefficient |
| JC | Jaccard Coefficient |
| IoU | Intersection and Union |
| INO | National Institute of Oncology |
| MSD | Medical Segmentation Decathlon |
| CNN | Convolutional Neural Network |
References
- Chi, Y.; Liu, J.; Venkatesh, S.K.; Huang, S.; Zhou, J.; Tian, Q.; Nowinski, W.L. Segmentation of liver vasculature from contrast enhanced CT images using context-based voting. IEEE Trans. Biomed. Eng. 2011, 58, 2144–2153. [Google Scholar]
- Lim, K.Y.; Ko, J.E.; Hwang, Y.N.; Lee, S.G.; Kim, S.M. TransRAUNet: A Deep Neural Network with Reverse Attention Module Using HU Windowing Augmentation for Robust Liver Vessel Segmentation in Full Resolution of CT Images. Diagnostics 2025, 15, 118. [Google Scholar] [CrossRef] [PubMed]
- Awais, M.; Al Taie, M.; O’Connor, C.S.; Castelo, A.H.; Acidi, B.; Tran Cao, H.S.; Brock, K.K. Enhancing Surgical Guidance: Deep Learning-Based Liver Vessel Segmentation in Real-Time Ultrasound Video Frames. Cancers 2024, 16, 3674. [Google Scholar] [CrossRef]
- Katagiri, H.; Nitta, H.; Kanno, S.; Umemura, A.; Takeda, D.; Ando, T.; Amano, S.; Sasaki, A. Safety and Feasibility of Laparoscopic Parenchymal-Sparing Hepatectomy for Lesions with Proximity to Major Vessels in Posterosuperior Liver Segments 7 and 8. Cancers 2023, 15, 2078. [Google Scholar] [CrossRef]
- Svobodova, P.; Sethia, K.; Strakos, P.; Varysova, A. Automatic Hepatic Vessels Segmentation Using RORPO Vessel Enhancement Filter and 3D V-Net with Variant Dice Loss Function. Appl. Sci. 2023, 13, 548. [Google Scholar] [CrossRef]
- Brunese, M.C.; Rocca, A.; Santone, A.; Cesarelli, M.; Brunese, L.; Mercaldo, F. Explainable and Robust Deep Learning for Liver Segmentation Through U-Net Network. Diagnostics 2025, 15, 878. [Google Scholar] [CrossRef]
- Prencipe, B.; Altini, N.; Cascarano, G.D.; Brunetti, A.; Guerriero, A.; Bevilacqua, V. Focal Dice Loss-Based V-Net for Liver Segments Classification. Appl. Sci. 2022, 12, 3247. [Google Scholar] [CrossRef]
- Deshpande, R.R.; Heaton, N.D.; Rela, M. Surgical anatomy of segmental liver transplantation. Br. J. Surg. 2002, 89, 1078–1088. [Google Scholar] [CrossRef]
- Montgomery, A.E.; Rana, A. Current state of artificial intelligence in liver transplantation. Transplant. Rep. 2025, 10, 100173. [Google Scholar] [CrossRef]
- Zimmermann, C.; Michelmann, A.; Daniel, Y.; Enderle, M.D.; Salkic, N.; Linzenbold, W. Application of Deep Learning for Real-Time Ablation Zone Measurement in Ultrasound Imaging. Cancers 2024, 16, 1700. [Google Scholar] [CrossRef]
- Ahn, J.C.; Qureshi, T.A.; Singal, A.G.; Li, D.; Yang, J.D. Deep learning in hepatocellular carcinoma: Current status and future perspectives. World J. Hepatol. 2021, 13, 2039–2051. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Aravazhi, P.S.; Gunasekaran, P.; Benjamin, N.Z.Y.; Thai, A.; Chandrasekar, K.K.; Kolanu, N.D.; Prajjwal, P.; Tekuru, Y.; Brito, L.V.; Inban, P. The integration of artificial intelligence into clinical medicine: Trends, challenges, and future directions. Disease-a-Month 2025, 71, 101882. [Google Scholar] [CrossRef] [PubMed]
- Ciecholewski, M.; Kassjański, M. Computational Methods for Liver Vessel Segmentation in Medical Imaging: A Review. Sensors 2021, 21, 2027. [Google Scholar] [CrossRef] [PubMed]
- Jenssen, H.B.; Nainamalai, V.; Pelanis, E.; Kumar, R.P.; Abildgaard, A.; Kolrud, F.K.; Edwin, B.; Jiang, J.; Vettukattil, J.; Elle, O.J.; et al. Challenges and artificial intelligence solutions for clinically optimal hepatic venous vessel segmentation. Biomed. Signal Process. Contro. 2025, 106, 107822. [Google Scholar] [CrossRef]
- Zhao, Z.; Li, W.; Ding, X.; Sun, J.; Xu, L.X. TTGA U-Net: Two-stage two-stream graph attention U-Net for hepatic vessel connectivity enhancement. Comput. Med Imaging Graph. 2025, 122, 102514. [Google Scholar] [CrossRef]
- Affane, A.; Chetoui, M.A.; Lamy, J.; Lienemann, G.; Peron, R.; Beaurepaire, P.; Dollé, G.; Lebre, M.-A.; Magnin, B.; Merveille, O.; et al. The R-Vessel-X Project. IRBM 2025, 46, 100876. [Google Scholar] [CrossRef]
- Yang, J.; Fu, M.; Hu, Y. Liver vessel segmentation based on inter-scale V-Net. Math. Biosci. Eng. 2021, 18, 4327–4340. [Google Scholar] [CrossRef]
- Alirr, O.I.; Rahni, A.A.A. Hepatic vessels segmentation using deep learning and preprocessing enhancement. J. Appl. Clin. Med Phys. 2023, 24, e13966. [Google Scholar] [CrossRef]
- Yu, W.; Fang, B.; Liu, Y.; Gao, M.; Zheng, S.; Wang, Y. Liver vessels segmentation based on 3D Residual U-Net in ICIP. In Proceedings of the 2019 IEEE International Conference on Image Processing (ICIP), Taipei, Taiwan, 22–25 September 2019; IEEE: Piscataway, NJ, USA, 2019. [Google Scholar]
- Thomson, B.R.; Nijkamp, J.; Ivashchenko, O.; van der Heijden, F.; Smit, J.N.; Kok, N.F.; Kuhlmann, K.F.D.; Ruers, T.J.M.; Fusaglia, M. Hepatic vessel segmentation using a reduced filter 3D U-Net in Ultrasound imaging in MIDL 2019 Medical Imaging with Deep Learning. arXiv 2019, arXiv:1907.12109. [Google Scholar]
- Liu, F.; Zheng, Q.; Tian, X.; Shu, F.; Jiang, W.; Wang, M.; Elhanashi, A.; Saponara, S. Rethinking the multi-scale feature hierarchy in object detection transformer (DETR). Appl. Soft Comput. 2025, 175, 113081. [Google Scholar] [CrossRef]
- Zhao, L.; Li, J.; Xu, X.; Zhu, C.; Cheng, W.; Liu, S.; Zhao, M.; Zhang, L.; Zhang, J.; Yin, J.; et al. A Deep Learning-Based Ocular Structure Segmentation for Assisted Myasthenia Gravis Diagnosis from Facial Images. Tsinghua Sci. Technol. 2025, 30, 2592–2605. [Google Scholar] [CrossRef]
- Jiang, Z.; Cheng, D.; Qin, Z.; Gao, J.; Lao, Q.; Ismoilovich, A.B.; Gayrat, U.; Elyorbek, Y.; Habibullo, B.; Tang, D.; et al. TV-SAM: Increasing Zero-Shot Segmentation Performance on Multimodal Medical Images Using GPT-4 Generated Descriptive Prompts Without Human Annotation. Big Data Min. Anal. 2024, 7, 1199–1211. [Google Scholar] [CrossRef]
- Antonelli, M.; Reinke, A.; Bakas, S.; Farahani, K.; Kopp-Schneider, A.; Landman, B.A.; Litjens, G.; Menze, B.; Ronneberger, O.; Summers, R.M.; et al. The Medical Segmentation Decathlon. Nat. Commun. 2022, 13, 4128. [Google Scholar] [CrossRef]
- Roy, S.; Carass, A.; Bazin, P.-L.; Prince, J.L.; Dawant, B.M.; Haynor, D.R. Intensity Inhomogeneity Correction of Magnetic Resonance Images using Patches. In Proceedings of the 2011 SPIE Medical Imaging, Lake Buena Vista, FL, USA, 14–16 February 2011; Volume 7962, p. 79621F. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Masoudi, S.; Harmon, S.A.; Mehralivand, S.; Walker, S.M.; Raviprakash, H.; Bagci, U.; Choyke, P.L.; Turkbey, B. Quick guide on radiology image pre-processing for deep learning applications in prostate cancer research. J. Med. Imaging 2021, 8, 010901. [Google Scholar] [CrossRef]
- Benfares, A.; Mourabiti, A.Y.; Alami, B.; Boukansa, S.; El Bouardi, N.; Lamrani, M.Y.A.; El Fatimi, H.; Amara, B.; Serraj, M.; Mohammed, S.; et al. Non-invasive, fast, and high-performance EGFR gene mutation prediction method based on deep transfer learning and model stacking for patients with Non-Small Cell Lung Cancer. Eur. J. Radiol. Open 2024, 13, 100601. [Google Scholar] [CrossRef]
- Yushkevich, P.A.; Gao, Y.; Gerig, G. ITK-SNAP: An interactive tool for semi-automatic segmentation of multi-modality biomedical images. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 16–20 August 2016; pp. 3342–3345. [Google Scholar] [CrossRef]
- Egger, J.; Kapur, T.; Fedorov, A.; Pieper, S.; Miller, J.V.; Veeraraghavan, H.; Freisleben, B.; Golby, A.J.; Nimsky, C.; Kikinis, R. GBM volumetry using the 3D slicer-medical image computing platform. Sci. Rep. 2013, 3, 1364. [Google Scholar] [CrossRef]
- Velazquez, E.R.; Parmar, C.; Jermoumi, M.; Mak, R.H.; van Baardwijk, A.; Fennessy, F.M.; Lewis, J.H.; De Ruysscher, D.; Kikinis, R.; Lambin, P.; et al. Volumetric CT-based segmentation of NSCLC using 3D-Slicer. Sci. Rep. 2013, 3, 3529. [Google Scholar] [CrossRef]
- Benfares, A.; Mourabiti, A.y.; Alami, B.; Boukansa, S.; Benomar, I.; El Bouardi, N.; Alaoui Lamrani, M.Y.; El Fatimi, H.; Amara, B.; Serraj, M.; et al. Nomogram Based on the Most Relevant Clinical, CT, and Radiomic Features, and a Machine Learning Model to Predict EGFR Mutation Status in Non-Small Cell Lung Cancer. J. Respir. 2025, 5, 11. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Yang, L.; Zhang, Z.; Cai, X.; Dai, T. Attention-Based Personalized Encoder-Decoder Model for Local Citation Recommendation. Comput. Intell. Neurosci. 2019, 2019, 1232581. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the 3rd International Conference on Learning Representations (ICLR 2015), San Diego, CA, USA, 7–9 May 2015; Computational and Biological Learning Society: Leesburg, Virginia, 2015; pp. 1–14. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 4510–4520. [Google Scholar]
- Mangai, U.; Samanta, S.; Das, S.; Chowdhury, P.; Ug, M. A survey of decision fusion and feature fusion strategies for pattern classification. IETE Tech. Rev. 2010, 27, 293–307. [Google Scholar] [CrossRef]
- Giannakakis, G.; Pediaditis, M.; Manousos, D.; Kazantzaki, E.; Chiarugi, F.; Simos, P.; Marias, K.; Tsiknakis, M. Stress and anxiety detection using facial cues from videos. Biomed. Signal Process. Control 2017, 31, 89–101. [Google Scholar] [CrossRef]
- Zhou, H.; Geng, Z.; Sun, M.; Wu, L.; Yan, H. Context-Guided SAR Ship Detection with Prototype-Based Model Pretraining and Check–Balance-Based Decision Fusion. Sensor 2025, 25, 4938. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, Q.; Wang, Y. Road Extraction by Deep Residual U-Net. IEEE Geosci. Remote Sens. Lett. 2017, 15, 749–753. [Google Scholar] [CrossRef]
- Kittler, J.; Hatef, M.; Duin, R.P.; Matas, J. On combining classifiers. IEEE Trans. Pattern Anal. Mach. Intell. 1998, 20, 226–239. [Google Scholar] [CrossRef]
- Yang, J.; Lu, Y.; Zhang, Z.; Wei, J.; Shang, J.; Wei, C.; Tang, W.; Chen, J. A Deep Learning Method Coupling a Channel Attention Mechanism and Weighted Dice Loss Function for Water Extraction in the Yellow River Basin. Water 2025, 17, 478. [Google Scholar] [CrossRef]
- Ibragimov, B.; Toesca, D.; Chang, D.; Koong, A.; Xing, L. Combining deep learning with anatomical analysis for segmentation of the portal vein for liver SBRT planning. Phys. Med. Biol. 2017, 62, 8943. [Google Scholar] [CrossRef]
- Zhang, R.; Zhou, Z.; Wu, W.; Lin, C.C.; Tsui, P.H.; Wu, S. An improved fuzzy connectedness method for automatic three-dimensional liver vessel segmentation in CT images. J. Healthc. Eng. 2018, 2018, 2376317. [Google Scholar] [CrossRef]
- Huang, Q.; Sun, J.; Ding, H.; Wang, X.; Wang, G. Robust liver vessel extraction using 3D U-Net with variant dice loss function. Comput. Biol. Med. 2018, 101, 153–162. [Google Scholar] [CrossRef]
- Thomson, B.R.; Smit, J.N.; Ivashchenko, O.V.; Kok, N.F.; Kuhlmann, K.F.; Ruers, T.J.; Fusaglia, M. MR-to-US Registration Using Multiclass Segmentation of Hepatic Vasculature with a Reduced 3D U-Net. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; pp. 275–284. [Google Scholar]
- Yan, Q.; Wang, B.; Zhang, W.; Luo, C.; Xu, W.; Xu, Z.; Zhang, Y.; Shi, Q.; Zhang, L.; You, Z. An Attention-guided Deep Neural Network with Multi-scale Feature Fusion for Liver Vessel Segmentation. IEEE J. Biomed. Health Inf. 2020, 25, 2629–2642. [Google Scholar] [CrossRef]
- Xu, M.; Wang, Y.; Chi, Y.; Hua, X. Training liver vessel segmentation deep neural networks on noisy labels from contrast CT imaging. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 1552–1555. [Google Scholar]
- Su, J.; Liu, Z.; Zhang, J.; Sheng, V.S.; Song, Y.; Zhu, Y.; Liu, Y. DV-Net: Accurate liver vessel segmentation via dense connection model with D-BCE loss function. Knowl.-Based Syst. 2021, 232, 107471. [Google Scholar] [CrossRef]
- Soler, L.; Hostettler, A.; Agnus, V.; Charnoz, A.; Fasquel, J.; Moreau, J.; Osswald, A.; Bouhadjar, M.; Marescaux, J. 3D Image Reconstruction for Comparison of Algorithm Database: A Patient Specific Anatomical and Medical Image Database. 2010. Available online: https://www-sop.inria.fr/geometrica/events/wam/abstract-ircad.pdf (accessed on 20 October 2024).



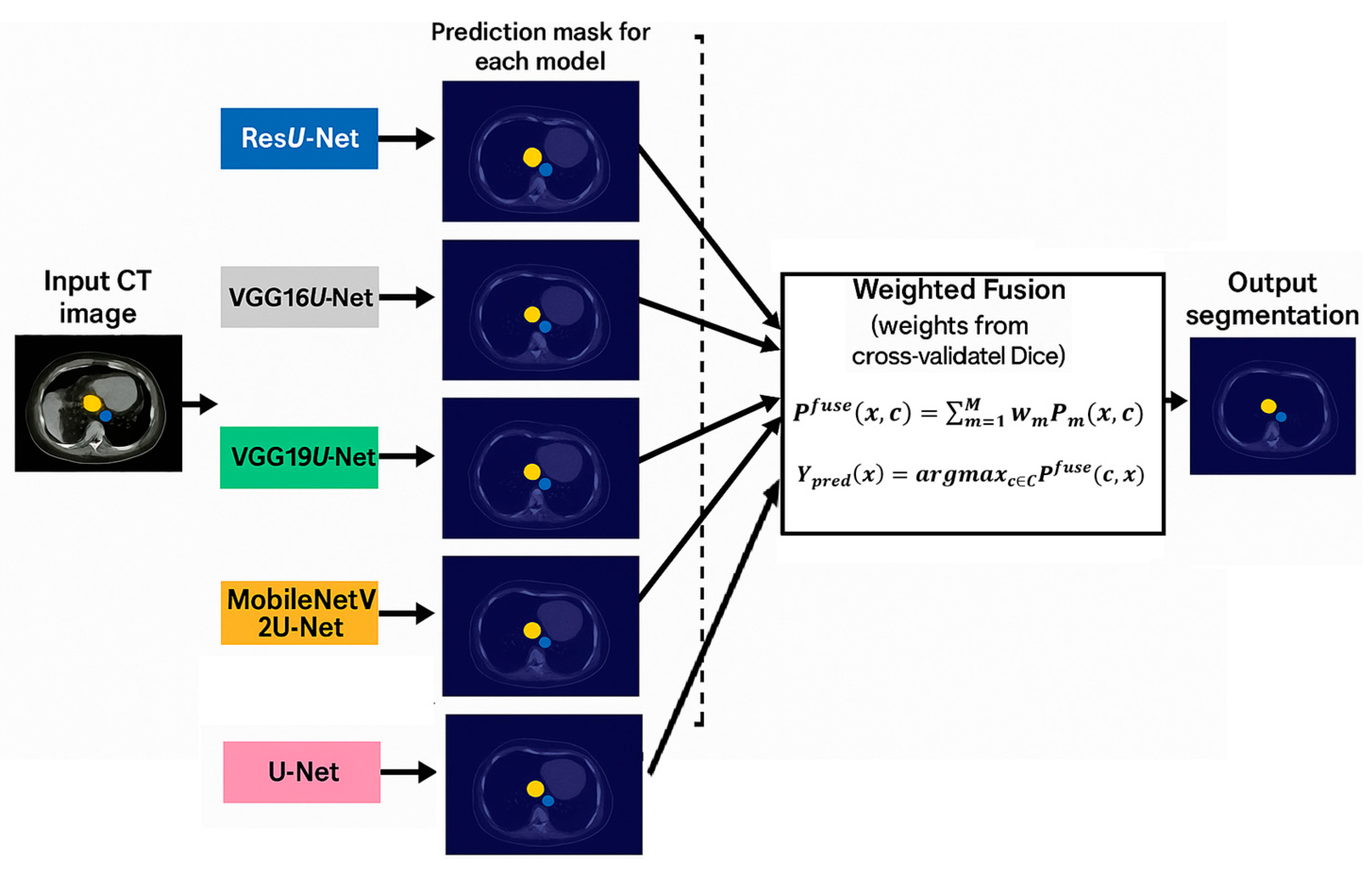
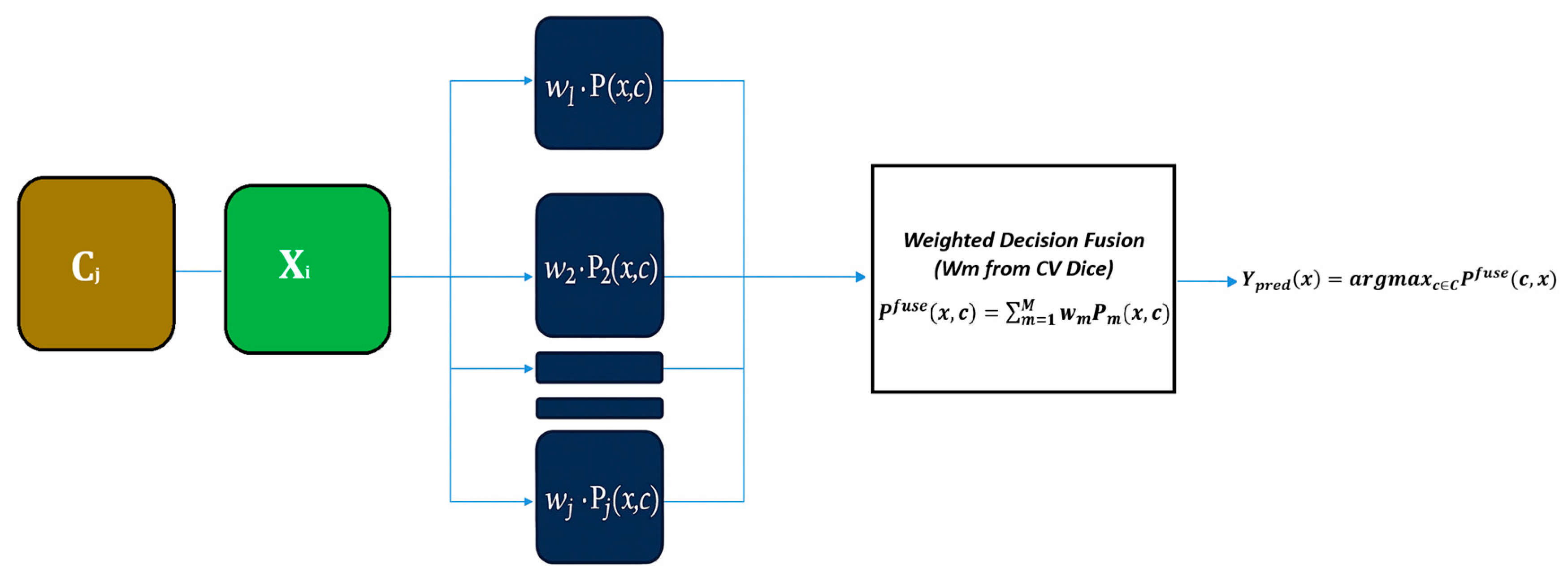
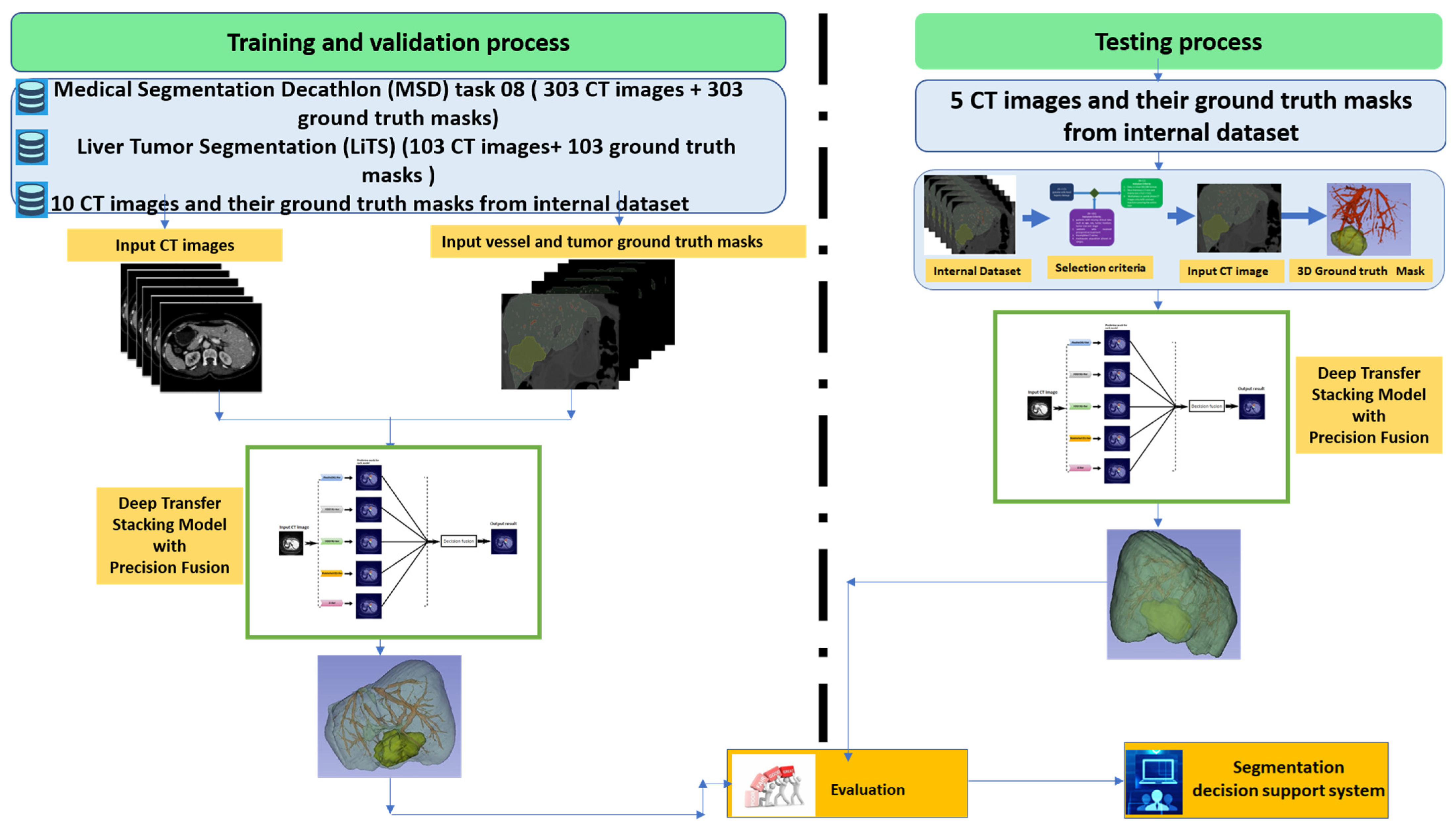

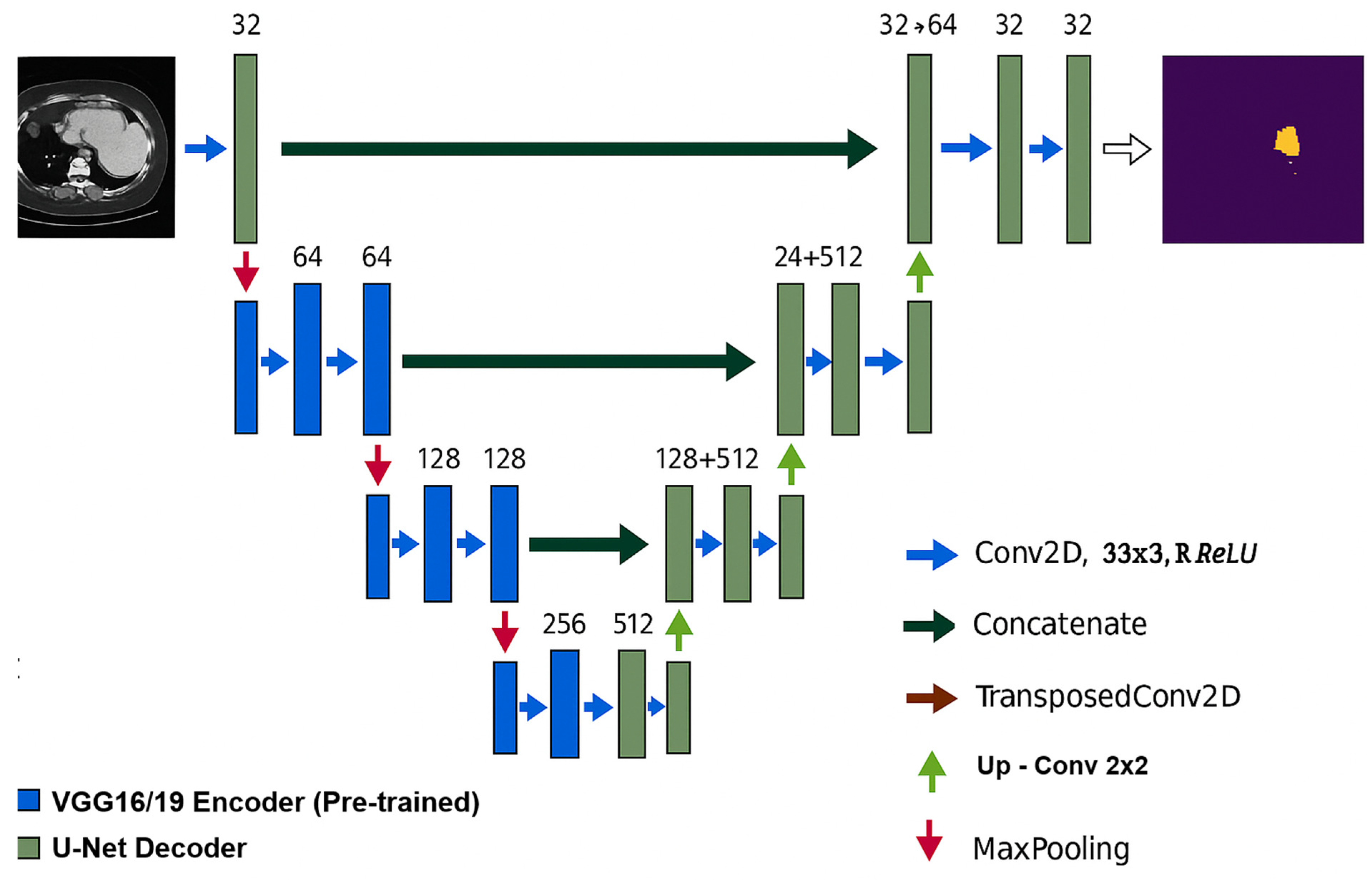



| Step | Description |
|---|---|
| Inputs | Training dataset train = {()} Base learners = {U-Net, VGG16U-Net, VGG19U-Net, ResU-Net, MobileNetV2U-Net} Classes = {background, vessel, tumor} |
| 1. Preprocessing | Convert CT (DICOM) → Hounsfield Units Normalize intensities, inhomogeneity correction Alignment (Elastix), resize to 226 × 226 z-score normalization, optional CLAHE |
| 2. Cross-Validated Training (K = 5) | For each model m:
|
| 3. Weight Estimation | Compute weights: with Ensure |
| 4. Inference (Per New CT) | Preprocess input (Step 1) Run each model m → probability map |
| 5. Weighted Fusion | For each pixel x and class c: Final label: |
| 6. Postprocessing | 3D connected-component filtering (min size) Morphological closing of vessel gaps Topology-aware pruning to preserve thin vessels |
| Parameter | Value/Strategy | Rationale |
|---|---|---|
| Optimizer | Adam | Widely used for medical image segmentation; balances convergence and stability |
| Initial Learning Rate | 0.001 | Standard starting point for fine-tuning CNNs |
| Learning Rate Schedule | Reduce by factor of 0.1 after 5 epochs without improvement | Prevents stagnation and supports stable convergence |
| Batch Size | 8 | Compromise between GPU memory (4× GTX 1080 Ti) and stable gradients |
| Dropout Rate | 0.5 (decoder layers) | Prevents overfitting and improve generalization |
| Normalization | Batch normalization (all conv. blocks) | Stabilizes training and accelerates convergence |
| Early Stopping | Patience of 10 epochs | Prevents overfitting and unnecessary computation |
| Epochs (Maximum) | 200 | Ensures convergence with safeguard by early stopping |
| Loss Function | Dice + cross-entropy (Equations (6)–(8)) | Balances overlap accuracy (Dice) and voxel-wise classification (CE) |
| Data Augmentation | Random rotation, zoom, translation, flipping | Improves robustness to acquisition variability |
| Model | Vessel and Tumor Liver Segmentation | |
|---|---|---|
| U-Net | 65.82 ± 0.08 | 49.05 ± 0.09 |
| VGG16U-Net | 66.66 ± 0.04 | 50.00 ± 0.05 |
| VGG19U-Net | 67.53 ± 0.03 | 50.98 ± 0.03 |
| ResU-Net | 71.58 ± 0.07 | 55.73 ± 0.08 |
| MobileNetV2U-Net | 73.64 ± 0.03 | 58.28 ± 0.04 |
| Proposed Model | 84.67 ± 0.05 | 73.41 ± 0.07 |
| Model | Vessel and Tumor Liver Segmentation | |
|---|---|---|
| U-Net | 60.87 ± 0.07 | 43.74 ± 0.07 |
| VGG16U-Net | 61.95 ± 0.02 | 64.38 ± 0.09 |
| VGG19U-Net | 65.72 ± 0.04 | 44.87 ± 0.02 |
| ResU-Net | 69.85 ± 0.06 | 53.68 ± 0.07 |
| MobileNetV2U-Net | 71.91 ± 0.04 | 56.14 ± 0.05 |
| Proposed Model | 83.21 ± 0.03 | 72.76 ± 0.04 |
| Author | DSC (%) |
|---|---|
| Ibragimov et al. [42] | 83.01 |
| Zhang et al. [43] | 67.12 |
| Huang et al. [44] | 75.45 |
| Thomson et al. [45] | 66.25 |
| Yan et al. [46] | 80.19 |
| Xu et al. [47] | 68.08 |
| Yu et al. 2019 [19] | 74.56 |
| Best DSC in DSM Decathlon [24] | 71.42 |
| V-net (Su et al.) [48] | 75.46 |
| Proposed Model | 83.21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qjidaa, M.; Benfares, A.; El Azami El Hassani, M.A.; Benkabbou, A.; Souadka, A.; Majbar, A.; El Moatassim, Z.; Oumlaz, M.; Lahnaoui, O.; Mouhcine, R.; et al. High-Precision, Automatic, and Fast Segmentation Method of Hepatic Vessels and Liver Tumors from CT Images Using a Fusion Decision-Based Stacking Deep Learning Model. BioMedInformatics 2025, 5, 53. https://doi.org/10.3390/biomedinformatics5030053
Qjidaa M, Benfares A, El Azami El Hassani MA, Benkabbou A, Souadka A, Majbar A, El Moatassim Z, Oumlaz M, Lahnaoui O, Mouhcine R, et al. High-Precision, Automatic, and Fast Segmentation Method of Hepatic Vessels and Liver Tumors from CT Images Using a Fusion Decision-Based Stacking Deep Learning Model. BioMedInformatics. 2025; 5(3):53. https://doi.org/10.3390/biomedinformatics5030053
Chicago/Turabian StyleQjidaa, Mamoun, Anass Benfares, Mohammed Amine El Azami El Hassani, Amine Benkabbou, Amine Souadka, Anass Majbar, Zakaria El Moatassim, Maroua Oumlaz, Oumayma Lahnaoui, Raouf Mouhcine, and et al. 2025. "High-Precision, Automatic, and Fast Segmentation Method of Hepatic Vessels and Liver Tumors from CT Images Using a Fusion Decision-Based Stacking Deep Learning Model" BioMedInformatics 5, no. 3: 53. https://doi.org/10.3390/biomedinformatics5030053
APA StyleQjidaa, M., Benfares, A., El Azami El Hassani, M. A., Benkabbou, A., Souadka, A., Majbar, A., El Moatassim, Z., Oumlaz, M., Lahnaoui, O., Mouhcine, R., Lakhssassi, A., & Cherkaoui, A. (2025). High-Precision, Automatic, and Fast Segmentation Method of Hepatic Vessels and Liver Tumors from CT Images Using a Fusion Decision-Based Stacking Deep Learning Model. BioMedInformatics, 5(3), 53. https://doi.org/10.3390/biomedinformatics5030053






