A Tutorial Toolbox to Simplify Bioinformatics and Biostatistics Analyses of Microbial Omics Data in an Island Context
Abstract
1. Introduction
2. Results
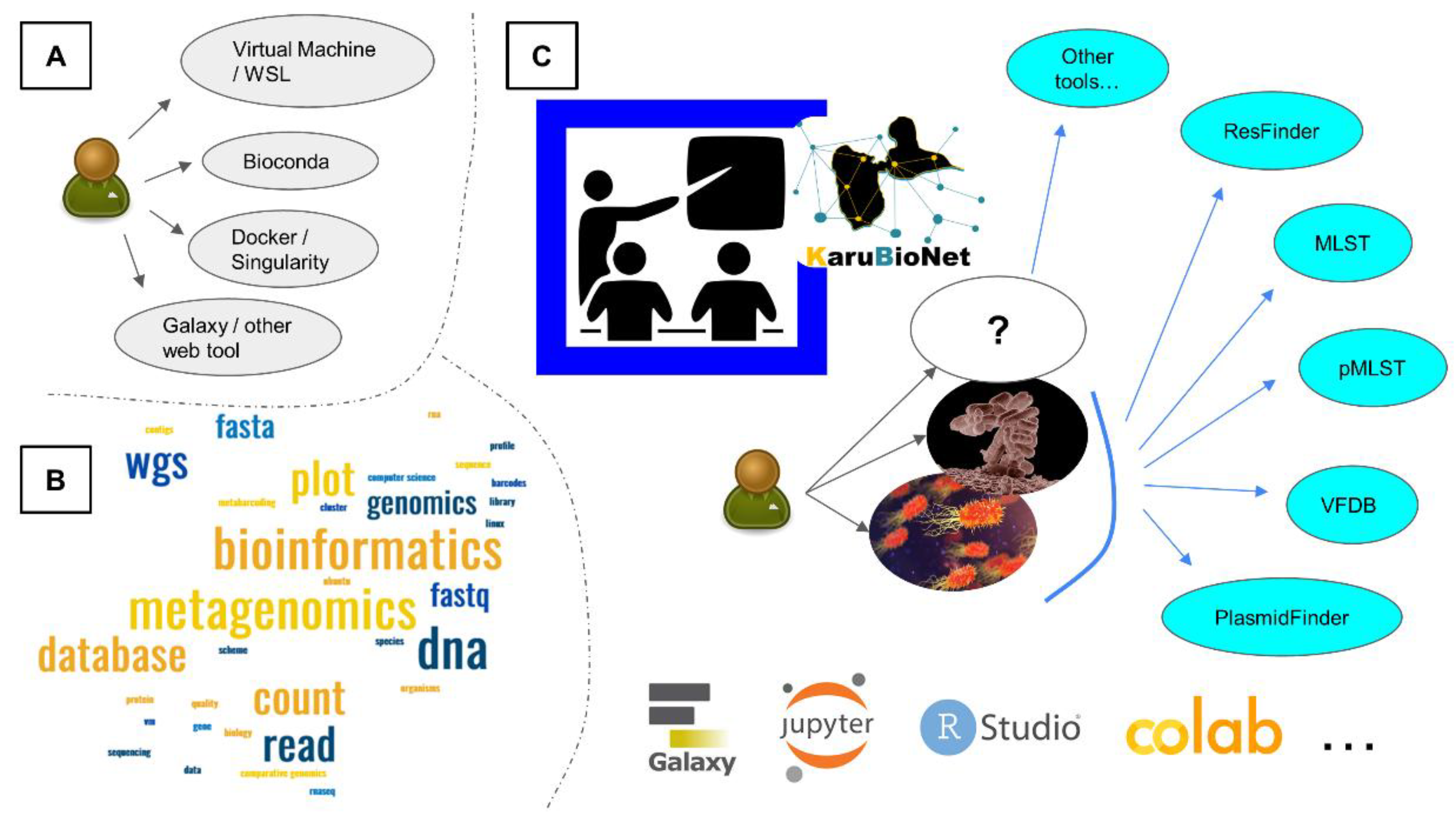
2.1. Trained Bioinformatics Users
2.1.1. High Performance Computing in Bioinformatics
2.1.2. Programming Languages
2.1.3. Virtual Machines and Windows Subsystem for Linux
2.1.4. Jupyter Notebook
2.1.5. Containers and Package Managers
2.2. Untrained Bioinformatics Users
2.2.1. Galaxy Web Interface
2.2.2. EPI2ME
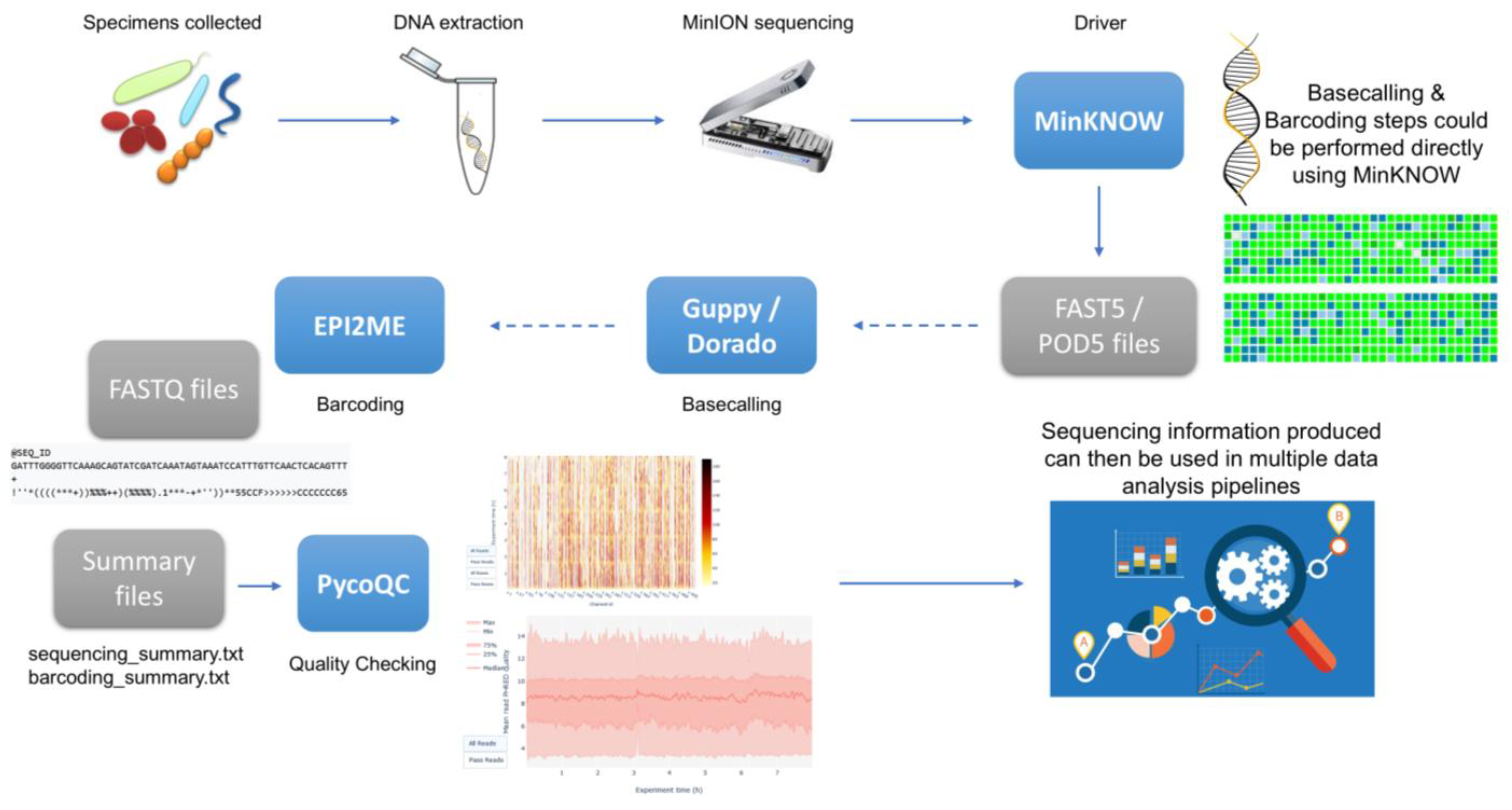
2.3. Genomics and Metagenomics Workflows: Example of MinION Sequencing
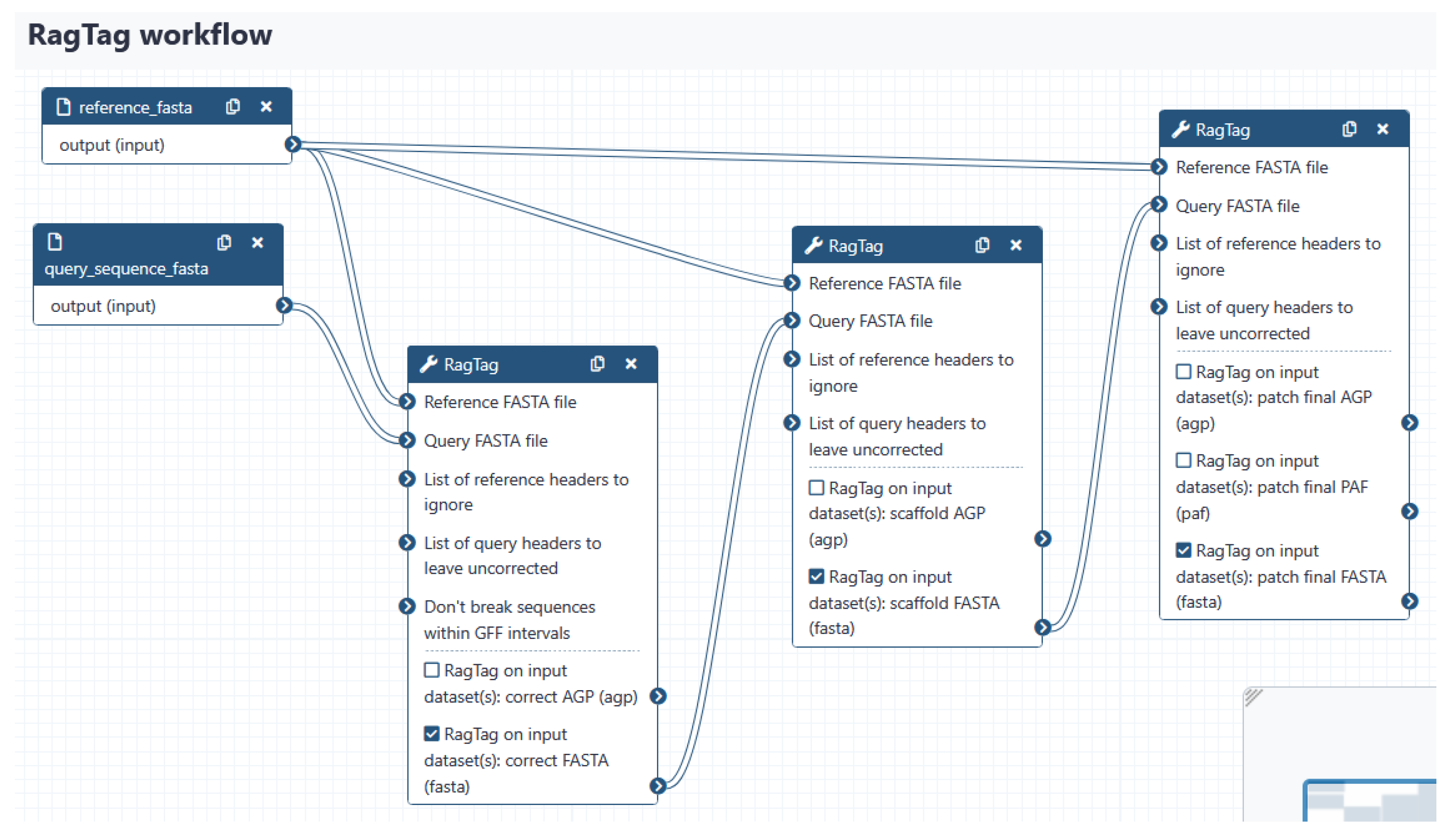
2.3.1. Genome De Novo Assembly, Scaffolding, and Annotation
2.3.2. Basic Local Alignment Search Tool (BLAST)
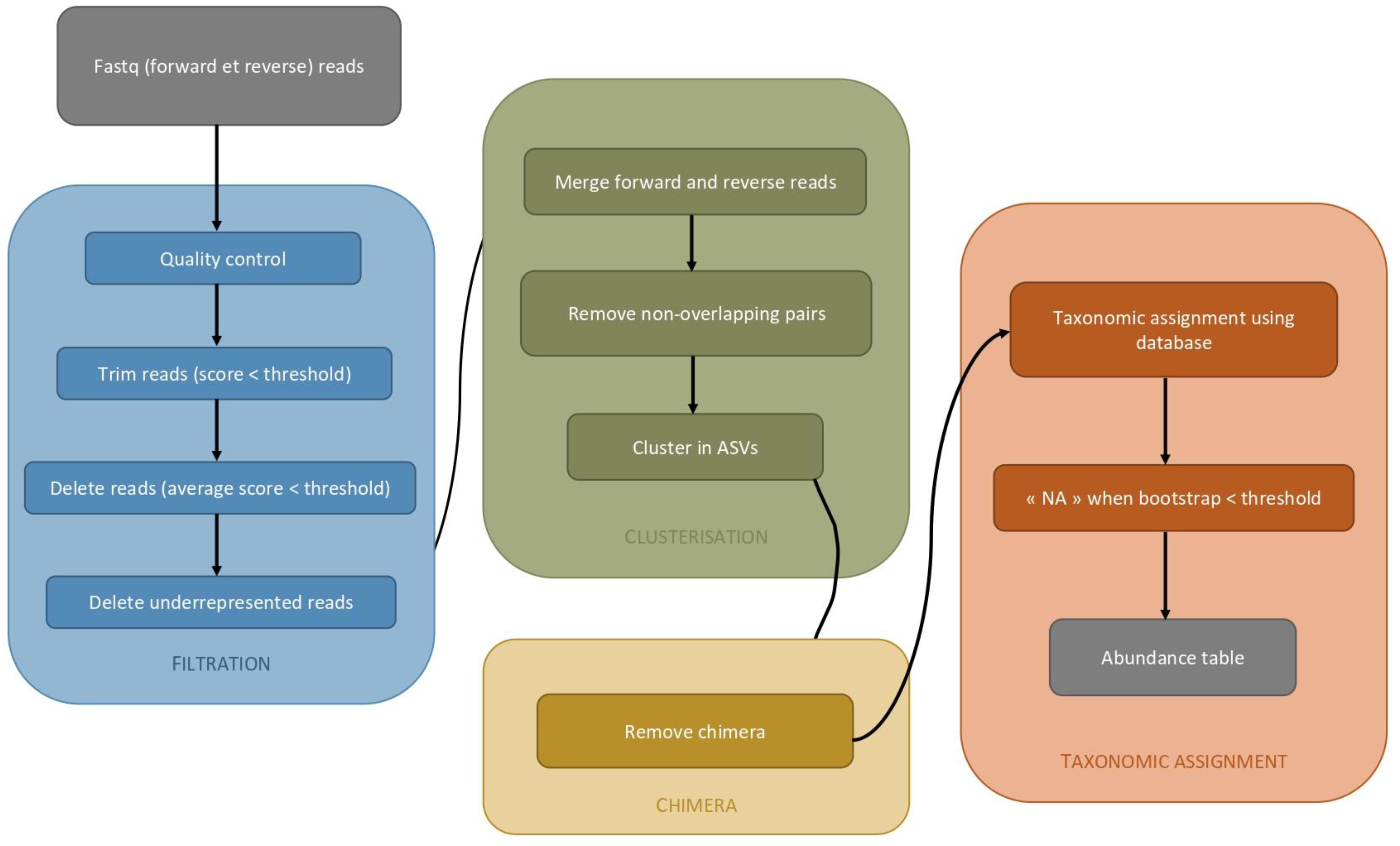
2.3.3. Metabarcoding Analysis with DADA2, Phyloseq, and Vegan
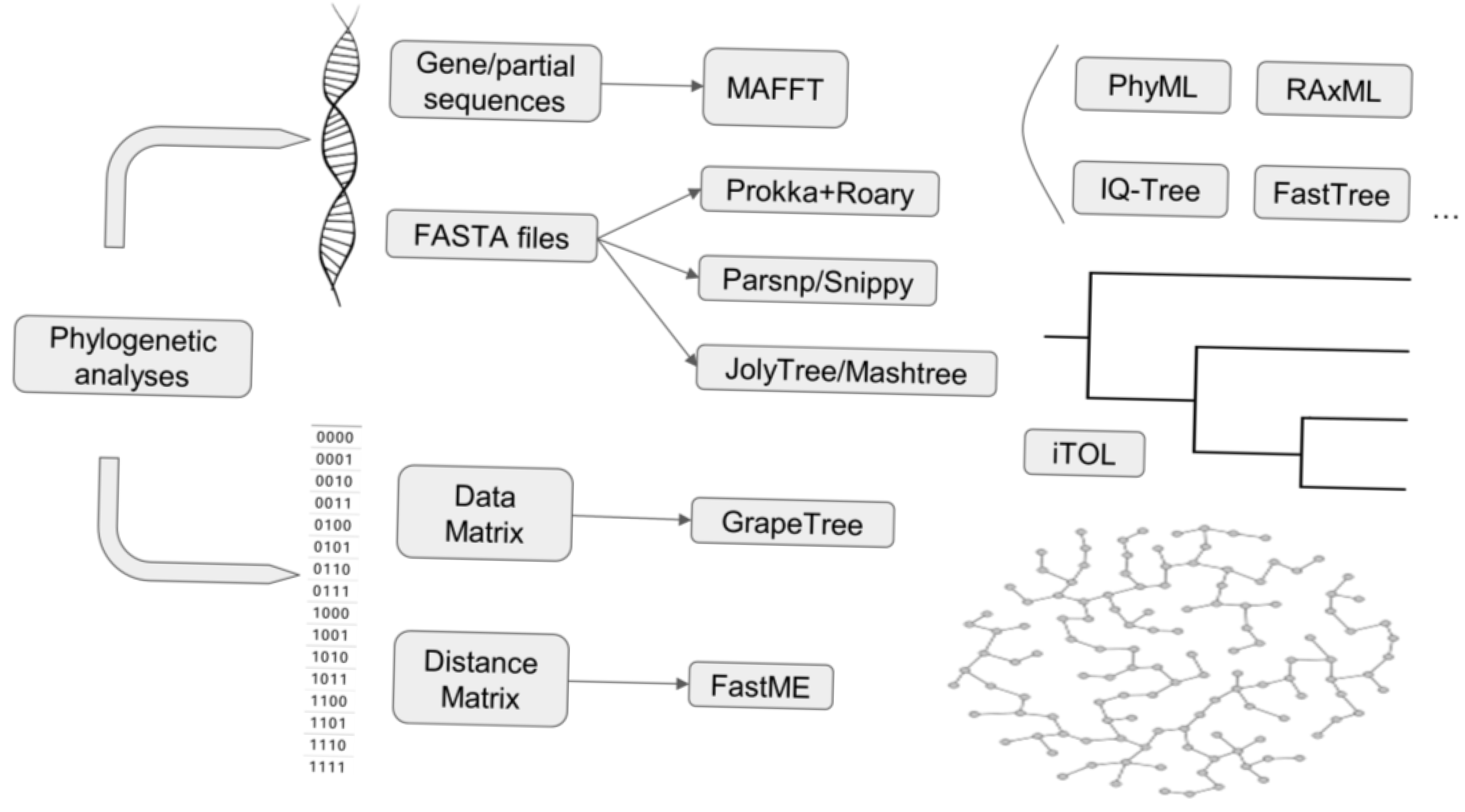
2.3.4. Core Genome/SNP and Other Phylogenomic Analyses
2.4. Reproducibility of Pipelines and Workflow Management Systems
2.5. Limitations of Proposed Workflows and Tools
3. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pot, M.; Reynaud, Y.; Couvin, D.; Dereeper, A.; Ferdinand, S.; Bastian, S.; Foucan, T.; Pommier, J.-D.; Valette, M.; Talarmin, A.; et al. Emergence of a Novel Lineage and Wide Spread of a blaCTX-M-15/IncHI2/ST1 Plasmid among Nosocomial Enterobacter in Guadeloupe. Antibiotics 2022, 11, 1443. [Google Scholar] [CrossRef] [PubMed]
- Dereeper, A.; Gruel, G.; Pot, M.; Couvin, D.; Barbier, E.; Bastian, S.; Bambou, J.-C.; Gelu-Simeon, M.; Ferdinand, S.; Guyomard-Rabenirina, S.; et al. Limited Transmission of Klebsiella pneumoniae among Humans, Animals, and the Environment in a Caribbean Island, Guadeloupe (French West Indies). Microbiol. Spectr. 2022, 10, e0124222. [Google Scholar] [CrossRef] [PubMed]
- Mauri, M.; Elli, T.; Caviglia, G.; Uboldi, G.; Azzi, M. RAWGraphs: A visualisation platform to create open outputs. In Proceedings of the 12th Biannual Conference on Italian SIGCHI Chapter, Cagliari, Italy, 18–20 September 2017; ACM: New York, NY, USA, 2017; pp. 28:1–28:5. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- The Galaxy Community; Afgan, E.; Nekrutenko, A.; Blankenberg, D.; Goecks, J.; Schatz, M.C.; Ostrovsky, A.E.; Mahmoud, A.; Lonie, A.J.; Syme, A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 2022, 50, W345–W351. [Google Scholar] [CrossRef]
- Arkin, A.P.; Cottingham, R.W.; Henry, C.S.; Harris, N.L.; Stevens, R.L.; Maslov, S.; Dehal, P.; Ware, D.; Perez, F.; Canon, S.; et al. KBase: The United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol. 2018, 36, 566–569. [Google Scholar] [CrossRef]
- Schatz, M.C.; Philippakis, A.A.; Afgan, E.; Banks, E.; Carey, V.J.; Carroll, R.J.; Culotti, A.; Ellrott, K.; Goecks, J.; Grossman, R.L.; et al. Inverting the model of genomics data sharing with the NHGRI Genomic Data Science Analysis, Visualization, and Informatics Lab-space. Cell Genom. 2022, 2, 100085. [Google Scholar] [CrossRef]
- Eren, A.M.; Kiefl, E.; Shaiber, A.; Veseli, I.; Miller, S.E.; Schechter, M.S.; Fink, I.; Pan, J.N.; Yousef, M.; Fogarty, E.C.; et al. Community-led, integrated, reproducible multi-omics with anvi’o. Nat. Microbiol. 2021, 6, 3–6. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Grüning, B.; Dale, R.; Sjödin, A.; Chapman, B.A.; Rowe, J.; Tomkins-Tinch, C.H.; Valieris, R.; Köster, J.; Bioconda Team. Bioconda: Sustainable and comprehensive software distribution for the life sciences. Nat. Methods 2018, 15, 475–476. [Google Scholar] [CrossRef] [PubMed]
- Ison, J.; Rapacki, K.; Ménager, H.; Kalaš, M.; Rydza, E.; Chmura, P.; Anthon, C.; Beard, N.; Berka, K.; Bolser, D.; et al. Tools and data services registry: A community effort to document bioinformatics resources. Nucleic Acids Res. 2016, 44, D38–D47. [Google Scholar] [CrossRef] [PubMed]
- Di Tommaso, P.; Chatzou, M.; Floden, E.W.; Barja, P.P.; Palumbo, E.; Notredame, C. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 2017, 35, 316–319. [Google Scholar] [CrossRef]
- Mölder, F.; Jablonski, K.P.; Letcher, B.; Hall, M.B.; Tomkins-Tinch, C.H.; Sochat, V.; Forster, J.; Lee, S.; Twardziok, S.O.; Kanitz, A.; et al. Sustainable data analysis with Snakemake. F1000Research 2021, 10, 33. [Google Scholar] [CrossRef]
- Couvin, D.; Dereeper, A.; Meyer, D.F.; Noroy, C.; Gaete, S.; Bhakkan, B.; Poullet, N.; Gaspard, S.; Bezault, E.; Marcelino, I.; et al. KaruBioNet: A network and discussion group for a better collaboration and structuring of bioinformatics in Guadeloupe (French West Indies). Bioinform. Adv. 2022, 2, vbac010. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 11 December 2023).
- Cock, P.J.A.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Giordano, F.; Ning, Z. Oxford Nanopore MinION Sequencing and Genome Assembly. Genom. Proteom. Bioinform. 2016, 14, 265–279. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Holt, K.E. Deepbinner: Demultiplexing barcoded Oxford Nanopore reads with deep convolutional neural networks. PLoS Comput. Biol. 2018, 14, e1006583. [Google Scholar] [CrossRef]
- Leger, A.; Leonardi, T. pycoQC, interactive quality control for Oxford Nanopore Sequencing. J. Open Source Softw. 2019, 4, 1236. [Google Scholar] [CrossRef]
- Lanfear, R.; Schalamun, M.; Kainer, D.; Wang, W.; Schwessinger, B. MinIONQC: Fast and simple quality control for MinION sequencing data. Bioinformatics 2018, 35, 523–525. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Zimin, A.V.; Marçais, G.; Puiu, D.; Roberts, M.; Salzberg, S.L.; Yorke, J.A. The MaSuRCA genome assembler. Bioinformatics 2013, 29, 2669–2677. [Google Scholar] [CrossRef]
- Gabriel, L.; Brůna, T.; Hoff, K.J.; Ebel, M.; Lomsadze, A.; Borodovsky, M.; Stanke, M. BRAKER3: Fully automated genome annotation using RNA-Seq and protein evidence with GeneMark-ETP, AUGUSTUS and TSEBRA. Genome Res. 2024, 34, 769–777. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Wei, Y.; Lyu, M.; Wu, Z.; Liu, X.; Luo, H.; Yan, C. A comprehensive review of scaffolding methods in genome assembly. Briefings Bioinform. 2021, 22, bbab033. [Google Scholar] [CrossRef]
- Alonge, M.; Lebeigle, L.; Kirsche, M.; Jenike, K.; Ou, S.; Aganezov, S.; Wang, X.; Lippman, Z.B.; Schatz, M.C.; Soyk, S. Automated assembly scaffolding using RagTag elevates a new tomato system for high-throughput genome editing. Genome Biol. 2022, 23, 1–19. [Google Scholar] [CrossRef]
- Boratyn, G.M.; Camacho, C.; Cooper, P.S.; Coulouris, G.; Fong, A.; Ma, N.; Madden, T.L.; Matten, W.T.; McGinnis, S.D.; Merezhuk, Y.; et al. BLAST: A more efficient report with usability improvements. Nucleic Acids Res. 2013, 41, W29–W33. [Google Scholar] [CrossRef]
- Van Nguyen, H.; Lavenier, D. PLAST: Parallel local alignment search tool for database comparison. BMC Bioinform. 2009, 10, 329. [Google Scholar] [CrossRef]
- Mirdita, M.; Steinegger, M.; Breitwieser, F.; Söding, J.; Karin, E.L. Fast and sensitive taxonomic assignment to metagenomic contigs. Bioinformatics 2021, 37, 3029–3031. [Google Scholar] [CrossRef] [PubMed]
- Yim, W.C.; Cushman, J.C. Divide and Conquer (DC) BLAST: Fast and easy BLAST execution within HPC environments. PeerJ 2017, 5, e3486. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Buchfink, B.; Reuter, K.; Drost, H.-G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 2021, 18, 366–368. [Google Scholar] [CrossRef]
- Steinegger, M.; Söding, J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 2017, 35, 1026–1028. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Kopylova, E.; Navas-Molina, J.A.; Mercier, C.; Xu, Z.Z.; Mahé, F.; He, Y.; Zhou, H.-W.; Rognes, T.; Caporaso, J.G.; Knight, R. Open-Source Sequence Clustering Methods Improve the State of the Art. mSystems 2016, 1, e00003-15. [Google Scholar] [CrossRef]
- Westcott, S.L.; Schloss, P.D. De novo clustering methods outperform reference-based methods for assigning 16S rRNA gene sequences to operational taxonomic units. PeerJ 2015, 12, e1487. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R package version 2.5-7. Preprint, 2020. [Google Scholar]
- Katz, L.; Griswold, T.; Morrison, S.; Caravas, J.; Zhang, S.; Bakker, H.; Deng, X.; Carleton, H. Mashtree: A rapid comparison of whole genome sequence files. J. Open Source Softw. 2019, 4, 1762. [Google Scholar] [CrossRef]
- Criscuolo, A. A fast alignment-free bioinformatics procedure to infer accurate distance-based phylogenetic trees from genome assemblies. Res. Ideas Outcomes 2019, 5, e36178. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Treangen, T.J.; Ondov, B.D.; Koren, S.; Phillippy, A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014, 15, 524. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.-F.; Sergeant, M.J.; Luhmann, N.; Vaz, C.; Francisco, A.P.; Carriço, J.A.; Achtman, M. GrapeTree: Visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018, 28, 1395–1404. [Google Scholar] [CrossRef]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A Comprehensive, Accurate, and Fast Distance-Based Phylogeny Inference Program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef] [PubMed]
- Kurtzer, G.M.; Sochat, V.; Bauer, M.W. Singularity: Scientific containers for mobility of compute. PLoS ONE 2017, 12, e0177459. [Google Scholar] [CrossRef] [PubMed]
- Leprevost, F.d.V.; Grüning, B.A.; Aflitos, S.A.; Röst, H.L.; Uszkoreit, J.; Barsnes, H.; Vaudel, M.; Moreno, P.; Gatto, L.; Weber, J.; et al. BioContainers: An open-source and community-driven framework for software standardization. Bioinformatics 2017, 33, 2580–2582. [Google Scholar] [CrossRef] [PubMed]
- Ewels, P.A.; Peltzer, A.; Fillinger, S.; Patel, H.; Alneberg, J.; Wilm, A.; Garcia, M.U.; Di Tommaso, P.; Nahnsen, S. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 2020, 38, 276–278. [Google Scholar] [CrossRef]
| Feature | Conventional Laptop | HPC System |
|---|---|---|
| Processing Power | Limited to a single CPU with few cores | Multiple nodes with many CPUs and cores |
| Memory (RAM) | Typically 8–32 gigabyte (GB) | Hundreds to thousands of GBs distributed across nodes |
| Storage | Limited storage (gigabyte—terabyte) | Large-scale distributed storage systems (e.g., petabytes) |
| CPU Type | General-purpose CPUs (e.g., Intel i5/i7, AMD Ryzen) | High-end server-grade CPUs (e.g., Intel Xeon, AMD EPYC) |
| GPU | Optional, usually for basic graphics tasks | Often includes powerful GPUs for parallel processing |
| Networking | Basic Wi-Fi/Ethernet connectivity | High-speed interconnects (e.g., InfiniBand) for fast data transfer |
| Power Consumption | Low, suitable for personal use | High, requires dedicated cooling and power supply |
| Cost | Affordable, consumer-level pricing | High-cost, enterprise-level investment |
| Use Cases | Everyday tasks, basic software development | Scientific simulations, big data analysis, Artificial Intelligence (AI) training |
| Scalability | Limited to hardware constraints | Highly scalable, can add more nodes as needed |
| Maintenance | Minimal, user-level maintenance | Requires dedicated informaticians for management and upkeep |
| Operating System (OS) | General-purpose OS (e.g., Windows, macOS, Linux) | Often runs Linux-based OS optimized for HPC tasks |
| Software | General productivity and entertainment apps | Specialized scientific and engineering software |
| Language | Execution Speed | Ease of Use | Main Applications | Paradigm(s) | Community & Support |
|---|---|---|---|---|---|
| Python | Medium (interpreted) | Very easy, clear syntax | Data Science, AI, Web, Automation | Object-oriented, Functional | Very large, excellent support |
| JavaScript | Medium | Easy to learn for the Web | Web Development (Frontend/Backend), Mobile | Object-oriented, Functional | Very large, strong web support |
| Java | Fast (compiled to bytecode) | Moderate, strict syntax | Web Apps, Mobile (Android), Enterprise Software | Object-oriented | Large, strong enterprise support |
| C | Very fast (compiled) | Complex (manual memory management) | Embedded Systems, OS, Low-Level Software | Procedural | Large, but more technical |
| C++ | Very fast (compiled) | More complex than C but powerful | Game Development, Heavy Software, AI, Embedded Systems | Object-oriented, Procedural | Large, performance-focused |
| C# | Fast (compiled to bytecode) | Moderate, inspired by Java | Windows Apps, Game Development (Unity) | Object-oriented | Large, Microsoft-backed |
| Swift | Fast (compiled) | Easy for beginners | iOS/macOS Development | Object-oriented, Functional | Large, Apple-focused |
| Go (Golang) | Very fast (compiled) | Simple, clean syntax | Cloud Applications, Backend, Networking | Procedural, Concurrent | Growing, strong support |
| PHP | Medium (interpreted) | Easy for web development | Web Backend Development | Procedural, Object-oriented | Large, primarily for web |
| Rust | Very fast (compiled) | Complex (strict memory management) | Embedded Systems, Security, Performance-Critical Applications | Functional, System | Expanding rapidly |
| R | Medium (interpreted) | Steeper learning curve | Statistics, Data analysis, machine learning | Functional, with some support for Object-oriented programming | Large and active community |
| Category | Tool | Computational Efficiency | Usability | Accuracy |
|---|---|---|---|---|
| Genome Assembly | SPAdes | Moderate; optimized for short-read data | User-friendly with extensive documentation | High accuracy for small genomes |
| Canu | Lower efficiency due to intensive long-read correction algorithms | Moderate; may require parameter tuning | High accuracy for long-read assemblies | |
| Flye | High; particularly efficient with long reads | Relatively easy to use with sensible default settings | Good accuracy; performance can vary with dataset quality | |
| Nanopore Basecalling | Dorado | High; leverages GPU acceleration for faster processing | Highly usable; streamlined interface with regular updates | High accuracy with continuous improvements |
| Bonito | Moderate; designed for research with deep learning frameworks | Requires command-line familiarity; active development | Competitive accuracy; often used in experimental settings | |
| DeepNano-blitz | Variable; experimental approaches may affect speed | Lower usability; fewer support resources available | Moderate accuracy; not as widely validated in the community | |
| Phylogenetic | RAxML | Computationally intensive, especially with large datasets | Steep learning curve; primarily command-line based | High accuracy using maximum likelihood |
| IQ-TREE | High; optimized algorithms for rapid inference | User-friendly; offers both GUI and command-line options | High accuracy with integrated model selection | |
| FastTree | Extremely efficient; ideal for very large datasets | Very easy to use with minimal configuration required | Good accuracy; some compromises on precision | |
| PhyML | Moderate; performs well on small to medium datasets | Reasonably user-friendly; includes GUI and web interfaces | High accuracy for likelihood-based tree estimation |
| User Level | Dataset Size | Model | Why? | Benefits | Drawbacks |
|---|---|---|---|---|---|
| Beginner | Small (<1 GB) | Galaxy | Easy GUI, no installation needed, available online | Intuitive interface, built-in tools, no coding | Limited customization, slower for large data |
| Intermediate | Medium (1–10 GB) | Galaxy or Laptop Command Line | Galaxy if no CLI experience; CLI for learning | Galaxy easy to use; CLI builds skills | Galaxy job limits; CLI setup can be tough |
| Expert | Large (>10 GB) | HPC Command Line | Galaxy may fail or queue jobs too long | HPC handles big data well | Requires technical skills, setup time |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quétel, I.; Tirera, S.; Cazenave, D.; Allouch, N.; Baum, C.; Reynaud, Y.; Batantou Mabandza, D.; Nerrière, V.; Vedy, S.; Pot, M.; et al. A Tutorial Toolbox to Simplify Bioinformatics and Biostatistics Analyses of Microbial Omics Data in an Island Context. BioMedInformatics 2025, 5, 27. https://doi.org/10.3390/biomedinformatics5020027
Quétel I, Tirera S, Cazenave D, Allouch N, Baum C, Reynaud Y, Batantou Mabandza D, Nerrière V, Vedy S, Pot M, et al. A Tutorial Toolbox to Simplify Bioinformatics and Biostatistics Analyses of Microbial Omics Data in an Island Context. BioMedInformatics. 2025; 5(2):27. https://doi.org/10.3390/biomedinformatics5020027
Chicago/Turabian StyleQuétel, Isaure, Sourakhata Tirera, Damien Cazenave, Nina Allouch, Chloé Baum, Yann Reynaud, Degrâce Batantou Mabandza, Virginie Nerrière, Serge Vedy, Matthieu Pot, and et al. 2025. "A Tutorial Toolbox to Simplify Bioinformatics and Biostatistics Analyses of Microbial Omics Data in an Island Context" BioMedInformatics 5, no. 2: 27. https://doi.org/10.3390/biomedinformatics5020027
APA StyleQuétel, I., Tirera, S., Cazenave, D., Allouch, N., Baum, C., Reynaud, Y., Batantou Mabandza, D., Nerrière, V., Vedy, S., Pot, M., Breurec, S., Lavergne, A., Ferdinand, S., Guerlais, V., & Couvin, D. (2025). A Tutorial Toolbox to Simplify Bioinformatics and Biostatistics Analyses of Microbial Omics Data in an Island Context. BioMedInformatics, 5(2), 27. https://doi.org/10.3390/biomedinformatics5020027






