1. Introduction
Parkinson’s Disease (PD) is a neurological condition that worsens with time and mostly affects movement. Tremors, stiffness, bradykinesia (slowness of movement), and postural instability are some of its symptoms. Non-motor symptoms are also common and include sadness, sleep difficulties, and cognitive impairment [
1,
2]. The illness is brought on by the death of dopamine-producing neurons in the midbrain area known as the substantia nigra. One neurotransmitter that is essential for coordinating balanced and fluid muscular actions is dopamine. Although the precise origin of neuron degeneration is unknown, a mix of environmental and genetic variables is thought to be involved. After Alzheimer’s Disease, PD is the second most common neurodegenerative condition, affecting about 1% of the over-60 population [
3]. PD is more common in males than in women, and its prevalence rises with age. PD cannot be definitively diagnosed; instead, the condition is managed with a combination of physical and neurological examinations, medical history, symptoms, and dopamine replacement medication response. While imaging tests such as Magnetic Resonance Imaging (MRI) and DaT scans are not commonly used to diagnose PD, they can be useful in ruling out other illnesses. Significant motor function impairments caused by PD make daily tasks including eating, dressing, and walking difficult. Patients may experience progressively worsening disabilities as the illness worsens. Patients’ and their families’ emotional well-being are significantly impacted by the prevalent conditions of depression, anxiety, and cognitive impairment.
Parkinson’s motor symptoms manifest when 60% to 80% of these cells are lost due to the inadequate production of dopamine. Recent research has identified a close connection between speech impairment and PD, leading to the development of classification algorithms for PD identification. Artificial Intelligence (AI) is leading the way in next-generation computing for data analytics, particularly in predictive edge analytics for high-risk diseases including PD. Deep Learning (DL) techniques play a crucial role in facilitating edge AI applications for real-time and enhanced data processing. In contrast to conventional methods, which mostly depend on the dynamic (kinematic and spatio-temporal) aspects of handwriting, we objectively assess the visual characteristics in this work in order to characterize graphomotor samples of PD patients. Even with Machine Learning (ML) advances, choosing the best attributes that greatly improve PD diagnostic accuracy continues to be a major difficulty. High dimensionality and superfluous or unnecessary characteristics are common problems with existing models, which can cause overfitting and decreased generalizability. Furthermore, a lot of research has concentrated on static feature sets rather than investigating dynamic strategies that may adjust to various patient data distributions. Research is still being done to find biomarkers for early diagnosis, improve our understanding of the mechanism of PD, and create medicines that alter the condition. DL and other AI advances have the potential to improve diagnosis, forecast the course of disease, and customize therapy regimens. Owing to the intricacy and variability of PD, feature selection (FS) and categorization provide a number of difficulties. The primary difficulties consist of:
High Dimensionality: Clinical, genetic, imaging, and biochemical indicators are only a few of the many aspects that biomedical data frequently contain. It is challenging to determine which features are most important for precise categorization because of this high dimensionality.
Heterogeneity of Data: Patients with PD might have very different symptoms and rates of progression, which results in heterogeneous data that makes feature selection more difficult.
Unbalanced Data: The distribution of PD datasets is frequently unbalanced, with fewer positive cases than healthy controls. This can cause bias in the classification models.
Redundancy and Noise: Biomedical data may have duplicate information and be noisy, which can impair the effectiveness of classification systems.
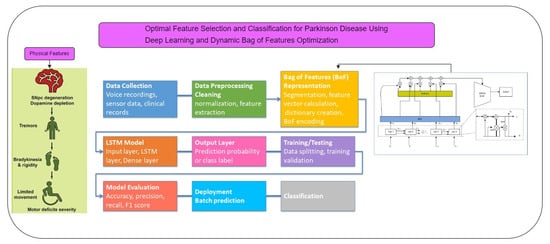
The proposed method makes use of the Dynamic Bag of Features Optimization Technique (DBOFOT) to overcome the shortcomings of previous studies in FS and classification for PD. By streamlining the classification process and dynamically choosing the most pertinent features, this technique seeks to improve overall performance. DBOFOT uses a dynamic FS procedure that changes depending on the particulars of the dataset. Long Short-Term Memory-DBOFOT (LSTM-DBOFOT) continuously modifies the feature set according to each feature’s relevance and contribution to classification accuracy, in contrast to static FS approaches that could ignore significant characteristics or keep unimportant ones [
4]. The model can more efficiently manage the high complexity and variability of biomedical data dueto its adaptability. LSTM-DBOFOT improves classification accuracy by reducing noise and redundancy in the data by dynamically choosing the most relevant characteristics. By limiting the usage of only the most informative variables, the optimization strategy improves the model’s capacity to discriminate between PD patients and healthy controls. The shortcomings of earlier study [
1,
2] that had trouble with noisy and redundant data are addressed by this method. LSTM-DBOFOT uses optimization methods to make the FS process more efficient and less time-consuming. For managing large-scale biomedical datasets, which are typical in PD research, this efficiency is essential. By reducing the computing overhead of conventional FS techniques, the method enables more rapid and scalable analysis. This method improves the model’s interpretability by illuminating the chosen characteristics and how they contribute to the classification goal. For clinical applications, where knowing the importance of traits can help with improved diagnosis and treatment planning, this transparency is essential. Furthermore, by being dynamic, LSTM-DBOFOT addresses the variability problems in earlier studies and enhances the model’s generalizability across various datasets. Multimodal data sources including clinical, genetic, imaging, and gait data can be integrated with DBOFOT. A more complete picture of the patient’s state is provided by this integration, which results in more reliable and accurate categorization models. Studies that concentrate on a single data type have limitations; nevertheless, the model’s ability to handle many data kinds guarantees that it can utilize the entire range of information available.
Our model can effectively capture the temporal dependencies included in the data by integrating LSTM cells, which makes it a suitable fit for the intricate patterns linked to PD. By real-time feature input optimization, this hybrid methodology overcomes the drawbacks of conventional FS techniques and improves the performance of DL models. The major contribution includes:
The proposed method integrates an LSTM network with the BoF representation, offering a novel approach for PD prediction. It focuses on robust feature selection, and dynamic feature sets, improving classification accuracy.
The use ofDBOFOT has been used to handwriting-based PD identification. By allowing for adaptive feature combinations based on the significance of the data, it enhances sensitivity to minute handwriting differences that are common in individuals with Parkinson’s Disease.
The proposed work employs optimization techniques of transfer learning, cross-validation, or regularization to counter overfitting and ensure robust performance even with smaller datasets.
Handwriting patterns are subtle indications of motor deficits, and the Parkinson’s Disease handwriting (PaHaW) collection used in proposed analysis offers thorough information on handwriting patterns.
The deployment and continuous monitoring aspects of the proposed method are designed to maintain high accuracy and relevance over time.
The remaining contribution of this paper is structured as follows:
Section 2 reviews various prediction methods.
Section 3 details the implementation of the LSTM–BoF model and the dataset used, while
Section 4 covers the evaluation procedure and analysis of the results on two different datasets. Finally,
Section 5 concludes the work.
2. Literature Review
DL and ML models have been used more and more in the diagnosis of PD in recent years. Conventional ML methods, including Random Forest (RF) and Support Vector Machine (SVM), have demonstrated some effectiveness but are frequently limited in their scalability by the need for human feature extraction. By automatically learning hierarchical representations of data, DL models—in particular, CNN and LSTM networks—offer notable advances in feature extraction and classification tasks. The study of computer programs that use statistical models and algorithms to infer information through patterns and inference without explicit programming is known as ML [
1]. As ML algorithms learning by training and they improve over time. These algorithms use methods for training models, then use the information they have learned to determine outputs on their own [
2]. Moreover, ML systems are capable of adjusting to changing surroundings. According to the study by [
3], the three most well-known signs of PD that may be seen in handwriting are “tremor”, “brachykinesia”, and “micrographia”. Maintaining the size and alignment of the generated graphomotor impressions becomes challenging for a patient with micrographia [
4]. A supervised ML model that applies classification techniques to two-group classification issues is called an SVM. SVM is well-known for being quick and dependable, and it performs well when processing small datasets [
3,
4,
5]. A prospective PD patient may take longer than normal to finish a graphomotor task if they have bradykinesia, or slowness of movement (caused by either motor or cognitive impairment) [
6]. Uncontrollably moving back and forth, “tremors” are represented by asymmetrical character and drawing forms. The coexistence or independence of various Parkinsonian disorders is contingent upon the nature and course of the ailment.
The Decision Tree (DT) approach in ML systems partitions data into subsets with the training data divided into the smallest tree. In order to check the target classes in leaf nodes, this supervised classification approach uses split tests in internal nodes [
7]. Because of their consistency and adaptability, decision trees are frequently employed in categorization [
8,
9,
10,
11,
12]. A supervised learning technique that is both flexible and easy to use, RF frequently produces good results without requiring a lot of hyperparameter tweaking [
13]. An RF is made up of randomly generated DTs that use majority vote to decide the expected output. Even if DTs alone might be less precise and dependable, the RF makes up for these drawbacks [
14]. Unlike Logistic Regression (LR), which can only handle linear situations, SVM is superior at managing nonlinear problems. SVM determines maximum margin solutions, which efficiently handles outliers. When it comes to handling collinearity, DTs perform better than LR, particularly for categorical values. Compared to individual DTs, RF shave higher accuracy thanks to their ensemble of DTs. While DTs use hyperrectangles in input space to solve problems, SVM use kernel approaches to address nonlinear problems. SVM typically performs better than RF in classification problems [
15].
Medical diagnosis has made extensive use of ML models [
16,
17,
18,
19]. This research examines different performances of ML for Alzheimer’s diagnosis. A genetic and irreversible brain disorder called Alzheimer’s syndrome gradually impairs critical abilities such as memory and reasoning [
20]. To categorize PD using speech data, for example, in reference [
21] the author used a CNN-based model, demonstrating a high accuracy rate by capturing subtle patterns that are frequently ignored by traditional ML approaches [
21,
22,
23,
24]. In a similar way, ref. [
25] showed how well LSTM networks handle time-series biological data and performs better in the early stages of Parkinson’s Disease-identification. The symptoms usually appear in the mid-30s and mid-60s and range from altered sleep patterns to despair and anxiety. Alzheimer’s Disease causes memory loss and reduced cognitive ability, and its symptoms take ten to twenty years to manifest. The primary cause of Alzheimer’s Disease is dementia, which affects 40–50 million people worldwide and is predicted to increase to 131.5 million by 2050. Remarkably, 70% of dementia sufferers live in low-income nations [
26].
Salmanpour et al. [
27] used ML methods predict the cognitive effects of PI. A comprehensive review of the literature on FS and ML was carried out by Wan et al. [
28]. ML is actively used in brain surgery to pinpoint the exact area that has to be treated. The post-diagnosis aspects of PD were also examined by the researchers.
Cavallo et al. [
29] used motion data from patients’ upper limbs to create a predictive model for PD. Two groups participated in the study: those with PD and those who did not. The subjects had a device attached to their upper limbs, and they were given instructions on how to carry out different tasks. Frequency and spatiotemporal analyses were performed during data analysis. Different learning strategies were then used to complete the classification process. An alternate method [
30] used different ML and feature extraction approaches to identify PD. The most useful criterion for diagnosing the illness, according to the researchers, was phonation. Optimum Path Forest, K-Nearest Neighbor (KNN), SVM, and Multilayer Perceptron (MLP) classifiers were used. Artificial Neural Networks (ANNs) were employed to reduce linguistic impairments, while SVM contributed to the classification process, facilitating the ML-based identification of PD.
Researchers presented an unsupervised method for PD identification in [
31]. For clustering and prediction, they used an incremental Support Vector Regression (SVR) and a Self-Organizing Map (SOM), respectively. After dimension reduction using Partial Least Squares (PLS), the Unified PD Rating Scale (UPDRS) is predicted and SOM and SVR approaches are implemented in the suggested method. In a different work [
32], authors used the KNN classifier technique for PD identification and investigated a Fuzzy-based C-means clustering algorithm to evaluate feature weights. To find the ideal K value, the KNN classifier made use of a weighted PD dataset with a range of K values.
In order to handle unbalanced data, Wang et al. [
33,
34] used sophisticated learning techniques to diagnose PD. These techniques included a weighted scheme and a nonlinear mapping of the kernel function. Features were chosen and variables were optimized using the Artificial Bee Colony (ABC) algorithm. Fisher Discriminant Ratio (FDR), Principal Component Analysis (PCA), and SVM were utilized for FS, dimension optimization, and classification, respectively, in a different method put forward by PI. Abdulhay et al. [
35] used sensors beneath patients’ feet to collect physical signals and looked at tremor and gait symptoms for PD detection. With an accuracy rate of over 92%, gait features were extracted from the raw data in the PhysioNet database by analyzing the electrical pulses to identify anomalous peaks. Using voice inputs from a shared dataset, Yaman et al. [
36] applied feature augmentation to find salient features for illness detection. A total of sixty-six characteristics were chosen to categorize PD. Instead of depending on MRI, motion, or voice data, a noteworthy study in [
37,
38,
39,
40,
41,
42] investigated the possibility of using handwriting inspection to detect PD.
High classification rates are demonstrated by ML-based techniques [
43,
44,
45,
46,
47] for determining PD, according to a thorough evaluation of the literature. Although ML is useful foridentifying PD, its application is fraught with difficulties. While considering a lot of features improves identification rates, it also uses more resources in terms of computing cost and execution time. On the other hand, utilizing fewer features could make results less reliable.
According to recent research, computation time can be greatly decreased by using a simpler classifier, a lightweight feature extraction model, and a smaller number of features. Moreover, it has been noted that feature extraction from speech signals is comparatively easier than approaches based on motion [
39] or MRI. To increase classification rates, a significant number of vocal features have been chosen by several researchers [
40,
41]. In contrast, speech analysis is a simpler process compared to the extraction of features from MRI images, which yields higher classification accuracy. A summarization of the existing works is given in
Table 1.
PD is a progressive neurological disorder that impairs movement and significantly impacts quality of life, necessitating timely and accurate diagnosis for effective management. Traditional diagnostic methods, reliant on clinical examination, can be subjective and delay diagnosis. This work introduces a novel approach using advanced ML techniques to improve the early detection of PD by analyzing diverse patient data sources. The proposed system employs dynamic FS to enhance classification accuracy by reducing noise and redundancy, making it particularly effective for large-scale biomedical datasets. By integrating LSTM networks with the BoF technique, the model captures temporal patterns and complex correlations in data, improving interpretability and generalizability. The LSTM–BoF PD model is designed to enhance the discriminative power of features across various modalities, such as gait, imaging, and genetic data, offering a more accurate and comprehensive understanding of PD dynamics. This integrated approach aims to advance healthcare technologies by enabling a more reliable and efficient system for early PD diagnosis.
In the proposed system, we extend existing methods by combining a DL classification framework with an optimization method called DBoF. This hybrid technique overcomes the drawbacks of previous models that just use ML algorithms by ensuring appropriate feature selection and improved classification accuracy. Furthermore, by incorporating DL, the model can recognize more intricate patterns in the data, increasing its versatility and scalability across various datasets. Predictions become more accurate as a result of the DBoF approach, which improves the system’s capacity to eliminate noise and choose the most pertinent features. The model is highly suited forPDdiagnosis because of method’s special ability to handle large-scale and heterogeneous biological datasets. Moreover, it enables dynamic feature selection for ongoing improvement, guaranteeing the system’s sustained high performance.
4. Performance Evaluation
This section presents the performance evaluation of the proposed model by evaluating various metrics such as accuracy, precision, recall, and F1-score, which identify the model’s accuracy in identifying PD, and performance is evaluated using a dataset that included both patients and healthy persons.
4.3. Analysis of Parkinson Using Handwriting
We also used the PaHaW dataset [
31] in our tests to examine the resilience and efficacy of our approach. The PD handwriting database includes several handwriting/drawing samples from 38 healthy controls (HC) and 33 PD patients.
Throughout the assessment procedure, we used stratified 10-fold cross-validation. Ten equal-sized subsets that were mutually exclusive were created from the dataset. The training data for each subset werethe union of the remaining subsets. Furthermore, we used data augmentation, producing thirty additional versions for every training data sample that was first used. Ten iterations of the entire process were performed until one test set was used for each fold. To calculate the overall accuracy, we then summed the accuracies of the various subgroups.
To prepare it for batch processing, the image is first enlarged to
pixels and then turned into an array with an extra dimension. Matplotlib is used to display the batches of augmented photos generated by an image data generator (Train_Generator), as shown in
Figure 6. By subjecting machine learning models to a greater range of picture transformations, this data augmentation strategy contributes to the improvement of the training dataset and may increase the robustness and performance of the models.
Further, we can also check on detection using hand drawings. Although there is no known cure for PD, early identification and appropriate treatment can greatly reduce symptoms and enhance quality of life. For this reason, the condition is a valuable study subject, particularly when developing novel diagnostic methods. The study [
44] discovered that asking a patient to draw a spiral and then tracking might be used to identify PD.
Table 11 gives the model’s learning progress over time by capturing the model’s accuracy and loss for both training and validation sets across each of the 65 periods. Drawing speed and pressure can be used for detection. Research [
45] discovered that patients with PD drew more slowly and used less pressure when writing; this difference was particularly noticeable in those with more severe or advanced stages of the illness. Using this fact, tremors and muscular rigidity, two of the most prevalent Parkinson’s symptoms, have an immediate effect on how a hand-drawn image appears visually, as shown in
Figure 7.
Figure 7 shows a sequence of spirals that were made by hand and are used to assess motor control skills. This technique is very helpful in differentiating between people who have problems with their motor control and people who do not. People are asked to draw spirals on paper or a digital device as part of the process. After that, these spirals are examined for particular patterns that point to the existence of problems with motor control. The spiral features that have been made by hand are each labeled with a forecast. These hypotheses probably match the output of a classification model that determines whether the drawing points to PD or shows normal motor control. A prediction, which seems to be a tuple of values, is labeled for each image. These numbers reflects the probability that the model allocated to the various classes (e.g., 1 for Parkinson’s, 0 for healthy).
A label such as PREDICTION: [0.50145745, 0.49854255], for instance, indicates that the model is marginally more certain that the picture belongs to the first class (healthy) than the second class (Parkinson’s). The spirals themselves serve as illustrations of motor control in pictures. Smooth Spirals a sign of good motor control and is usually connected to well people. An assessment of the model’s ability to discriminate between normal and abnormal motor control can be made by looking at the spirals as well as the predictions. When a spiral’s forecast is near [0.5, 0.5], it means that the model is unsure of its classification. More extreme prediction spirals (such [0.7, 0.3] or [0.3, 0.7]) indicate a higher level of categorization confidence, as shown in
Figure 8. The forecasts’ distribution and the spirals that go along with it can reveal information about how well the model works. A robust model would be suggested by consistently accurate predictions with high confidence; on the other hand, frequent misclassifications or uncertainty might point to the need for improvement.
With the AUC-ROC of 0.86 as shown in
Figure 9, the ANN model produced an accuracy of 86%. In terms of itsperformance, however, there wasalmost similar accuracy received for plain LSTM, but less sensitivity and specificity than the suggested LSTM–BoF. The 82% accuracy and 0.87 AUC-ROC of the conventional LSTM model indicated competitive performance, but in terms of sensitivity it was not as good as the suggested LSTM–BoF. Among the compared methods, the Bayesian classifier had the lowest AUC-ROC score of 0.81 and the highest accuracy of 79%. SVM performed exceptionally well, with the greatest AUC-ROC score of 0.91 and an accuracy of 86%. But in terms of sensitivity, it was not as good as the suggested LSTM–BoF.
One important indicator is how smooth the spiral lines are. Generally speaking, smooth, continuous lines indicate strong motor control. Uneven, jagged, or wavering lines frequently signify the existence of tremors, which are typical in a number of neurological disorders. Measurements are made of the tremor waves’ height (amplitude) and frequency (number of waves per unit length). Elevated frequency and amplitude are suggestive of more serious problems with motor control. Repeatedly deviating from the planned spiral trajectory indicates a hand movement incapacity, which is a typical sign in several circumstances. The precision, recall, F1-score, and support for each class are included in the classification report. These are crucial metrics for assessing a classifier’s effectiveness. An easy-to-understand indicator called the accuracy score determines how many of the model’s overall predictions were accurate. When combined, these three components offer a thorough assessment of the classifier’s performance using the test dataset.
The model’s performance in classifying PD is shown by the confusion matrix. The model demonstrated a high recall rate by correctly identifying 42 (true positives) out of 47 real Parkinson’s cases. However, it affected the precision by incorrectly predicting Parkinson’s in three healthy persons (false positives), as shown in
Figure 10. Furthermore, five genuine Parkinson’s cases were overlooked by the model (false negatives), which is important for a precise diagnosis. Nine out of twelve examples (true negatives) were properly detected by the model for healthy persons.
The classification report indicates the performance of the model in distinguishing between healthy individuals (class 0) and those with Parkinson’s (class 1). The model has a precision of 0.64 and a recall of 0.75 for class 0, meaning it correctly identifies 64% of the predictions for healthy individuals and captures 75% of actual healthy cases. For class 1, the precision is 0.93 and recall is 0.89, indicating the model accurately predicts 93% of Parkinson’s cases and correctly identifies 89% of actual Parkinson’s instances. The overall accuracy of the model is 86.44%, as shown in
Table 12, which suggests that it performs well when classifying the images with an overall balanced precision, recall, and F1-score, especially for identifying PD. Out of all the positive predictions the model makes, precision indicates the percentage of real positive predictions (erroneously classified as Parkinson’s). For example, our model’s accuracy for the Parkinson’s class was 0.93, which means that 93% of the people who were predicted to have the disease were really diagnosed.
Recall measures how effectively the model detects Parkinson’s Disease patients by dividing the number of true positives by the total number of actual positives. With a recall of 0.89 for the Parkinson’s class, 89% of the real Parkinson’s patients were correctly recognized by the model.
When working with unbalanced classes, the F1-Score offers a balanced statistic as it is the harmonic mean of precision and recall. The model shows a solid balance between accurately identifying people with the condition and reducing false positives, as seen by its F1-score of 0.91 for the Parkinson’s class.
As the ratio of accurate predictions (both for healthy and Parkinson’s patients) to the total number of occurrences, accuracy measures the overall correctness of the model for both groups. With an accuracy of 0.86, our model was able to classify 86% of the cases correctly.