Impact of Data Pre-Processing Techniques on XGBoost Model Performance for Predicting All-Cause Readmission and Mortality Among Patients with Heart Failure
Abstract
1. Introduction
- Evaluate and compare the effectiveness of different imputation methods (Mean, Multivariate Imputation by Chained Equation (MICE), k-nearest Neighbors (kNNs), and Random Forest) in enhancing model performance. While Mean imputation offers simplicity, MICE considers variable relations, kNN captures local data structures, and Random Forest imputation offers robustness against data complexity.
- Assess the impact of feature encoding and standardization on prediction accuracy.
- Examine the robustness of these models through comprehensive cross-validation.
2. Materials and Methods
2.1. Dataset
2.2. Pre-Processing Techniques Applied
2.2.1. Handling Missing Values and Imputation
2.2.2. Label Encoding and Standardization
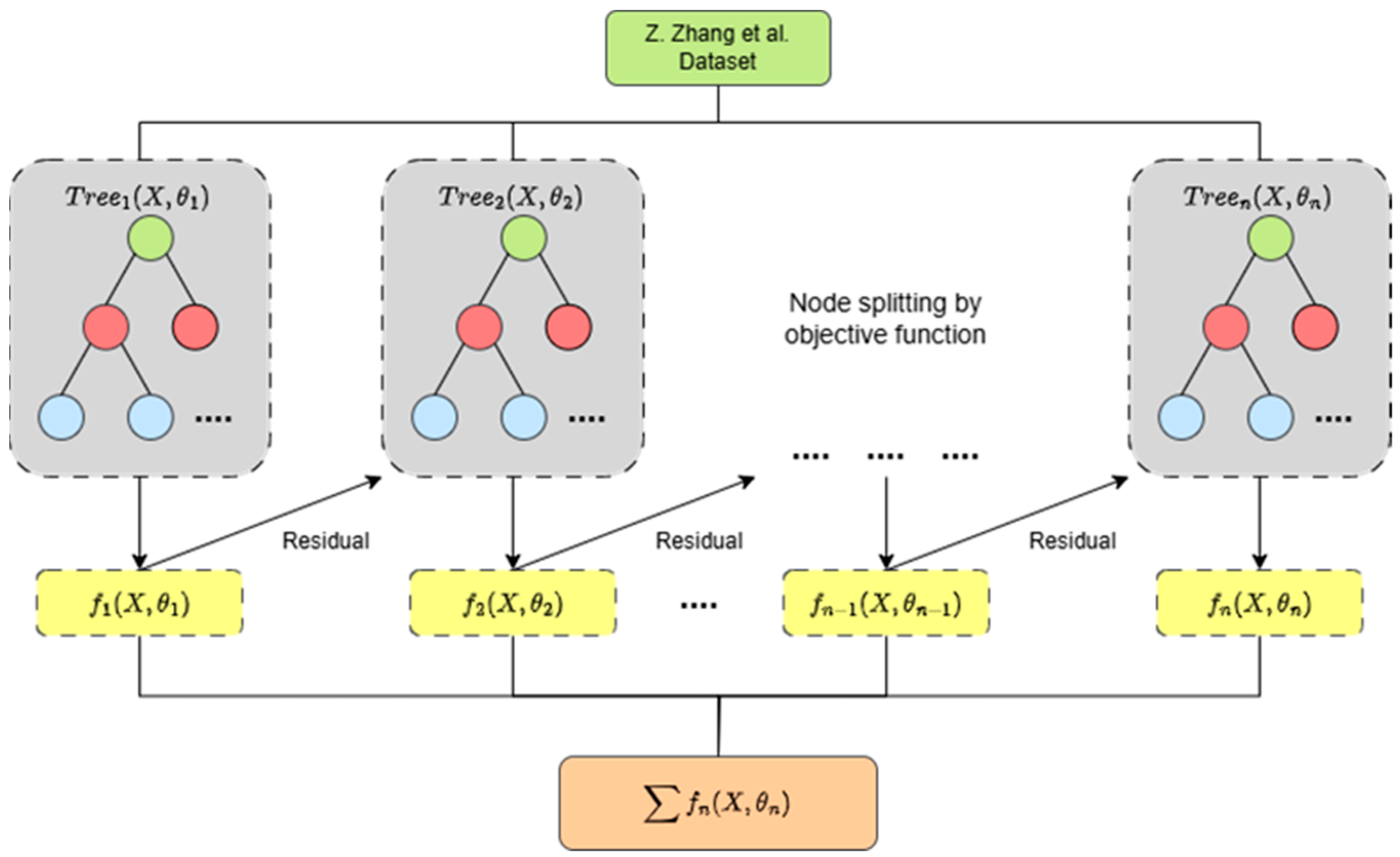
2.3. XGBoost Model Building
2.4. Model Evaluation and Validation
2.5. Cross-Validation
3. Results
3.1. Model Performance Comparison
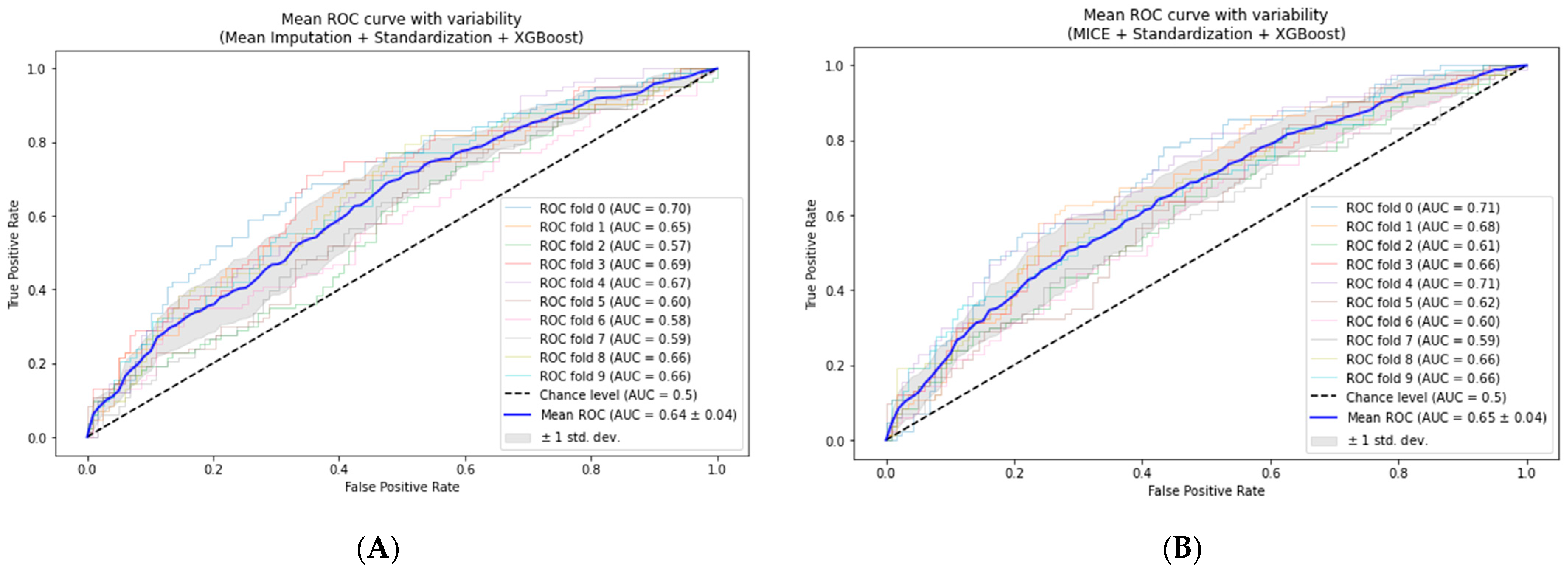
3.2. Cross-Validation of the Models
- Mean-based imputation model: The AUC increased to 0.64 ± 0.04, suggesting a slight enhancement in predictive power and generalizability.
- MICE-based model: The mean of the AUC slightly increased from 0.647 to 0.65 ± 0.04, demonstrating stability across different data subsets.
- kNN-based imputation model: We observed improvement from 0.619 to 0.63 ± 0.06, indicating a modest increase in model performance.
- Random Forest-based imputation: This method generated slight improvement, from 0.625 to 0.63 ± 0.06, showing stability in various data subsets.
3.3. Comparison to Related Studies
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shahim, B.; Kapelios, C.J.; Savarese, G.; Lund, L.H. Global Public Health Burden of Heart Failure: An Updated Review. Card. Fail. Rev. 2023, 9, e11. [Google Scholar] [CrossRef] [PubMed]
- Helm, J.E.; Alaeddini, A.; Stauffer, J.M.; Bretthauer, K.M.; Skolarus, T.A. Reducing Hospital Readmissions by Integrating Empirical Prediction with Resource Optimization. Prod. Oper. Manag. 2016, 25, 233–257. [Google Scholar] [CrossRef]
- Krittayaphong, R.; Chichareon, P.; Komoltri, C.; Sairat, P.; Lip, G.Y.H. Predicting Heart Failure in Patients with Atrial Fibrillation: A Report from the Prospective COOL-AF Registry. J. Clin. Med. 2023, 12, 1265. [Google Scholar] [CrossRef] [PubMed]
- Badawy, M.; Ramadan, N.; Hefny, H.A. Healthcare Predictive Analytics Using Machine Learning and Deep Learning Techniques: A Survey. J. Electr. Syst. Inf. Technol. 2023, 10, 40. [Google Scholar] [CrossRef]
- Sundararaman, A.; Valady Ramanathan, S.; Thati, R. Novel Approach to Predict Hospital Readmissions Using Feature Selection from Unstructured Data with Class Imbalance. Big Data Res. 2018, 13, 65–75. [Google Scholar] [CrossRef]
- Sharma, V.; Kulkarni, V.; Mcalister, F.; Eurich, D.; Keshwani, S.; Simpson, S.H.; Voaklander, D.; Samanani, S. Predicting 30-Day Readmissions in Patients With Heart Failure Using Administrative Data: A Machine Learning Approach. J. Card. Fail. 2022, 28, 710–722. [Google Scholar] [CrossRef]
- Zhang, X.; Yan, C.; Gao, C.; Malin, B.A.; Chen, Y. Predicting Missing Values in Medical Data Via XGBoost Regression. J. Healthc. Inform. Res. 2020, 4, 383–394. [Google Scholar] [CrossRef]
- Chen, Z.-Y.; Zhang, T.-H.; Zhang, R.; Zhu, Z.-M.; Yang, J.; Chen, P.-Y.; Ou, C.-Q.; Guo, Y. Extreme Gradient Boosting Model to Estimate PM2.5 Concentrations with Missing-Filled Satellite Data in China. Atmos. Environ. 2019, 202, 180–189. [Google Scholar] [CrossRef]
- Jing, L.; Ulloa Cerna, A.E.; Good, C.W.; Sauers, N.M.; Schneider, G.; Hartzel, D.N.; Leader, J.B.; Kirchner, H.L.; Hu, Y.; Riviello, D.M.; et al. A Machine Learning Approach to Management of Heart Failure Populations. JACC Heart Fail. 2020, 8, 578–587. [Google Scholar] [CrossRef]
- Luo, C.; Zhu, Y.; Zhu, Z.; Li, R.; Chen, G.; Wang, Z. A Machine Learning-Based Risk Stratification Tool for in-Hospital Mortality of Intensive Care Unit Patients with Heart Failure. J. Transl. Med. 2022, 20, 136. [Google Scholar] [CrossRef]
- Mallikharjuna Rao, K.; Saikrishna, G.; Supriya, K. Data Preprocessing Techniques: Emergence and Selection towards Machine Learning Models—A Practical Review Using HPA Dataset. Multimed. Tools Appl. 2023, 82, 37177–37196. [Google Scholar] [CrossRef]
- Zhang, Z.; Cao, L.; Chen, R.; Zhao, Y.; Lv, L.; Xu, Z.; Xu, P. Electronic Healthcare Records and External Outcome Data for Hospitalized Patients with Heart Failure. Sci. Data 2021, 8, 46. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Cao, L.; Zhao, Y.; Xu, Z.; Chen, R.; Lv, L.; Xu, P. Hospitalized Patients with Heart Failure: Integrating Electronic Healthcare Records and External Outcome Data. PhysioNet 2020, 101, e215–e220. [Google Scholar] [CrossRef] [PubMed]
- Cismondi, F.; Fialho, A.S.; Vieira, S.M.; Reti, S.R.; Sousa, J.M.C.; Finkelstein, S.N. Missing Data in Medical Databases: Impute, Delete or Classify? Artif. Intell. Med. 2013, 58, 63–72. [Google Scholar] [CrossRef]
- Dahouda, M.K.; Joe, I. A Deep-Learned Embedding Technique for Categorical Features Encoding. IEEE Access 2021, 9, 114381–114391. [Google Scholar] [CrossRef]
- Dzierżak, R. Comparison of the influence of standardization and normalization of data on the effectiveness of spongy tissue texture classification. Inform. Autom. Pomiary W Gospod. I Ochr. Sr. 2019, 9, 66–69. [Google Scholar] [CrossRef]
- Milligan, G.W.; Cooper, M.C. A Study of Standardization of Variables in Cluster Analysis. J. Classif. 1988, 5, 181–204. [Google Scholar] [CrossRef]
- Ali, Z.H.; Burhan, A.M. Hybrid Machine Learning Approach for Construction Cost Estimation: An Evaluation of Extreme Gradient Boosting Model. Asian J. Civ. Eng. 2023, 24, 2427–2442. [Google Scholar] [CrossRef]
- Guo, R.; Zhao, Z.; Wang, T.; Liu, G.; Zhao, J.; Gao, D. Degradation State Recognition of Piston Pump Based on ICEEMDAN and XGBoost. Appl. Sci. 2020, 10, 6593. [Google Scholar] [CrossRef]
- Vujovic, Ž.Ð. Classification Model Evaluation Metrics. Int. J. Adv. Comput. Sci. Appl. 2021, 12, 599–606. [Google Scholar] [CrossRef]
- Sokolova, M.; Japkowicz, N.; Szpakowicz, S. Beyond Accuracy, F-Score and ROC: A Family of Discriminant Measures for Performance Evaluation. In AI 2006: Advances in Artificial Intelligence; Sattar, A., Kang, B., Eds.; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2006; Volume 4304, pp. 1015–1021. ISBN 978-3-540-49787-5. [Google Scholar]
- Tharwat, A. Classification Assessment Methods. Appl. Comput. Inform. 2021, 17, 168–192. [Google Scholar] [CrossRef]
- Berrar, D. Cross-Validation. In Encyclopedia of Bioinformatics and Computational Biology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 542–545. ISBN 978-0-12-811432-2. [Google Scholar]
- Lasfar, R.; Tóth, G. The Difference of Model Robustness Assessment Using Cross-validation and Bootstrap Methods. J. Chemom. 2024, 38, e3530. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, Z.; Wittrup, E.; Gryak, J.; Najarian, K. Increasing Efficiency of SVMp+ for Handling Missing Values in Healthcare Prediction. PLoS Digit. Health 2023, 2, e0000281. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Hu, W.; Yang, Y.; Cai, J.; Luo, Y.; Gong, L.; Li, Y.; Si, A.; Zhang, Y.; Liu, S.; et al. Predicting Six-Month Re-Admission Risk in Heart Failure Patients Using Multiple Machine Learning Methods: A Study Based on the Chinese Heart Failure Population Database. J. Clin. Med. 2023, 12, 870. [Google Scholar] [CrossRef] [PubMed]
- Psychogyios, K.; Ilias, L.; Ntanos, C.; Askounis, D. Missing Value Imputation Methods for Electronic Health Records. IEEE Access 2023, 11, 21562–21574. [Google Scholar] [CrossRef]
- Pereira, R.C.; Abreu, P.H.; Rodrigues, P.P. Partial Multiple Imputation with Variational Autoencoders: Tackling Not at Randomness in Healthcare Data. IEEE J. Biomed. Health Inform. 2022, 26, 4218–4227. [Google Scholar] [CrossRef]



| Model 1 | Accuracy | Recall | Precision | F1 | AUC |
|---|---|---|---|---|---|
| Default 1 | 0.587 | 0.512 | 0.372 | 0.432 | 0.60 |
| Mean + Enc 2 + Std 3 | 0.595 | 0.525 | 0.379 | 0.44 | 0.626 |
| MICE + Enc 2 + Std 3 | 0.592 | 0.518 | 0.432 | 0.471 | 0.647 |
| kNN + Enc 2 + Std 3 | 0.614 | 0.551 | 0.444 | 0.492 | 0.619 |
| RF + Enc 2 + Std 3 | 0.587 | 0.511 | 0.420 | 0.461 | 0.625 |
| No | Author | Overview | Algorithm | AUC | F1 | Sensitivity |
|---|---|---|---|---|---|---|
| 1 | Y. Zhang et al. [25] | The study built an optimized SVM-based model, I2-SVMp+, to address missingness compared to various common imputation approaches. | SVM | 0.536 | 0.708 | |
| SVM + MeanImp | 0.546 | 0.703 | ||||
| SVM + MultiImp | 0.556 | 0.701 | ||||
| I2-SVMp+ | 0.596 | 0.714 | ||||
| 2 | S. Chen et al. [26] | The study implemented kNN imputation to address missing values and feature selection based on three different approaches: single- and multi-factor regression, LASSO, and Random Forest (RF) with various algorithms for building predictive models. | LR | 0.634 | 0.324 | |
| CART | 0.594 | 0.486 | ||||
| XGBoost | 0.547 | 0.387 | ||||
| NB | 0.586 | 0.617 | ||||
| SVM | 0.562 | 0.189 | ||||
| RF | 0.575 | 0.293 | ||||
| 3 | K. Psychogyios et al. [27] | The study utilized various imputation techniques to address missingness and introduced an improved neighborhood aware autoencoder (I-NAA) and improved generative adversarial imputation networks (I-GAINs). They built predictive models using a simple random forest (RF) algorithm. | RF + SimpleImp | 0.4489 | ||
| RF + kNN-Imp | 0.4567 | |||||
| RF + MICE-Imp | 0.4421 | |||||
| RF + MF-Imp | 0.4553 | |||||
| RF + NAA | 0.4632 | |||||
| RF + I-NAA | 0.4799 | |||||
| RF + GAIN | 0.4672 | |||||
| RF + I-GAIN | 0.4755 | |||||
| 4 | R. C. Pereira et al. [28] | The study introduced the use of partial multiple imputation with a variational autoencoder (PMIVAE) and a denoising autoencoder (DAE) for missing values. They built predictive models using various algorithms. | ANN + PMIVAE | 0.539 | ||
| kNN + kNN-Imp | 0.481 | |||||
| RF + PMIVAE | 0.534 | |||||
| SVM + DAE | 0.538 | |||||
| 5 | Our current study | The study utilized various imputation techniques with coordinated encoding and standardization. We then built predictive models using XGBoost. | XGBoost + MeanImp | 0.626 | 0.4399 | 0.525 |
| XGBoost + MICE | 0.647 | 0.455 | 0.545 | |||
| XGBoost + kNN-Imp | 0.619 | 0.476 | 0.521 | |||
| XGBoost + RF-Imp | 0.624 | 0.453 | 0.519 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hidayaturrohman, Q.A.; Hanada, E. Impact of Data Pre-Processing Techniques on XGBoost Model Performance for Predicting All-Cause Readmission and Mortality Among Patients with Heart Failure. BioMedInformatics 2024, 4, 2201-2212. https://doi.org/10.3390/biomedinformatics4040118
Hidayaturrohman QA, Hanada E. Impact of Data Pre-Processing Techniques on XGBoost Model Performance for Predicting All-Cause Readmission and Mortality Among Patients with Heart Failure. BioMedInformatics. 2024; 4(4):2201-2212. https://doi.org/10.3390/biomedinformatics4040118
Chicago/Turabian StyleHidayaturrohman, Qisthi Alhazmi, and Eisuke Hanada. 2024. "Impact of Data Pre-Processing Techniques on XGBoost Model Performance for Predicting All-Cause Readmission and Mortality Among Patients with Heart Failure" BioMedInformatics 4, no. 4: 2201-2212. https://doi.org/10.3390/biomedinformatics4040118
APA StyleHidayaturrohman, Q. A., & Hanada, E. (2024). Impact of Data Pre-Processing Techniques on XGBoost Model Performance for Predicting All-Cause Readmission and Mortality Among Patients with Heart Failure. BioMedInformatics, 4(4), 2201-2212. https://doi.org/10.3390/biomedinformatics4040118






