Drug Repurposing for Amyotrophic Lateral Sclerosis Based on Gene Expression Similarity and Structural Similarity: A Cheminformatics, Genomic and Network-Based Analysis
Abstract
1. Introduction
2. Materials and Methods:
2.1. Part-1: Selection of Microarray Datasets and Gene Expression Analysis
2.1.1. Inclusion Criteria
- Datasets satisfying all the following criteria were selected
- Datasets with controls and ALS
- Datasets with expressional arrays
- Datasets studied in “Homo sapiens”
- Datasets with minimum of 2 samples in each category i.e., control and ALS
- Datasets with blood/brain samples
2.1.2. Exclusion Criteria
- Datasets with following criteria were excluded.
- Drug treated datasets.
- Datasets from other organisms
- Datasets with no details about controls
- Mutation studies
2.1.3. Gene Expression Analysis
2.2. Part-2: Creation of Drug Library Based on Structurally and Gene Expression Similarity
2.3. Part-3: Construction of Drug-Target-Network (DTN) and Functional Enrichment Analysis
3. Results
3.1. Part 1: Gene Expression Analysis
3.2. Part 2: Creation of Drug Library Based on Structural and Gene Expression Similarity
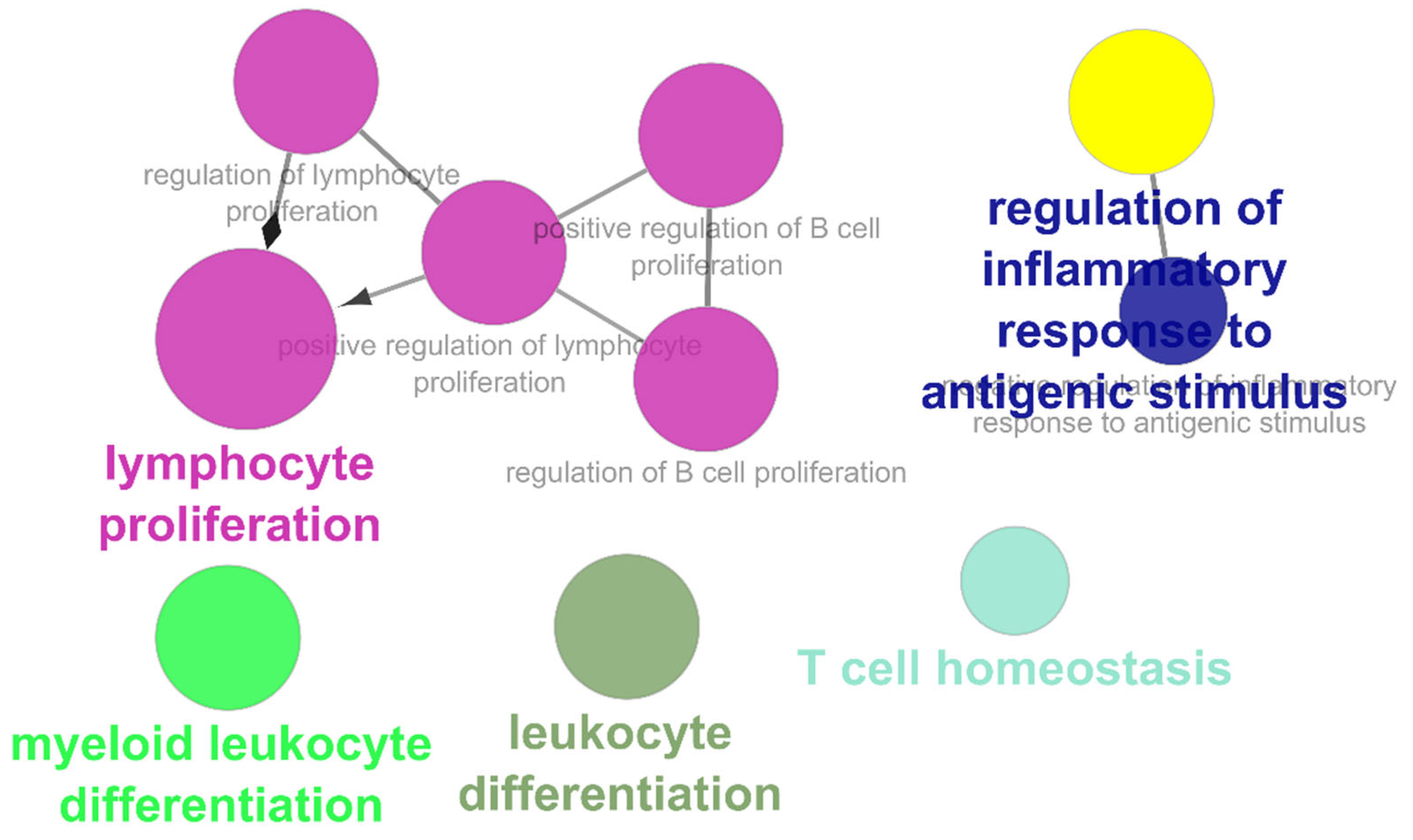
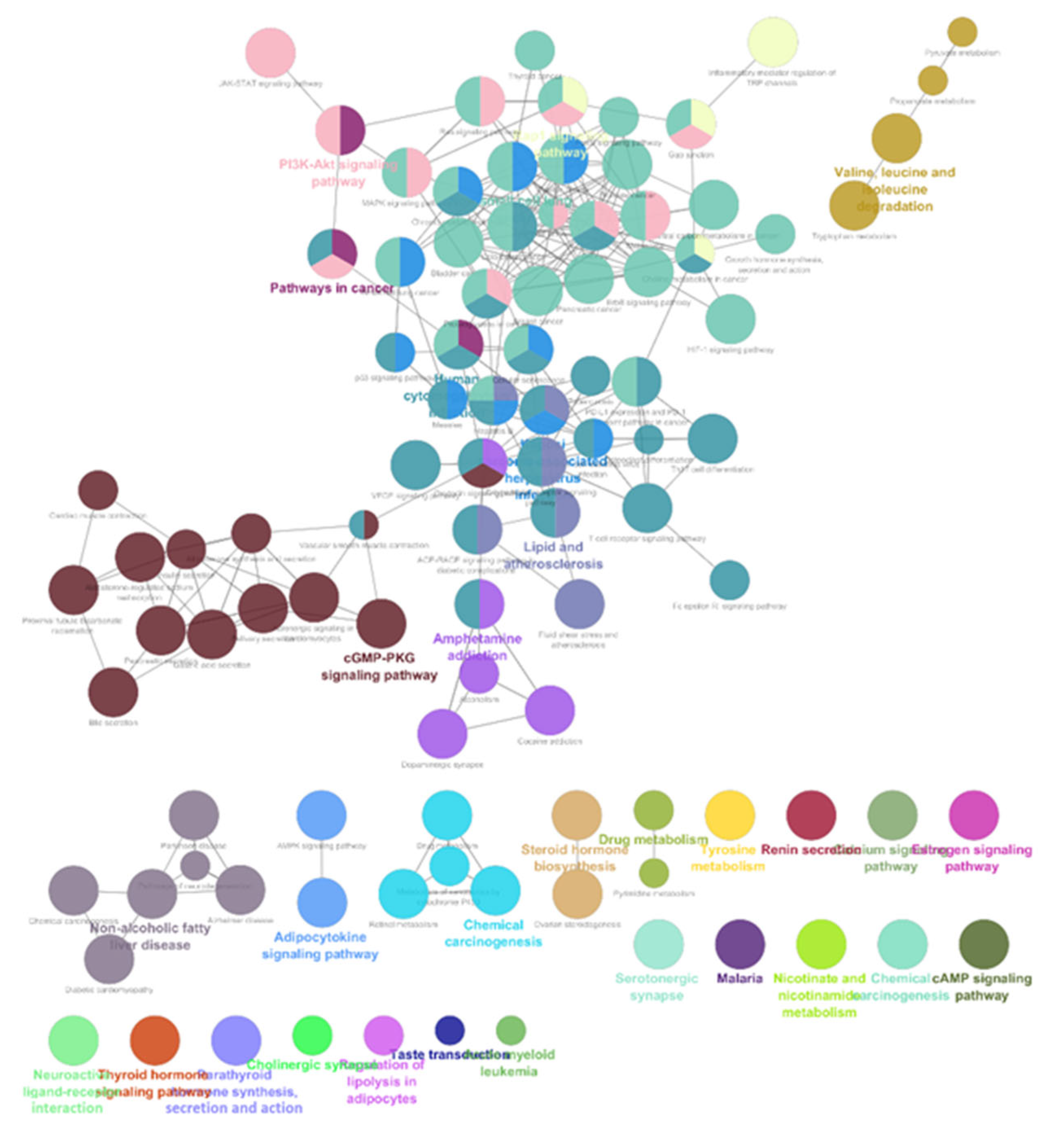
3.3. Part-3: Construction of DTN and Functional Enrichment Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lemos, J.P.; Tenório, L.P.G.; Mouly, V.; Butler-Browne, G.; Mendes-da-Cruz, D.A.; Savino, W.; Smeriglio, P. T cell biology in neuromuscular disorders: A focus on Duchenne Muscular Dystrophy and Amyotrophic Lateral Sclerosis. Front. Immunol. 2023, 14, 1202834. [Google Scholar] [CrossRef] [PubMed]
- Feldman, E.L.; Goutman, S.A.; Petri, S.; Mazzini, L.; Savelieff, M.G.; Shaw, P.J.; Sobue, G. Amyotrophic lateral sclerosis. Lancet 2022, 400, 1363–1380. [Google Scholar] [CrossRef] [PubMed]
- Van Es, M.A.; Hardiman, O.; Chio, A.; Al-Chalabi, A.; Pasterkamp, R.J.; Veldink, J.H.; Van den Berg, L.H. Amyotrophic lateral sclerosis. Lancet 2017, 390, 2084–2098. [Google Scholar] [CrossRef] [PubMed]
- Amyotrophic Lateral Sclerosis: Disease State Overview. Available online: https://www.ajmc.com/view/amyotrophic-lateral-sclerosis-disease-state-overview (accessed on 30 October 2023).
- Amyotrophic Lateral Sclerosis (ALS)—Symptoms and Causes—Mayo Clinic. Available online: https://www.mayoclinic.org/diseases-conditions/amyotrophic-lateral-sclerosis/symptoms-causes/syc-20354022 (accessed on 30 October 2023).
- Longinetti, E.; Fang, F. Epidemiology of amyotrophic lateral sclerosis: An update of recent literature. Curr. Opin. Neurol. 2019, 32, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Riancho, J.; Gonzalo, I.; Ruiz-Soto, M.; Berciano, J. Why do motor neurons degenerate? Actualization in the pathogenesis of amyotrophic lateral sclerosis. Neurologia 2019, 34, 27–37. [Google Scholar] [CrossRef] [PubMed]
- CenterWatch. Available online: https://www.centerwatch.com/search?utf8=✓&q=amyotrophic+lateral+sclerosis&taxonomy=FDA+Approved+Drugs&datatype=directory&button=&condition=&therapeutic=&companies=&approvalyear=&tax_id=1067 (accessed on 30 October 2023).
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.I.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Bioconductor—EnhancedVolcano. Available online: https://bioconductor.org/packages/release/bioc/html/EnhancedVolcano.html (accessed on 30 October 2023).
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, 1074–1082. [Google Scholar] [CrossRef] [PubMed]
- rdkit/rdkit: 2023_09_2 (Q3 2023) Release. Available online: https://zenodo.org/records/10099869 (accessed on 30 October 2023).
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Lerner, J.; Brunet, J.-P.; Subramanian, A.; Ross, K.A.; et al. The connectivity map: Using gene-expression signatures to connect small molecules, genes, and disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M.; von Mering, C.; Campillos, M.; Jensen, L.J.; Bork, P. STITCH: Interaction networks of chemicals and proteins. Nucleic Acids Res. 2008, 36, D684–D688. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Nakaya, A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002, 30, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Dangond, F.; Hwang, D.; Camelo, S.; Pasinelli, P.; Frosch, M.P.; Stephanopoulos, G.; Stephanopoulos, G.; Brown, R.H., Jr.; Gullans, S.R. Molecular signature of late-stage human ALS revealed by expression profiling of postmortem spinal cord gray matter. Physiol. Genom. 2004, 16, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Bakay, M.; Wang, Z.; Melcon, G.; Schiltz, L.; Xuan, J.; Zhao, P.; Sartorelli, V.; Seo, J.; Pegoraro, E.; Angelini, C.; et al. Nuclear envelope dystrophies show a transcriptional fingerprint suggesting disruption of Rb-MyoD pathways in muscle regeneration. Brain 2006, 129, 996–1013. [Google Scholar] [CrossRef] [PubMed]
- Dadgar, S.; Wang, Z.; Johnston, H.; Kesari, A.; Nagaraju, K.; Chen, Y.W.; Hill, D.A.; Partridge, T.A.; Giri, M.; Freishtat, R.J.; et al. Asynchronous remodeling is a driver of failed regeneration in Duchenne muscular dystrophy. J. Cell Biol. 2014, 207, 139–158. [Google Scholar] [CrossRef] [PubMed]
- Clinicaltrials.gov. 2018. Available online: https://clinicaltrials.gov/ct2/show/NCT02002819?term=levetiracetam&cond=Alzheimer+Disease&rank=2 (accessed on 30 October 2023).
- Zhang, J.J.; Zhou, Q.M.; Chen, S.; Le, W.D. Repurposing carbamazepine for the treatment of amyotrophic lateral sclerosis in SOD1-G93A mouse model. CNS Neurosci. Ther. 2018, 24, 1163–1174. [Google Scholar] [CrossRef] [PubMed]
- Chaytow, H.; Carroll, E.; Gordon, D.; Huang, Y.T.; Van Der Hoorn, D.; Smith, H.L.; Becker, T.; Becker, C.G.; Faller, K.M.E.; Talbot, K.; et al. Targeting phosphoglycerate kinase 1 with terazosin improves motor neuron phenotypes in multiple models of amyotrophic lateral sclerosis. EBioMedicine 2022, 83. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Li, M.; Ye, Z.; Hu, X.; He, X.; Wang, J.; Chen, G.; Zou, C.; Xu, D.; Zhang, H.; et al. Elevated peripheral levels of receptor-interacting protein kinase 1 (RIPK1) and IL-8 as biomarkers of human amyotrophic lateral sclerosis. Signal Transduct. Target. Ther. 2023, 8, 451. [Google Scholar] [CrossRef] [PubMed]


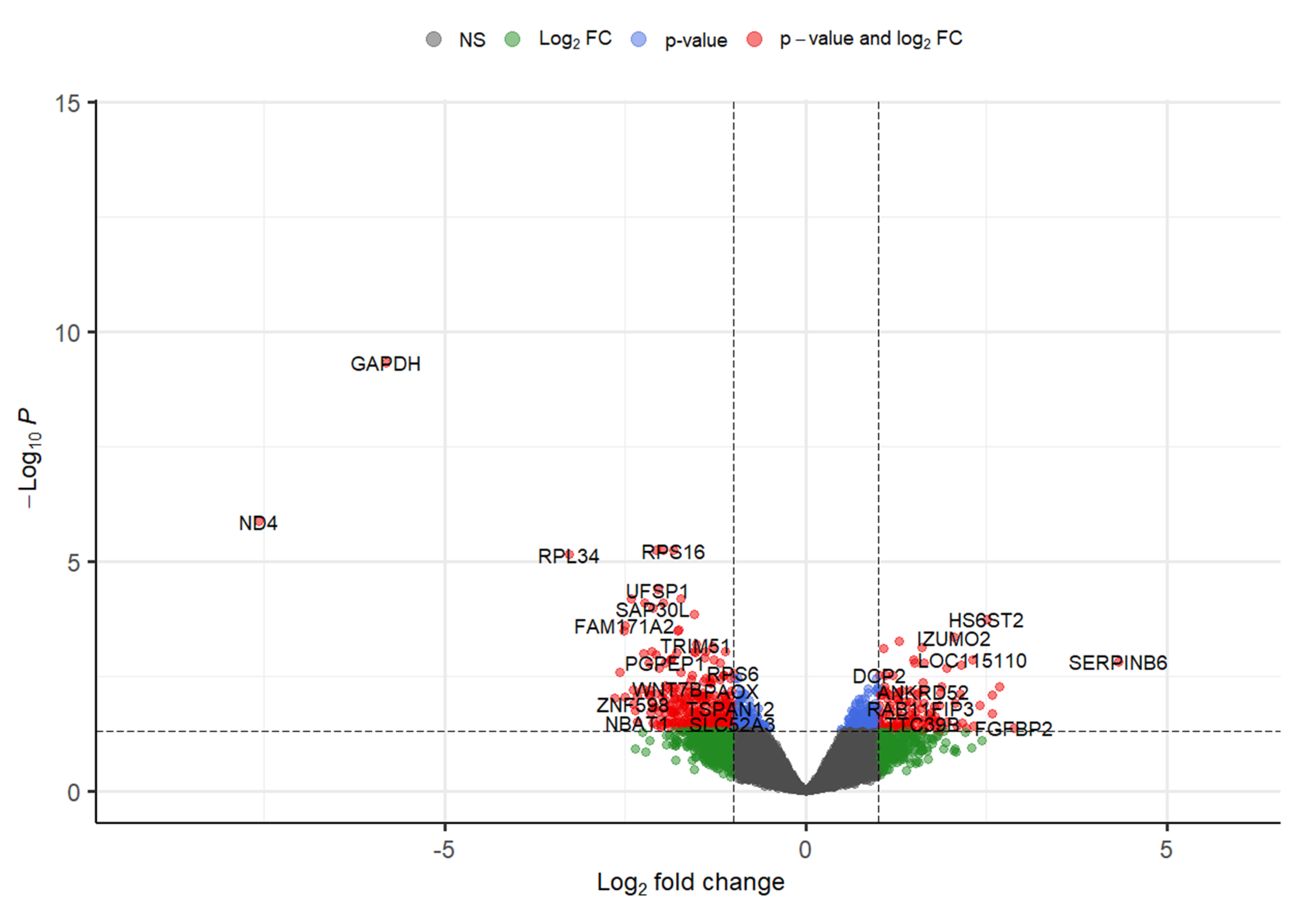
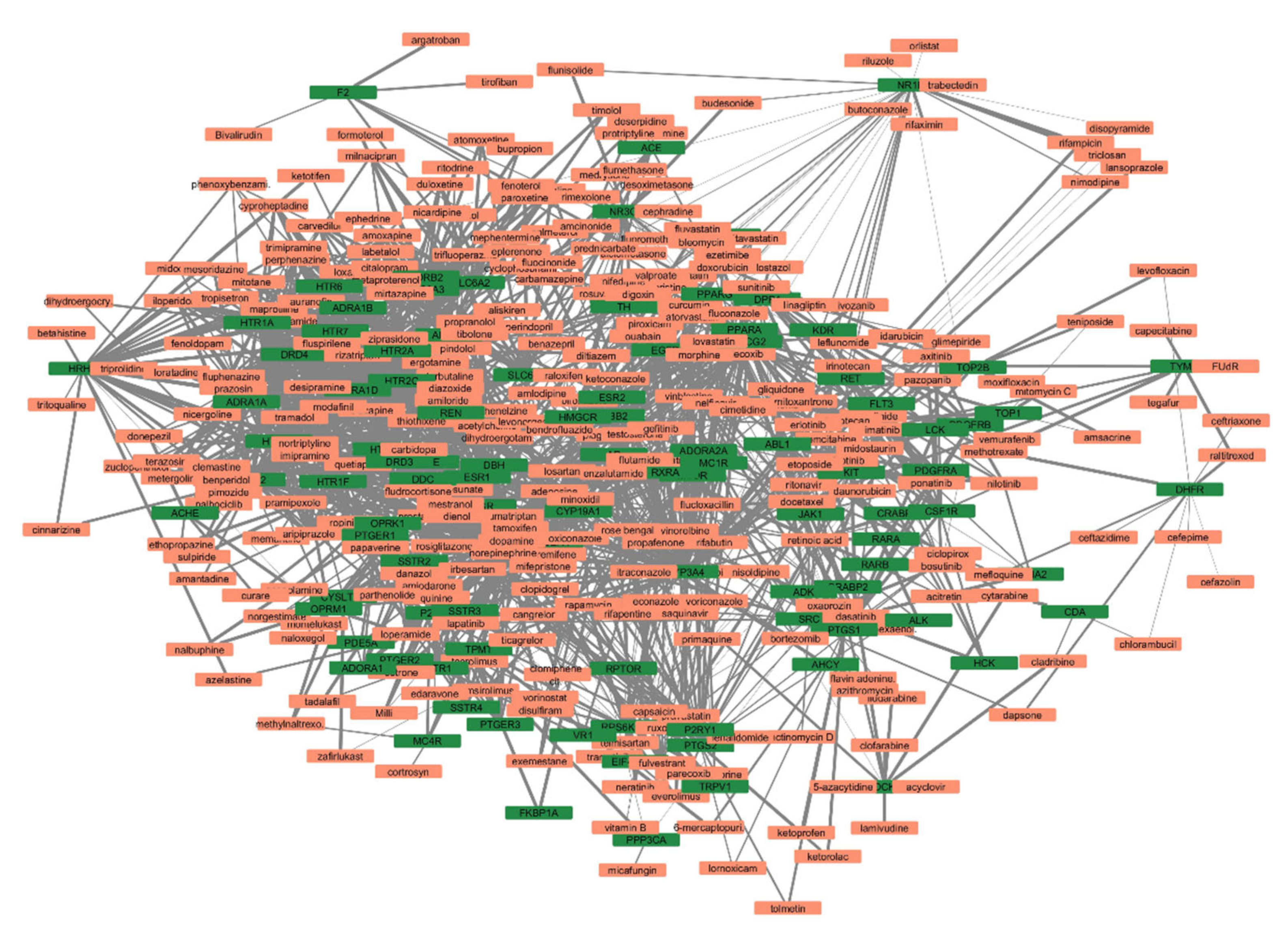

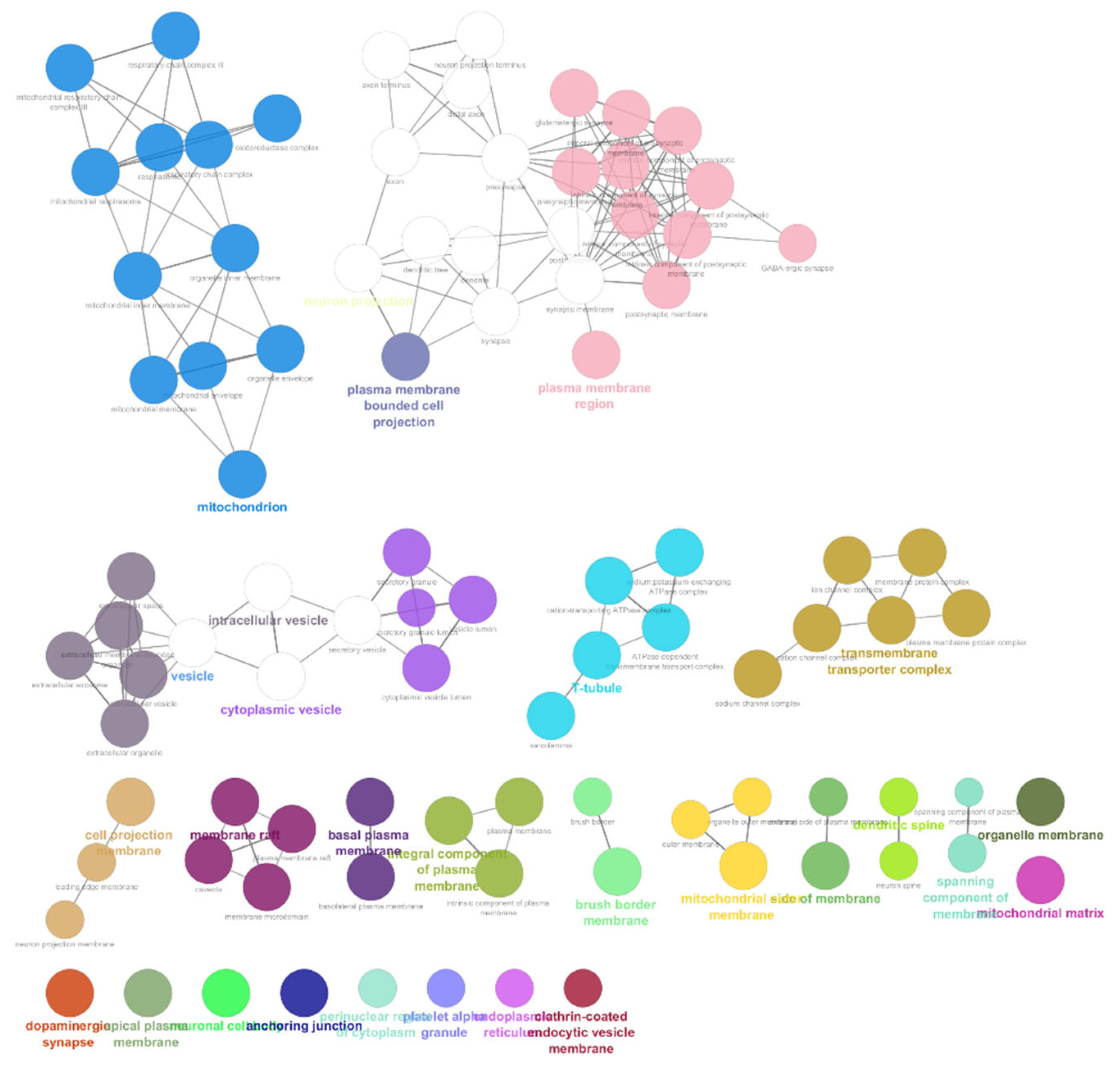
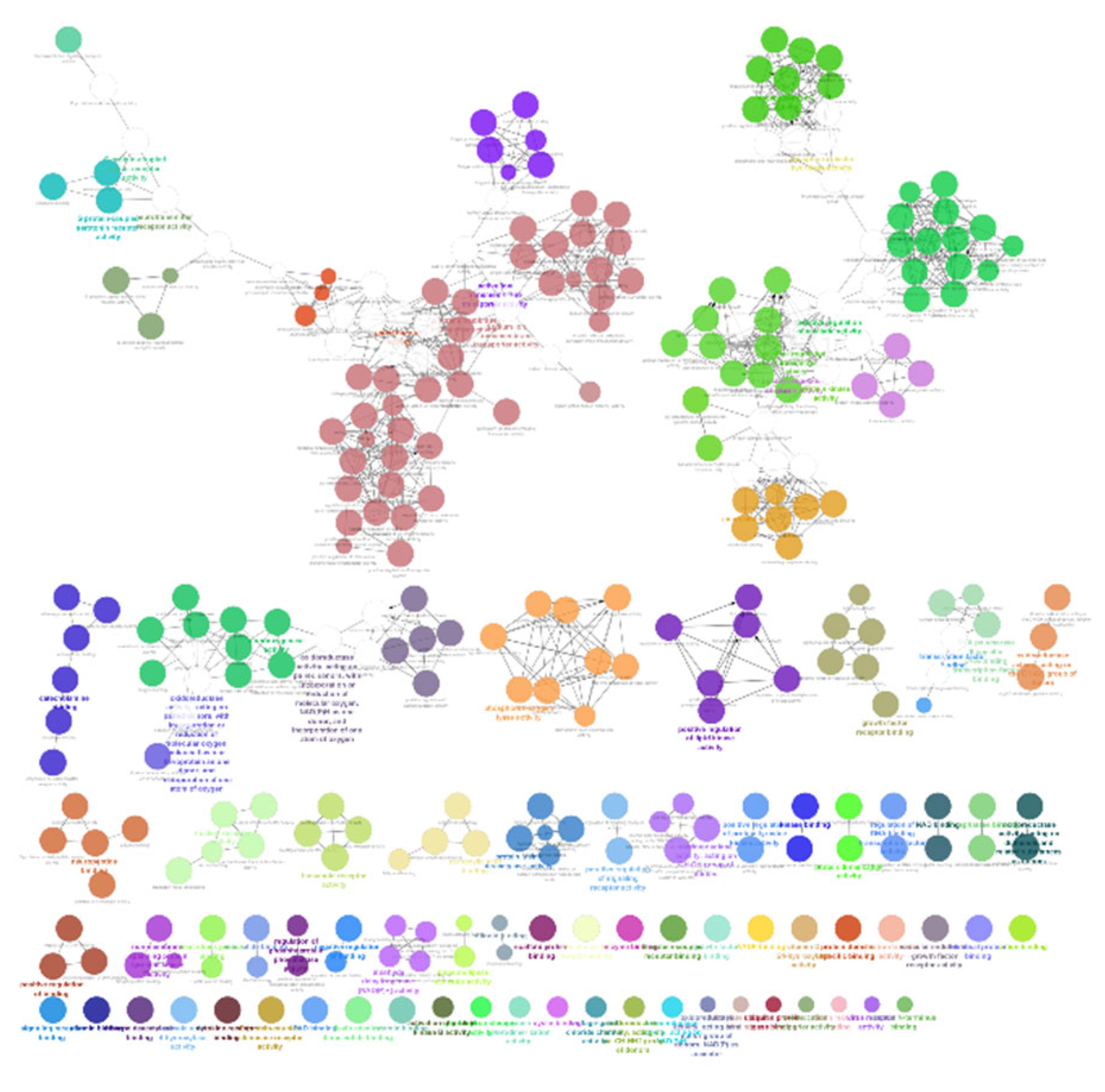
| Dataset Accession Number | Pubmed Reference | Number of Cases | Number of Controls | Genetic Source | Genotyping Platform | Genotyping Method |
|---|---|---|---|---|---|---|
| GSE833 [18] | 14645737 | 7 | 4 | Brain tissue | GPL80 | QRT-PCR |
| GSE3307 [19,20] | 16478798, 25313409 | 9 | 7 | Brain tissue | GPL97 | RT-PCR |
| Name of the Compound | Category | Phase of Clinical Trial | NCT Number |
|---|---|---|---|
| Riluzole | Approved | NA | NA |
| Edavarone | Approved | NA | NA |
| Ceftriaxone | Clinical trials | Phase 3 | NCT00349622 |
| TRO19622 | Clinical trials | Phase 2|3 | NCT01285583 |
| Escitalopram | Clinical trials | Phase 3 | NCT00965497 |
| Zilucoplan | Clinical trials | Phase 2|3 | NCT04436497 |
| Tirasemtiv | Clinical trials | Phase 3 | NCT02496767, NCT02936635 |
| Vitamin E | Clinical trials | Phase 3 | NCT00372879 |
| Arimoclomol | Clinical trials | Phase 3 | NCT00706147, NCT03491462 |
| Mexiletine | Clinical trials | Phase 4 | NCT01811355 |
| Dexpramipexole | Clinical trials | Phase 3 | NCT01281189 |
| Memantine (Ebixa) | Clinical trials | Phase 2|3 | NCT00353665 |
| Sodium Valproate | Clinical trials | Phase 3 | NCT00136110 |
| Levosimendan | Clinical trials | Phase 3 | NCT03505021 |
| MCI-186 | Clinical trials | Phase 3 | NCT00424463, NCT00415519, NCT01492686, NCT00330681 |
| E0302 (mecobalamin) | Clinical trials | Phase 2|3 | NCT00445172, NCT00444613 |
| Pridopidine | Clinical trials | Phase 2|3 | NCT04615923 |
| Minocycline | Clinical trials | Phase 3 | NCT00047723 |
| Verdiperstat | Clinical trials | Phase 2|3 | NCT04436510 |
| Compound Name | Compound Name | Compound Name |
|---|---|---|
| acitretin | ethopropazine | phenoxybenzamine |
| alfacalcidol | etodolac | pindolol |
| aliskiren | fenoldopam | piroxicam |
| alosetron | fenoterol | pitavastatin |
| alprazolam | fludrocortisone | prazosin |
| amiodarone | flunisolide | pregnenolone |
| amlexanox | fluocinonide | prochlorperazine |
| amodiaquine | fluorometholone | promazine |
| amoxapine | flupenthixol | propafenone |
| argatroban | fluspirilene | protriptyline |
| aripiprazole | formoterol | quetiapine |
| atomoxetine | fostamatinib | ranitidine |
| auranofin | glimepiride | remoxipride |
| azelastine | halcinonide | repaglinide |
| benazepril | heroin | rifabutin |
| bendrofluazide | homatropine | rifapentine |
| benperidol | iloperidone | rifaximin |
| bepridil | ipratropium bromide | ritodrine |
| betahistine | irbesartan | rizatriptan |
| biperiden | ketoprofen | ruxolitinib |
| budesonide | ketorolac | salbutamol |
| buprenorphine | labetalol | salmeterol |
| bupropion | lansoprazole | sulpiride |
| buserelin | levonorgestrel | sumatriptan |
| calcipotriol | linagliptin | tadalafil |
| cangrelor | loperamide | tegaserod |
| carvedilol | loratadine | telmisartan |
| chlorprothixene | lornoxicam | temsirolimus |
| cilostazol | losartan | terbutaline |
| cimetidine | loxapine | terfenadine |
| cinnarizine | maprotiline | thiothixene |
| citalopram | meloxicam | tibolone |
| cladribine | mesoridazine | ticagrelor |
| clofarabine | mestranol | timolol |
| clomiphene | metergoline | tirofiban |
| clomipramine | methylnaltrexone | tocainide |
| clopidogrel | miglitol | tolcapone |
| curare | milnacipran | tolmetin |
| dapsone | minoxidil | tramadol |
| desipramine | mirtazapine | triamcinolone |
| desoximetasone | mitotane | triamterene |
| diazoxide | mitoxantrone | trifluoperazine |
| diclofenac | moclobemide | triflupromazine |
| digitoxin | nadolol | trimeprazine |
| dihydroergocryptine | nalbuphine | trimipramine |
| dihydroergotamine | nalmefene | tripelennamine |
| diltiazem | nalorphine | triprolidine |
| diphenoxylate | nefazodone | troglitazone |
| diprenorphine | nisoldipine | tropisetron |
| disopyramide | orlistat | umeclidinium |
| domperidone | oxycodone | valdecoxib |
| droperidol | oxymorphone | yohimbine |
| duloxetine | papaverine | zafirlukast |
| elagolix | parecoxib | zileuton |
| ephedrine | parthenolide | ziprasidone |
| eplerenone | perhexiline | zuclopenthixol |
| ergotamine | perindopril | |
| estrone | phenelzine |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kadena, K.; Ouzounoglou, E. Drug Repurposing for Amyotrophic Lateral Sclerosis Based on Gene Expression Similarity and Structural Similarity: A Cheminformatics, Genomic and Network-Based Analysis. BioMedInformatics 2024, 4, 1713-1724. https://doi.org/10.3390/biomedinformatics4030093
Kadena K, Ouzounoglou E. Drug Repurposing for Amyotrophic Lateral Sclerosis Based on Gene Expression Similarity and Structural Similarity: A Cheminformatics, Genomic and Network-Based Analysis. BioMedInformatics. 2024; 4(3):1713-1724. https://doi.org/10.3390/biomedinformatics4030093
Chicago/Turabian StyleKadena, Katerina, and Eleftherios Ouzounoglou. 2024. "Drug Repurposing for Amyotrophic Lateral Sclerosis Based on Gene Expression Similarity and Structural Similarity: A Cheminformatics, Genomic and Network-Based Analysis" BioMedInformatics 4, no. 3: 1713-1724. https://doi.org/10.3390/biomedinformatics4030093
APA StyleKadena, K., & Ouzounoglou, E. (2024). Drug Repurposing for Amyotrophic Lateral Sclerosis Based on Gene Expression Similarity and Structural Similarity: A Cheminformatics, Genomic and Network-Based Analysis. BioMedInformatics, 4(3), 1713-1724. https://doi.org/10.3390/biomedinformatics4030093







