1. Introduction
Musculoskeletal ultrasound (MSK-US) is a type of medical imaging used to non-invasively assess the health of bones, muscles, tendons, and ligaments. MSK-US is preferable over alternatives such as magnetic resonance imaging given its lower-cost, bedside point of care, and safety [
1]. One prevalent application of MSK-US is the assessment of knee osteoarthritis (OA), a common degenerative joint disease affecting over 25% of adults in the United States [
2]. Effusion and thickened synovium have been identified as key indicators of OA, contributors to the disease’s development, and potential targets for its treatment. Effusion refers to the increase of fluid within the joint space. This may cause the joint recess to distend and inflammation (thickening) of the synovial membrane lining the joint space. The latter is known as synovitis, and may lead to further buildup of synovial fluid in the joint space, worsening the effusion [
3]. Joint recess distension and thickened synovium may occur together or separately. In
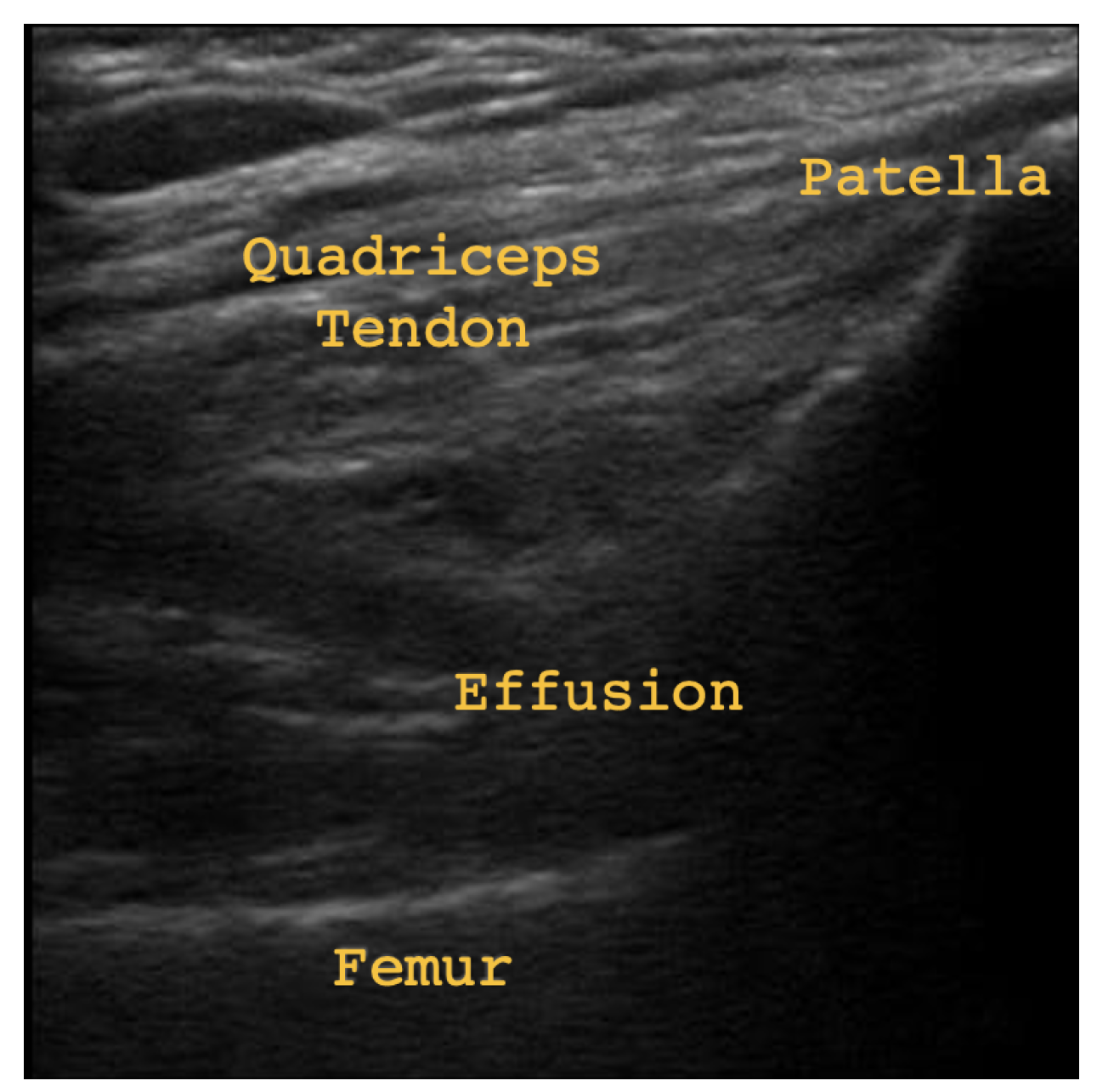
Figure 1 some subtle effusion may be discerned by darker patches (echo-free space) in the region labeled Effusion, indicating joint recess distension. In
Figure 2, thickened synovium and significantly more effusion may be discerned by the clear presence of echo-free space in the area labeled Effusion/Thickened Synovium. The presence of echo-free space is an indicator of fluid, since the waves emitted by the ultrasound probe are absorbed instead of reflected [
3]. Therefore,
Figure 2 provides evidence of both joint recess distension and thickened synovium.
Assessing thickened synovium in the presence of joint recess distension is very difficult [
4]. Hence, a new procedure to help clinicians more confidently assess joint recess distension and thickened synovium would help over 50 million patients suffering from OA within only the United States. This work develops such a procedure, incorporating recent advancements in machine learning (ML).
ML has become integral to the development of new diagnostic procedures in medical imaging [
5]. These models help to increase the accessibility of medicine, aid in the interpretation of clinical data, and expedite the rate at which diagnostic results are returned to patients. In the domain of medical imaging, convolutional neural networks (CNNs) have demonstrated success in the segmentation and classification of tumors, lesions, cancers, COVID-19, and many other medical conditions [
6]. These networks consist of a series of locally-connected layers (called convolutional layers), and fully-connected layers. Each convolutional layer applies a linear filter across the previous layer’s output, or across the input in the case of the first layer. These filters are learned during training, and can be thought of as feature detectors, where each filter tries to extract some defining characteristic of the image (edges, shapes, boundaries). Layers are connected by non-linear activation functions, increasing the complexity of the representations learnable by the network. Once the input moves through every convolutional layer, its final representation is passed to a sequence of fully-connected layers to obtain the output [
6]. The type of output depends on the task. In the diagnosis of cancer, for example, it is usually the probability that a given patient has cancer based on the input image.
CNNs are an example of supervised learning, whereby they are trained on input and output pairs. The performance and reliability of these models is highly dependent on the quality and quantity of their training data. Yet, large datasets are not feasibly obtained in medical imaging due to the ethical regulations protecting patient data, the rarity of certain medical conditions, and the costs of data labeling and healthcare [
5]. Therefore, numerous methods have been proposed to enrich datasets without requiring new patient data or annotations for that data. This paper will compare two such methods, using a MSK-US dataset to train a CNN for the diagnosis of thickened synovium and joint recess distension. The methodology and findings we present are relevant for other neural network architectures, such as vision transformers, and is not restricted to CNNs. We have decided to focus on neural networks and other supervised approaches over semi-supervised and unsupervised learning, which require less or no labeled data, due to their demonstrated success and fast growing prevalence in medical imaging, as well as other fields.
The first method is augmentation and involves enriching the model’s training set with transformations of existing images. Typically, these transformations include rotation, vertical and horizontal flipping, brightness and hue adjustment, and other similar geometric, intensity, and color transformations [
7]. With augmentation, the model learns potentially natural variations in the data to improve the robustness of its decision making.
Although augmentation is widely used, augmented images may be too similar to existing images, and not offer the model any additional information about the conditional distribution of outputs given inputs [
5]. Furthermore, augmented images may not be reflective of the inputs received by the model for inference. For example, in medical imaging, augmentation can suppress important biological markers in the training images, reducing the model’s reliability on un-augmented inputs from real patients. Hence, to do better than traditional augmentation, several generative ML models have been developed to synthesize new images as opposed to modifying existing ones. Ideally, synthetic images should contain the defining attributes of real images, while offering sufficient diversity to provide the model with new information about the distribution it must learn.
Generative ML models are characterized by their ability to sample from the unknown data generating distribution of the observed data. In some cases, it is possible to learn the parameters of this unknown distribution directly using maximum likelihood or maximum a posteriori estimation, and then a sampling procedure can be defined over the distribution. In the case of images, however, these distributions may be impossible to sample from directly by being extremely high dimensional. Alternatively, energy-based models learn the parameters of an un-normalized density function to avoid having to compute the normalizing constant (usually an intractable and high dimensional integral) [
8]. Finally, variational inference estimates the data generating distribution using a simpler distribution (such as a Gaussian), at the cost of any mathematical guarantees on the goodness of the approximation. These challenges with probabilistic models arising from the curse of dimensionality, have encouraged the development of numerous deep learning (DL) methods for generating synthetic images [
9]. One example is the generative adversarial network (GAN).
A GAN is a DL model capable of synthesizing high-quality images. It consists of a generator whose goal is to produce images that will fool the discriminator, a neural network trained to classify images as real or fake [
5]. Once the generator’s synthetic images are consistently classified as real by the discriminator, they can be used to enrich a dataset. In [
10], a GAN was used to generate MSK-US images of the gastrocnemius medialis muscle. They showed that synthetic images were similar in pixel intensity distribution to real images, and contained the desired muscle architecture. However, as pointed out by [
11], they did not train a DL model on the synthetic images to determine how these images might affect the performance of such a model in a real-world diagnostic task.
In [
11], they used a de-noising diffusion probabilistic model (DDPM) to generate MSK-US images opposed to a GAN. The advantage of DDPMs over GANs is their ability to generate equally high quality samples with greater diversity, ease of training, and avoidance of mode collapse, where the generator repeatedly produces the same set of images that have been found to fool the discriminator [
12]. DDPMs are trained via a forward process, where the model learns to predict the random Gaussian noise added to an image. New samples are then synthesized via a reverse process, beginning with an image of pure Gaussian noise, and incrementally de-noising it over a sequence of time steps until a noise-free image is obtained [
9]. In [
11], they synthesized images of muscle texture for the tibialis anterior, rectus femoris, gastrocnemius, and biceps brachii and brachialis anterior muscles using a DDPM trained on 1223 MSK-US images. Their synthetic images were found to be similar in pixel intensity distribution to the real images and depicted desired muscle textures. Finally, they trained a DL segmentation model on a mixture of real and synthetic images, resulting in a Dice coefficient and IoU 0.01 greater than the same model trained only on real images, indicating that the synthetic images marginally improved the model’s performance.
This work extends [
11], and goes beyond generating muscle texture by applying a DDPM to the significantly more difficult task of generating MSK-US images of synovial thickening and joint recess distension in the knee joint space. Moreover, it directly compares two methods to enrich a MSK-US dataset. First, we augment a subset of existing images using geometric and intensity transformations. Second, we train two DDPMs to generate brand-new synthetic images. The similarity between real, synthetic, and augmented images is assessed qualitatively and quantitatively. Then, the usefulness of the images is evaluated in a challenging clinical task, where we train a CNN using the enriched datasets to diagnose thickened synovium and compare its decision-making against human opinion.
To our knowledge, we are the first to compare augmentation and diffusion-based image synthesis for enriching medical imaging datasets. Our purpose is to determine the most appropriate methodology for enriching medical imaging datasets to train DL models in challenging clinical tasks. We therefore set the following objectives:
Train two DDPMs to generate MSK-US images of synovial thickening and joint recess distension.
Compare the performance of a DL model at diagnosing synovial thickening when trained on datasets of real images, augmented images, and synthetic images.
Offer conclusions regarding the usefulness of augmentation and diffusion-based image synthesis to train DL models within the context of medical imaging.
The contributions of this work are:
Provide evidence of the utility of dataset enrichment methods for deep learning in an important and challenging medical imaging problem.
Directly evaluate diffusion-based dataset enrichment against augmentation-based dataset enrichment in the context of medical imaging.
Facilitate the interpretability of deep learning models in medical imaging by comparing the decision-making heuristics of trained models against human opinion.
4. Discussion
Our work compares two methods to enrich a MSK-US dataset for thickened synovium and joint recess distension. First, we augment a subset of existing images using geometric and intensity transformations. Second, we train two DDPMs to generate brand-new synthetic images. The similarity between real, synthetic, and augmented images is assessed qualitatively and quantitatively. Then, the usefulness of the images is evaluated in a challenging clinical task, where we train a CNN using the enriched datasets to diagnose thickened synovium and compare its decision-making against human opinion.
Given the synthetic and augmented images, a qualitative analysis suggested that the synthetic images appear noticeably brighter than real and augmented images. This observation was reflected in the mean of the pixel intensity histogram for synthetic images being significantly greater than the means of the histograms for real and augmented images. Furthermore, synthetic images exhibited less contrast, which was again reflected in the standard deviation of the histogram for synthetic images being smaller than the standard deviations of the histograms for real and augmented images. These observations are consistent with [
11], where they also found the synthetic MSK-US images from a DDPM to be brighter than their real counterparts to some extent. In [
19], they found that generating extremely bright or extremely dark images is a general problem with diffusion models stemming from the fact that variance schedules do not produce pure noise by the last time step
T.
The near perfect correspondence between pixel intensity histograms for real and augmented images is due to many augmentations being geometric transformations. Unlike intensity or color transformations, geometric transformations do not affect the pixel intensity histogram of an image. Importantly, however, geometric transformations may reposition or conceal biological landmarks, meaning the augmented image becomes difficult to interpret and inconsistent with real images. This is particularly a concern for the CNN, which may then be encouraged to learn incorrect representations when trained on augmented images. In contrast, synthetic images portray more consistent anatomical structures than augmented images and contain resemblances to the three landmarks of the suprapatellar longitudinal view: The patella, femur, and quadriceps tendon. We emphasize, however, that the generation of anatomically accurate synthetic images was not a primary objective of our work and acknowledge that the synthetic images contain anatomical inaccuracies.
Next, similarity between images was assessed quantitatively using the PSNR, SSIM, and LPIPS metrics. Across all metrics, the similarity between augmented and real images was higher than between synthetic and real images. This was expected given that augmented images do not contain any new content. Of the three metrics, PSNR suggested that augmented images were approximately twice as similar to real images as synthetic images were to real images. This is because PSNR heavily penalized the brightness and contrast discrepancies across real and synthetic images. Even so, according to SSIM, augmented images were more similar by only 0.014 on average to real images than synthetic images were to real images. Likewise, according to LPIPS, augmented images were more similar by only 0.1183 on average to real images than synthetic images were to real images. Given that SSIM and LPIPS are most representative of human perceived similarity, these values should be attributed more importance than PSNR. It is worth noting, however, that MSK-US images consist mostly of black and gray pixels, and SSIM has been shown to severely underestimate similarity when comparing dark images [
20]. Furthermore, our PSNR, SSIM, and LPIPS all suggest significantly less similarity between real and synthetic images than in [
11]. We attribute this to the fact that suprapatellar longitudinal view is visually much more complex than their images of muscle textures. This complexity arises from the structures within the images, but also the greater variability across the images. Hence, the diffusion model is prone to performing worse than in [
11]. Nevertheless, applying diffusion models to a more challenging learning problem, even for humans, was one of the original motivators of our work. A final point is that a higher dissimilarity between real and synthetic images is not necessarily a limitation of synthetic images but may suggest that they are more diverse than augmented images. Capturing greater diversity was an original motivator for choosing DDPMs, since it provides the CNN with more information about the conditional distribution over labels given inputs.
The most important contribution of our work is the training of a CNN to perform the challenging clinical task of detecting synovial thickening in the presence of joint recess distension, with reasonable accuracy and precision. Five variants of this CNN were trained across our five experiments. In each experiment, the network was trained on a dataset of un-enriched or enriched images, using five-fold cross validation. This allowed us to directly compare its performance when trained on augmented and synthetic images. The results revealed that the CNN trained on an enriched dataset of synthetic images demonstrated maximal performance in all but one metric, across validation sets and test set. Moreover, the small value of the standard deviation for validation accuracy suggests that the model was consistent and reliable throughout the entire training set. This is a necessary condition for the model to be deployable in a clinical setting. Another desirable property demonstrated by our model trained with 20% synthetic images was its preference for sensitivity, which is vital in the medical imaging domain to ensure patients in need do not go undetected. In summary, our experimental results coincide with [
11], where their DL model trained on a mixture of synthetic and real images outperformed the base model (trained only on real images). As in [
11], the improvement offered by the synthetically enriched dataset is small (1%), however, the difficulty of detecting synovial thickening underscores the practical relevance of our results.
The fact that the synthetically enriched dataset yielded a better model than any augmented dataset can be attributed to the qualitative and quantitative comparisons made between synthetic and augmented images. To reiterate, synthetic images depict anatomical structures and landmarks with greater consistency than augmented images, exposing the model to more images representative of those it will receive from real patients. Therefore, the anatomical inaccuracies in synthetic images do not necessarily detract from their value in training a DL model, which was one of the primary objectives of our work. Moreover, synthetic images are more diverse, providing the model with more information about the conditional distribution of outputs given inputs. All of this is to say that synthetic images help the CNN achieve greater generalizability.
In our experiments, unlike the synthetic model which outperformed the base model across both validation and test sets, the augmented models performed unpredictably. To be exact, the model trained with 20% augmentation performed worse than the base model across both validation and test sets. Yet, it performed better on average across the validation sets than the model trained on images augmented at runtime, which in turn performed better than the base model over the test set. This unreliable performance makes augmentation an undesirable choice in high-risk clinical applications where lives are at stake. In our opinion, these observations are a result of the visual inconsistencies and distortions introduced by augmentation. The lack of realism in some of the augmented images may lead the model to learn incorrect representations, and thereby explain why the base model might outperform one trained with augmentation. In Ex. 5, the model was trained with both synthetic images and augmentation at runtime, and its performance decreased across validation and test sets compared to the models in Ex. 3 and Ex. 4. Augmentation at runtime likely decreased performance in these cases by devaluing both the real and synthetic images, as well as increasing the standard deviation of the model’s accuracy, whereby the model was unable to benefit from the synthetic images and experienced greater variability in its performance. In summary, through these observations we show that augmentation may degrade model performance in certain medical imaging tasks.
To rigorously understand and validate our CNN’s performance, heat maps were used to verify if points of interest identified by the model aligned with human opinion, and if these points were the same across real and synthetic images. The heat maps illustrated that the CNN attributed importance to relevant regions, near areas of noticeable effusion. Moreover, the regions identified in synthetic images matched those identified in real images for similar views. These observations suggest that the CNN’s decision-making is aligned with human opinion, and that the synthetic images maintain enough biological and visual fidelity for thickened synovium and joint recess distension to be identified consistently.
In summary, the reasons above indicate that diffusion-based image synthesis is preferable over traditional augmentation to enrich medical imaging datasets of DL models. Our results also substantiate the findings of [
11]. We proposed a successful methodology to enrich a MSK-US dataset for training a CNN in a diagnostic task, and demonstrated that traditional augmentation may actually degrade model performance at such a task. However, our study has limitations. Using augmentation at runtime, dictated by probabilities, means that it is impossible to verify the number of augmented images present in the training set. Although, this is the standard approach. Moreover, future work would incorporate attention block fine-tuning opposed to traditional fine-tuning to potentially reduce over-fitting [
21]. Furthermore, experimentation involving de-noising diffusion implicit models (DDIMs) would help compare the sampling quality between DDIMs and DDPMs, to determine the extent of the trade off between sample quality and improved sampling speed in the context of medical imaging [
22]. Our choice of 20% synthetic enrichment was influenced by the computational cost and time required to sample a DDPM. This slow iterative sampling process is a limitation of DDPMs addressed by DDIMs.