Bioinformatics-Based Identification of Human B-Cell Receptor (BCR) Stimulation-Associated Genes and Putative Promoters
Abstract
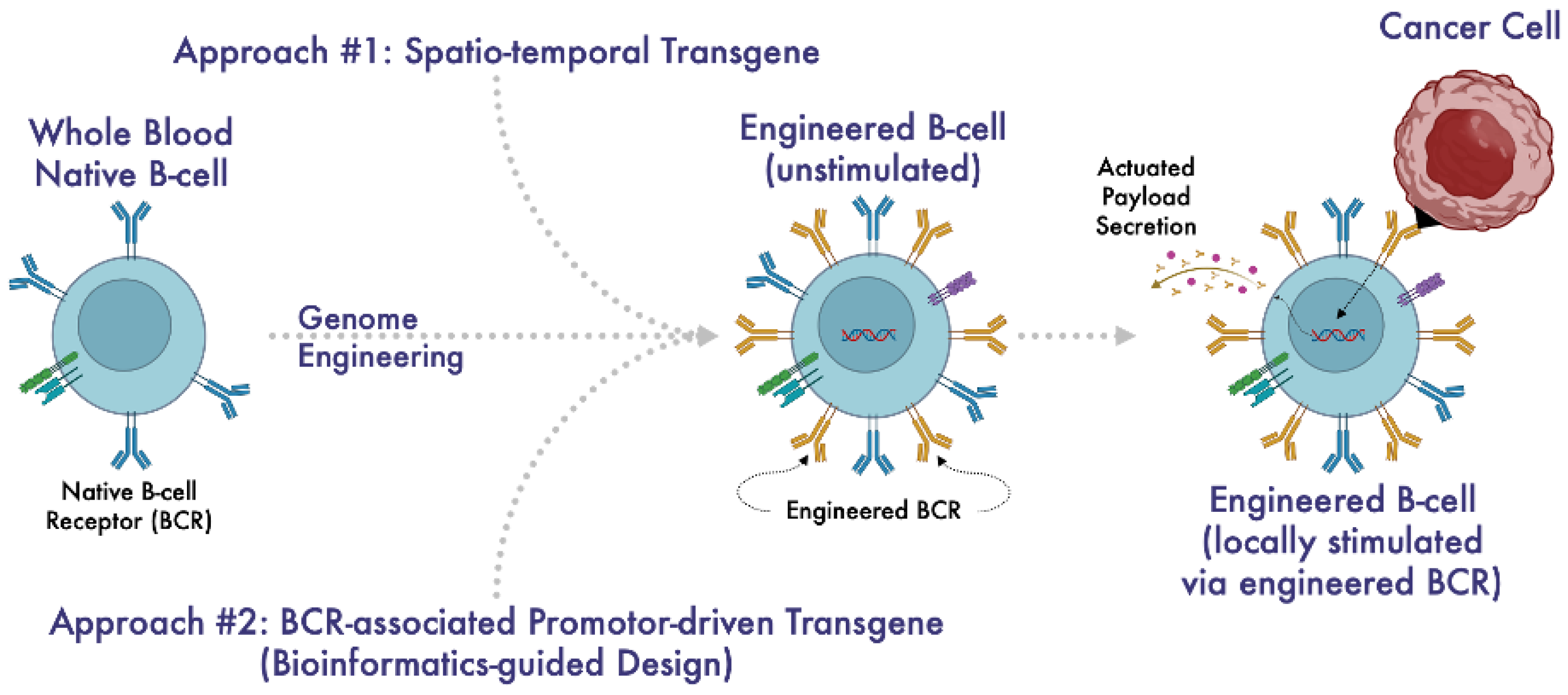
1. Introduction
2. Materials and Methods
3. Results
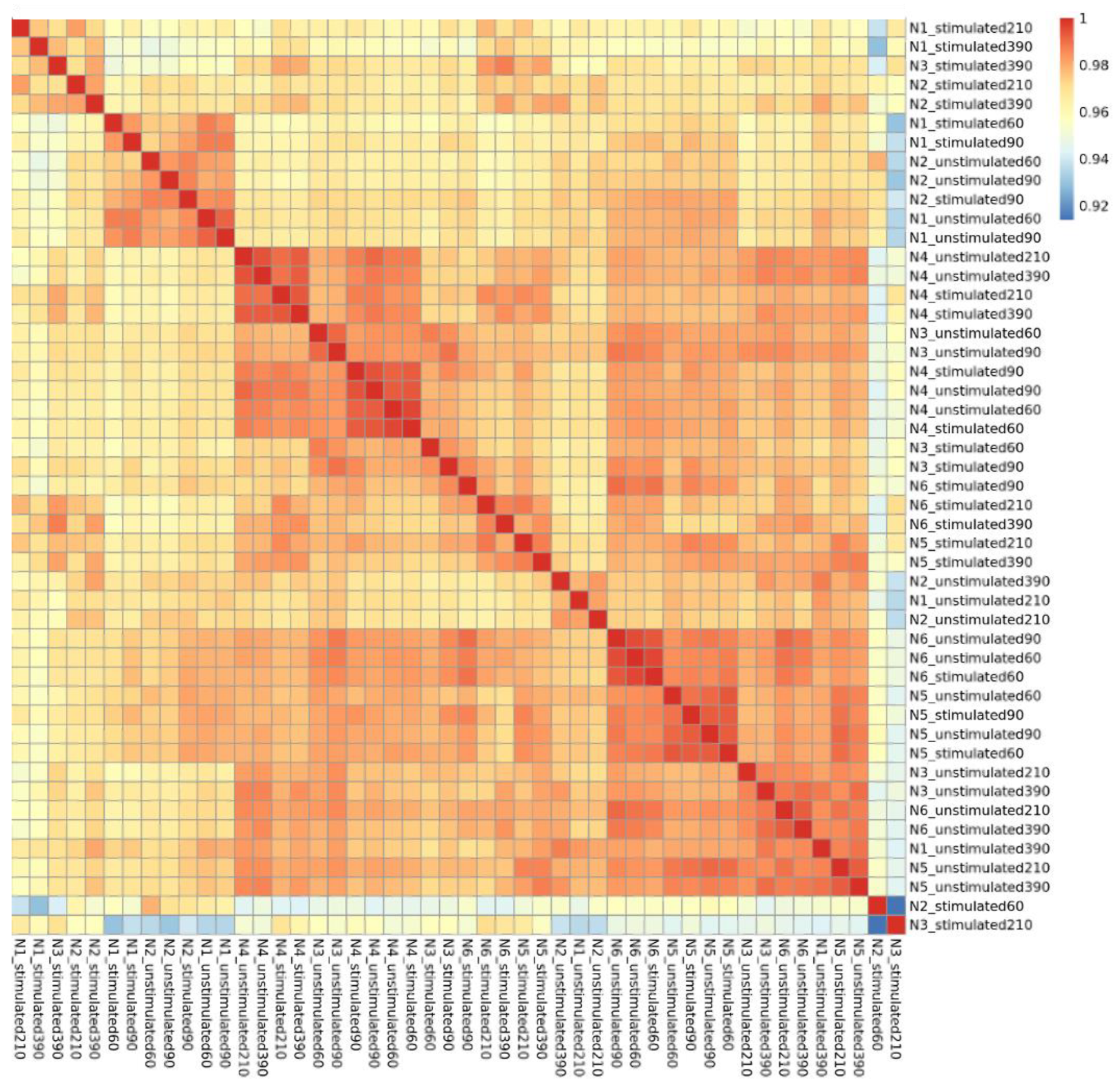
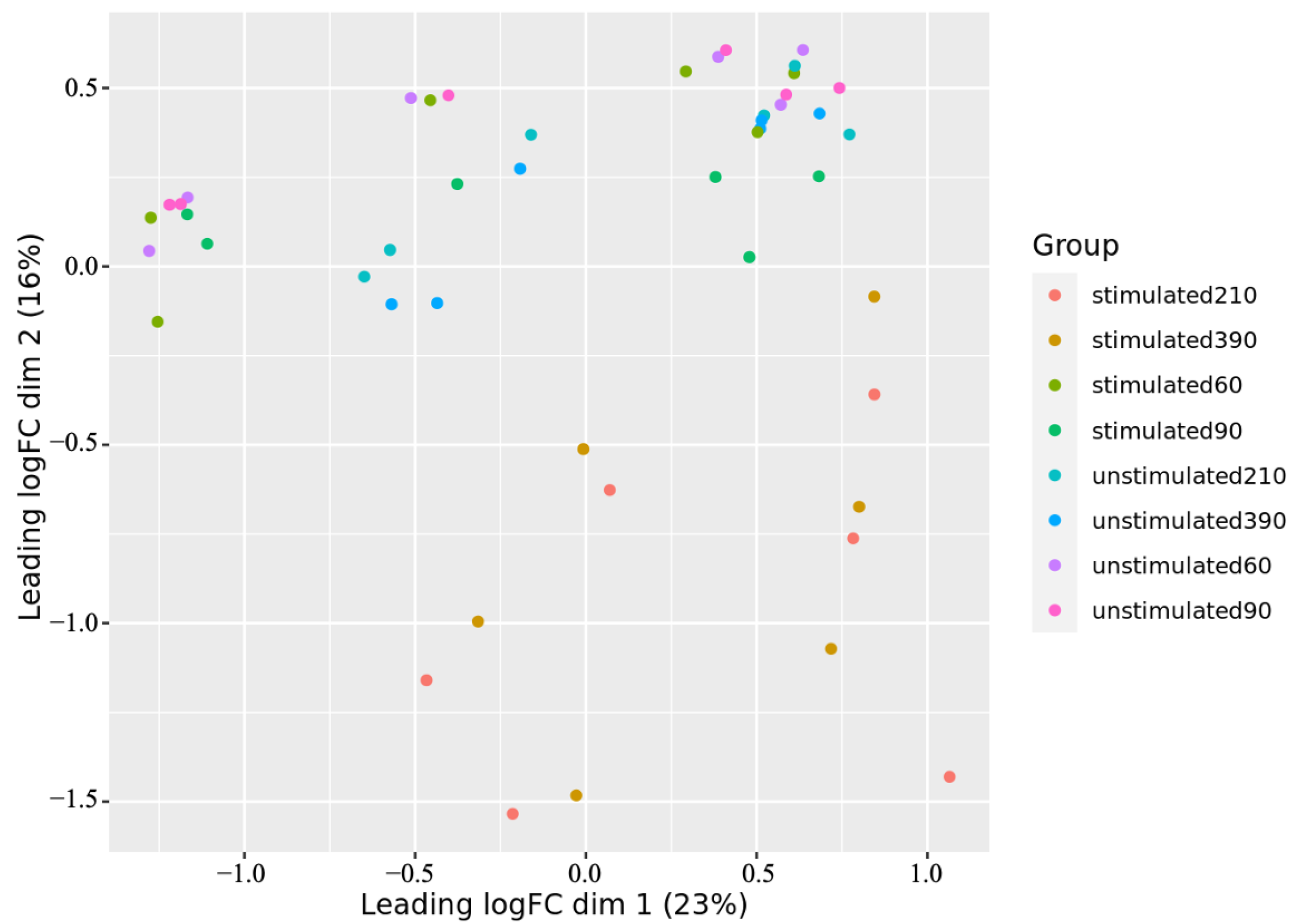
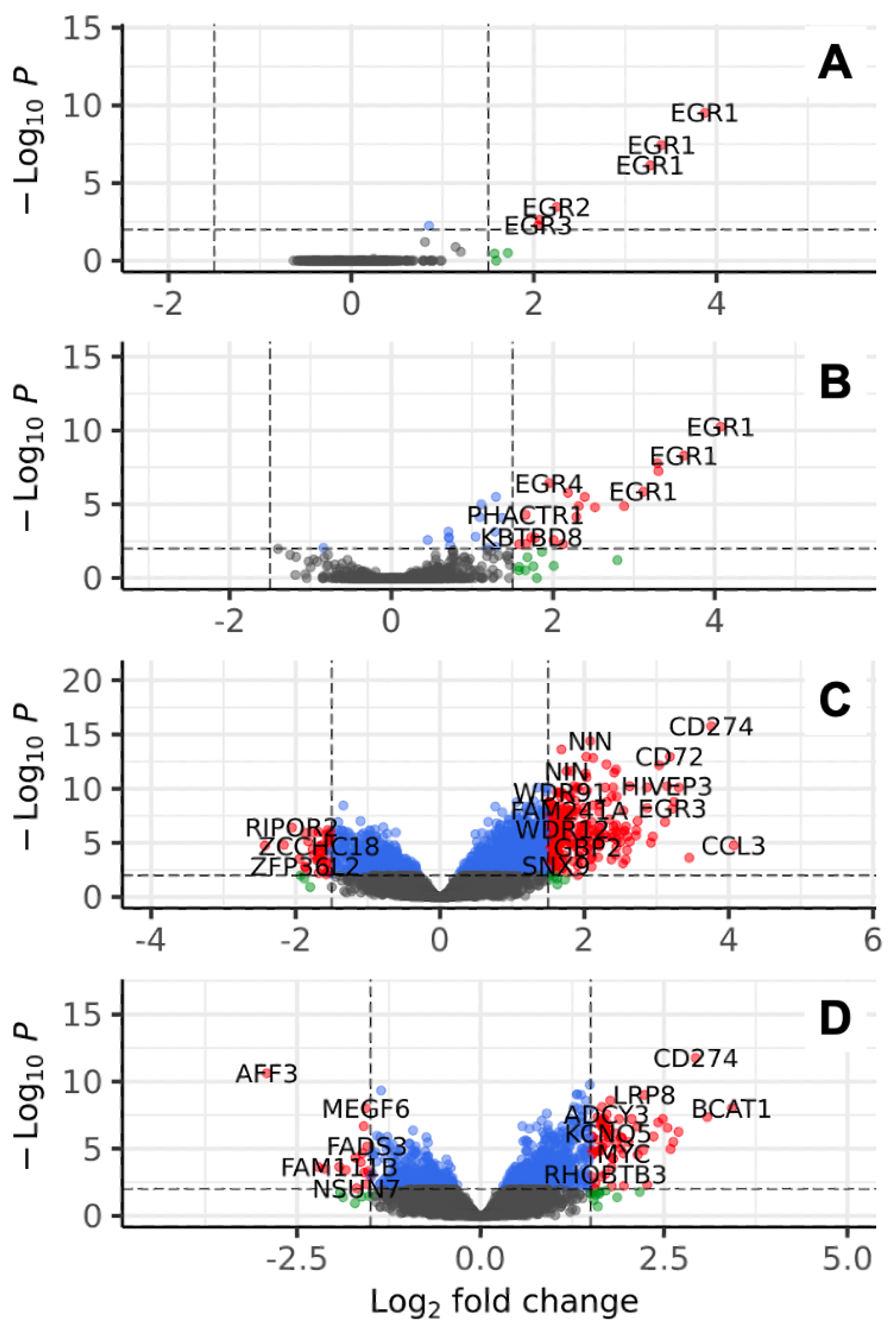
3.1. Gene Expression Analyses Revealed Several BCR Stimulation-Associated Upregulated Genes
3.2. Identification of Putative Gene Promoters for BCR Stimulation-Induced Payload Production
4. Discussion
- –
- BCAR3: AREB6, CBF-A, CBF-B, CBF-C, CP1A, HNF-3beta, Ik-2, NF-Y, and STAT5A.
- –
- BCAT1: AML1a, FOXO1, FOXO1a, and IRF-7A.
- –
- HIVEP3: AP-1, AREB6, ATF-2, c-Jun, c-Myc, Hlf, Max, Max1, Sox5, and SREBP-1b.
- –
- KCNQ5: AhR, aMEF-2, Arnt, c-Myb, Cart-1, MEF-2, NF-κB, NF-κB1, and RelA.
- –
- LRP8: RORalpha1, USF1, and USF2.
- –
- NETO1: AML1a, CP2, FOXJ2, and Nkx2-5.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Irvine, D.J.; Maus, M.V.; Mooney, D.J.; Wong, W.W. The future of engineered immune cell therapies. Science 2022, 378, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Cyster, J.G.; Allen, C.D.C. B cell responses—Cell interaction dynamics and decisions. Cell 2019, 177, 524–540. [Google Scholar] [CrossRef] [PubMed]
- Banchereau, J.; Bazan, F.; Blanchard, D.; Briè, F.; Galizzi, J.P.; Van Kooten, C.; Liu, Y.J.; Rousset, F.; Saeland, S. The CD40 antigen and its ligand. Ann. Rev. Immunol. 1994, 12, 881–922. [Google Scholar] [CrossRef]
- Pullen, S.S.; Dang, T.T.; Crute, J.J.; Kehry, M.R. CD40 signaling through tumor necrosis factor receptor-associated factors (TRAFs). Binding site specificity and activation of downstream pathways by distinct TRAFs. J. Biol. Chem. 1999, 274, 14246–14254. [Google Scholar] [CrossRef] [PubMed]
- Ranheim, E.A.; Kipps, T.J. Activated Tcells induce expression of B7/BB1 on normal or leukemic B cells through a CD40-dependent signal. J. Exp. Med. 1993, 177, 925–935. [Google Scholar] [CrossRef]
- Gricks, C.S.; Zahrieh, D.; Zauls, A.J.; Gorgun, G.; Drandi, D.; Mauerer, K.; Neuberg, D.; Gribben, J.G. Differential regulation of gene expression following CD40 activation of leukemic compared to healthy B cells. Blood 2004, 104, 4002–4009. [Google Scholar] [CrossRef][Green Version]
- Ruprecht, C.R.; Lanzavecchia, A. Toll-like receptor stimulation as a third signal required for activation of human naive B cells. Eur. J. Immunol. 2006, 36, 810–816. [Google Scholar] [CrossRef]
- Capolunghi, F.; Cascioli, S.; Giorda, E.; Rosado, M.M.; Plebani, A.; Auriti, C.; Seganti, G.; Zuntini, R.; Ferrari, S.; Cagliuso, M.; et al. CpG drives human transitional B cells to terminal differentiation and production of natural antibodies. J. Immunol. 2008, 180, 800–808. [Google Scholar] [CrossRef] [PubMed]
- Aranburu, A.; Ceccarelli, S.; Giorda, E.; Lasorella, R.; Ballatore, G.; Carsetti, R. TLR ligation triggers somatic hypermutation in transitional B cells inducing the generation of IgM memory B cells. J. Immunol. 2010, 185, 7293–7301. [Google Scholar] [CrossRef]
- Hung, K.L.; Meitlis, I.; Hale, M.; Chen, C.Y.; Singh, S.; Jackson, S.W.; Miao, C.H.; Khan, I.F.; Rawlings, D.J.; James, R.G. Engineering protein-secreting plasma cells by homology-directed repair in primary human B cells. Mol. Ther. 2018, 26, 456–467. [Google Scholar] [CrossRef]
- Moffett, H.F.; Harms, C.K.; Fitzpatrick, K.S.; Tooley, M.R.; Boonyaratanakornkit, J.; Taylor, J.J. B cells engineered to express pathogen-specific antibodies protect against infection. Sci. Immunol. 2019, 4, eaax0644. [Google Scholar] [CrossRef] [PubMed]
- Fusil, F.; Calattini, S.; Amirache, F.; Mancip, J.; Costa, C.; Robbins, J.B.; Douam, F.; Lavillette, D.; Law, M.; Defrance, T.; et al. A lentiviral vector allowing physiologically regulated membrane-anchored and secreted antibody expression depending on B-cell maturation status. Mol. Ther. 2015, 23, 1734–1747. [Google Scholar] [CrossRef]
- Huang, D.; Tran, J.T.; Olson, A.; Vollbrecht, T.; Tenuta, M.; Guryleva, M.V.; Fuller, R.P.; Schiffner, T.; Abadejos, J.R.; Couvrette, L.; et al. Vaccine elicitation of HIV broadly neutralizing antibodies from engineered B cells. Nat. Commun. 2020, 11, 5850. [Google Scholar] [CrossRef] [PubMed]
- National Institutes of Health, National Library of Medicine, National Center for Biotechnology Information, Gene Expression Omnibus. Available online: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE39411 (accessed on 1 April 2024).
- Vallat, L.D.; Park, Y.; Li, C.; Gribben, J.G. Temporal genetic program following B-cell receptor cross-linking: Altered balance between proliferation and death in healthy and malignant B cells. Blood 2007, 109, 3989–3997. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Carvalho, B.S.; Irizarry, R.A. A framework for oligonucleotide microarray preprocessing. Bioinformatics 2010, 26, 2363–2367. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, J.W. affycoretools: Functions Useful for Those Doing Repetitive Analyses with Affymetrix GeneChips. R Package Version 1.74.0. 2023. Available online: https://bioconductor.org/packages/affycoretools (accessed on 1 December 2022).
- Ritchie, M.E.; Phipson, B.; Wu, D.I.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Statist Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Azimuth: App for Reference-Based Single-Cell Analysis. Available online: https://azimuth.hubmapconsortium.org/references/human_pbmc/ (accessed on 1 February 2024).
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M.; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587. [Google Scholar] [CrossRef]
- Dal Bo, M.; D’Agaro, T.; Gobessi, S.; Zucchetto, A.; Dereani, S.; Rossi, D.; Zaja, F.; Pozzato, G.; Di Raimondo, F.; Gaidano, G.; et al. The SIRT1/TP53 axis is activated upon B-cell receptor triggering via miR-132 up-regulation in chronic lymphocytic leukemia cells. Oncotarget 2015, 6, 19102–19117. [Google Scholar] [CrossRef]
- Schleiss, C.; Carapito, R.; Fornecker, L.M.; Muller, L.; Paul, N.; Tahar, O.; Pichot, A.; Tavian, M.; Nicolae, A.; Miguet, L.; et al. Temporal multiomic modeling reveals a B-cell receptor proliferative program in chronic lymphocytic leukemia. Leukemia 2021, 35, 1463–1474. [Google Scholar] [CrossRef] [PubMed]
- Rivello, F.; van Buijtenen, E.; Matuła, K.; van Buggenum, J.A.; Vink, P.; van Eenennaam, H.; Mulder, K.W.; Huck, W.T. Single-cell intracellular epitope and transcript detection reveals signal transduction dynamics. Cell Rep. Methods 2021, 27, 100070. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Cerny, D.; Chua, E.; Duan, K.; Yi, J.T.J.; Shadan, N.B.; Lum, J.; Maho-Vaillant, M.; Zolezzi, F.; Wong, S.C.; et al. Human regulatory B cells combine phenotypic and genetic hallmarks with a distinct differentiation fate. J. Immunol. 2014, 193, 2258–2266. [Google Scholar] [CrossRef]
- Vallat, L.; Kemper, C.A.; Jung, N.; Maumy-Bertrand, M.; Bertrand, F.; Meyer, N.; Pocheville, A.; Fisher, J.W., III; Gribben, J.G.; Bahram, S. Reverse-engineering the genetic circuitry of a cancer cell with predicted intervention in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2013, 110, 459–464. [Google Scholar] [CrossRef] [PubMed]
- Kwak, K.; Akkaya, M.; Pierce, S.K. B cell signaling in context. Nat. Immunol. 2019, 20, 963–969. [Google Scholar] [CrossRef] [PubMed]
- Tolar, P.; Sohn, H.W.; Liu, W.; Pierce, S.K. The molecular assembly and organization of signaling active B-cell receptor oligomers. Immunol. Rev. 2009, 232, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Shinnakasu, R.; Kurosaki, T. Regulation of memory B and plasma cell differentiation. Curr. Opin. Immunol. 2017, 45, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Dal Porto, J.M.; Gauld, S.B.; Merrell, K.T.; Mills, D.; Pugh-Bernard, A.E.; Cambier, J. B cell antigen receptor signaling 101. Mol. Immunol. 2004, 41, 599–613. [Google Scholar] [CrossRef]
- Goodnow, C.C.; Vinuesa, C.G.; Randall, K.L.; Mackay, F.; Brink, R. Control systems and decision making for antibody production. Nat. Immunol. 2010, 11, 681–688. [Google Scholar] [CrossRef] [PubMed]
- Harwood, N.E.; Batista, F.D. Early events in B cell activation. Annu. Rev. Immunol. 2010, 28, 185–210. [Google Scholar] [CrossRef]
- Harwood, N.E.; Batista, F.D. New insights into the early molecular events underlying B cell activation. Immunity 2008, 28, 609–619. [Google Scholar] [CrossRef]
- Kurosaki, T.; Shinohara, H.; Baba, Y. B cell signaling and fate decision. Annu. Rev. Immunol. 2010, 28, 21–55. [Google Scholar] [CrossRef] [PubMed]
- Tullai, J.W.; Schaffer, M.E.; Mullenbrock, S.; Sholder, G.; Kasif, S.; Cooper, G.M. Immediate-early and delayed primary response genes are distinct in function and genomic architecture. J. Biol. Chem. 2007, 282, 23981–23995. [Google Scholar] [CrossRef] [PubMed]
- Sproul, D.; Gilbert, N.; Bickmore, W.A. The role of chromatin structure in regulating the expression of clustered genes. Nat. Rev. Genet. 2005, 6, 775–781. [Google Scholar] [CrossRef] [PubMed]
- Reményi, A.; Schöler, H.R.; Wilmans, M. Combinatorial control of gene expression. Nat. Struct. Mol. Biol. 2004, 11, 812–815. [Google Scholar] [CrossRef] [PubMed]
- Retis-Resendiz, A.M.; González-García, I.N.; León-Juárez, M.; Camacho-Arroyo, I.; Cerbón, M.; Vázquez-Martínez, E.R. The role of epigenetic mechanisms in the regulation of gene expression in the cyclical endometrium. Clin. Epigenetics 2021, 13, 116. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef]
- Page, A.; Delles, M.; Negre, D.; Costa, C.; Fusil, F.; Cosset, F.-L. Engineering B cells with customized therapeutic responses using a synthetic circuit. Mol. Ther. Nucleic Acids 2023, 33, 1–14. [Google Scholar] [CrossRef]

| Gene Name | Gene Description, Product, and/or Function | PBMC Predominant Distribution |
|---|---|---|
| BCAR3 | BCAR3 adaptor protein is a putative suppressor of breast cancer progression by inhibiting prometastatic TGFbeta/Smad signaling | B-cells |
| BCAT1 | Branched chain amino acid transaminase 1 (cytosolic form) | B-cells and Monocytes |
| HIVEP3 | HIVEP zinc finger 3 is a transcription factor that binds to Rss heptamer for somatic recombination of immunoglobulin and T cell receptor gene segments | Most PBMC |
| KCNQ5 | Potassium voltage-gated channel subfamily Q member 5 is expressed in subregions of the brain and in skeletal muscle | B-cells and T cells |
| LRP8 | LDL receptor-related protein 8 is a receptor for the cholesterol transport protein apolipoprotein E | Most PBMC |
| NETO1 | Neurolipin and talloid like 1 encodes a transmembrane protein containing two extracellular CUB domains followed by an LDL class A domain | B-cells |
| Organization Name | Initial Submission to NCBI GEO | NCBI GEO Accession Number(s) | B-Cell Stimulation | Human Cell Type |
|---|---|---|---|---|
| Dana-Farber Cancer Institute [6] | N/A | N/A | CD40 | Healthy and B-cell CLL cells |
| Helmholtz Zentrum München | 2010 | GSE25434 | BCR or inducible LMP2A | EBV immortalized lymphoblastoid cell lines |
| Strasbourg University Hospital [15,26] | 2012 | GSE39411 | BCR | Healthy and B-cell CLL cells |
| Singapore Immunology Network [25] | 2013 | GSE50895 | BCR and TLR9 | IL-10− and IL-10+ B-cells |
| CRO Aviano [22] | 2013 | GSE52774 GSE52775 | BCR | B-cell CLL |
| Strasbourg University Hospital [23] | 2019 | GSE130385 | BCR | B-cell CLL |
| Radboud University [24] | 2020 | GSE162461 | BCR | Burkitt lymphoma cells |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deitcher, E.; Trisler, K.; Moriarity, B.S.; Bostwick, C.J.; Leenen, F.A.D.; Deitcher, S.R. Bioinformatics-Based Identification of Human B-Cell Receptor (BCR) Stimulation-Associated Genes and Putative Promoters. BioMedInformatics 2024, 4, 1384-1395. https://doi.org/10.3390/biomedinformatics4020076
Deitcher E, Trisler K, Moriarity BS, Bostwick CJ, Leenen FAD, Deitcher SR. Bioinformatics-Based Identification of Human B-Cell Receptor (BCR) Stimulation-Associated Genes and Putative Promoters. BioMedInformatics. 2024; 4(2):1384-1395. https://doi.org/10.3390/biomedinformatics4020076
Chicago/Turabian StyleDeitcher, Ethan, Kirk Trisler, Branden S. Moriarity, Caleb J. Bostwick, Fleur A. D. Leenen, and Steven R. Deitcher. 2024. "Bioinformatics-Based Identification of Human B-Cell Receptor (BCR) Stimulation-Associated Genes and Putative Promoters" BioMedInformatics 4, no. 2: 1384-1395. https://doi.org/10.3390/biomedinformatics4020076
APA StyleDeitcher, E., Trisler, K., Moriarity, B. S., Bostwick, C. J., Leenen, F. A. D., & Deitcher, S. R. (2024). Bioinformatics-Based Identification of Human B-Cell Receptor (BCR) Stimulation-Associated Genes and Putative Promoters. BioMedInformatics, 4(2), 1384-1395. https://doi.org/10.3390/biomedinformatics4020076







