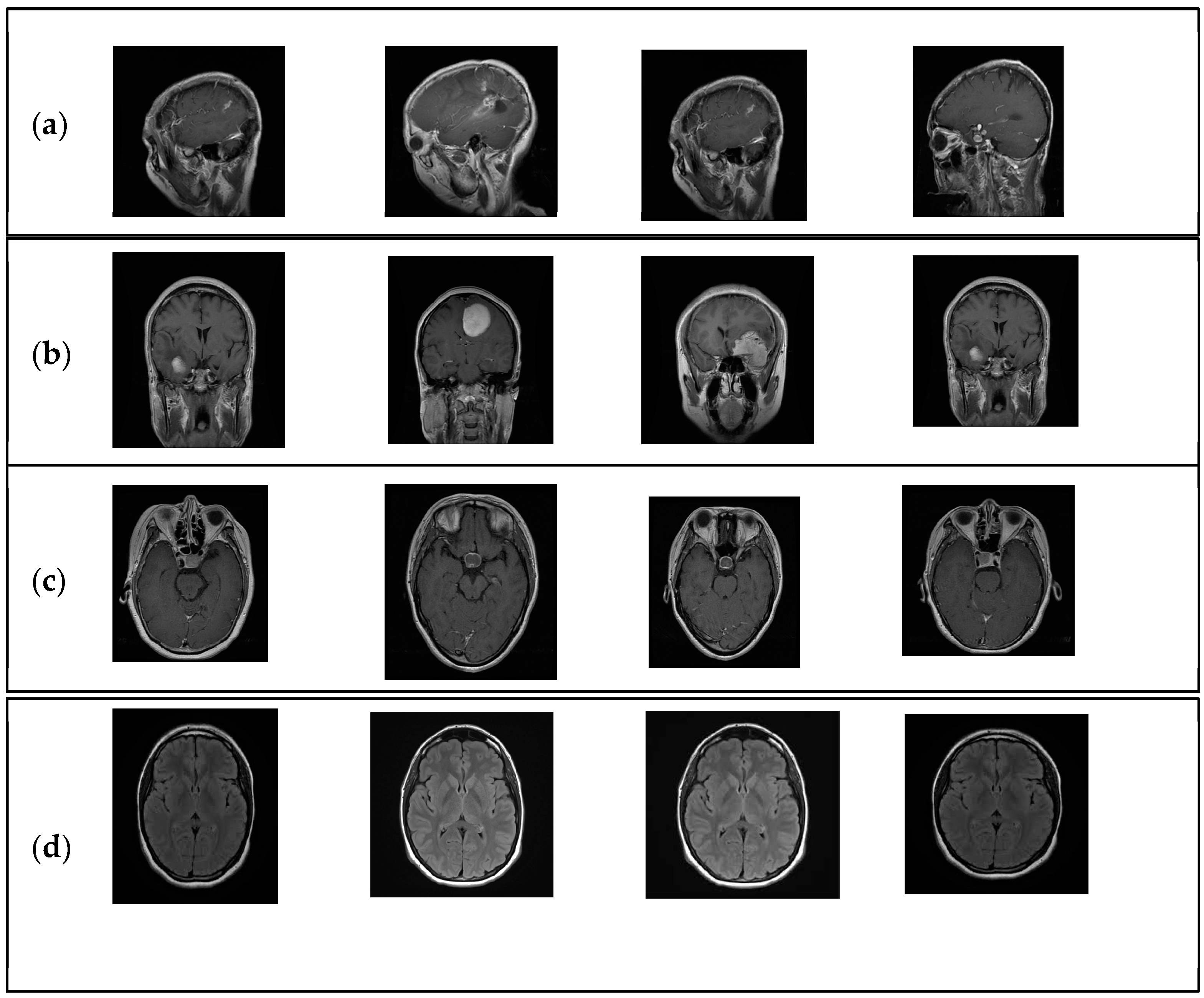
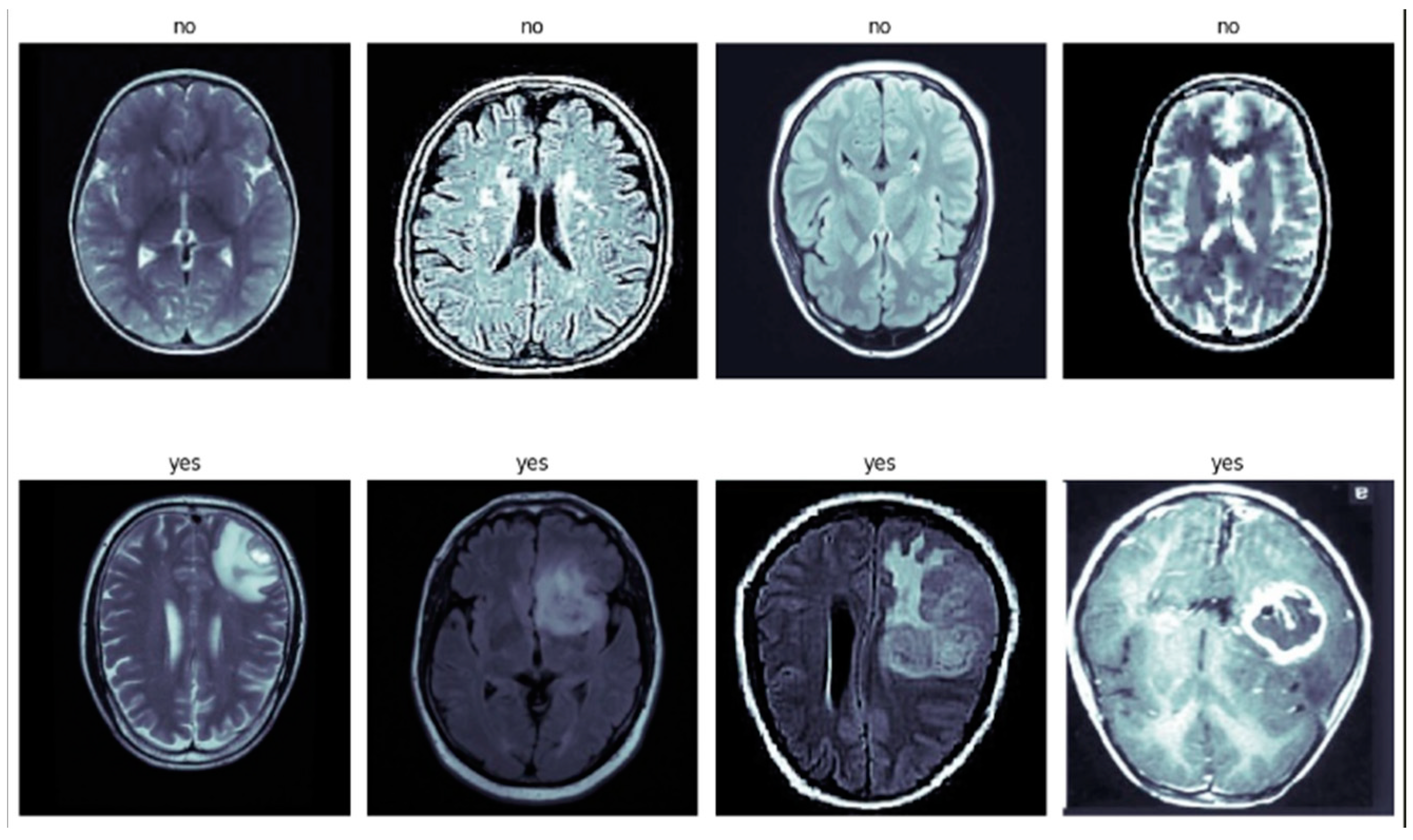
1. Introduction
There are approximately 200 different kinds of aberrant tissue growths that can occur in humans, and among these are tumors, which can be either malignant or benign. In particular, brain tumors are a dangerous condition that involves abnormal growths in brain tissue that impede the brain’s ability to operate. The fact that there has been a 300 percent increase in the number of deaths attributable to brain tumors over the past three decades demonstrates how critical it is to find a cure for this illness. Brain tumors have the potential to be lethal if they are not treated, highlighting the significance of early diagnosis and treatment in order to enhance patient survival rates. Despite the fact that biopsies of brain tumors can be difficult to perform because of the intricacy of the brain, magnetic resonance imaging (MRI) is frequently utilized as a diagnostic tool [
1,
2,
3,
4,
5,
6,
7,
8,
9,
10].
The majority of brain tumors, known as gliomas, begin in the glial cells of the brain. Gliomas are the most frequent type of brain tumor. They are responsible for approximately 30% of all tumors that can be found in the brain and central nervous system, and they account for 80% of all malignant brain tumors [
11]. According to the categorization used by the World Health Organization (WHO), gliomas can be broken down into one of four classes, ranging from grade I all the way up to grade IV. Grade I tumors are considered benign and have a texture that is very similar to that of normal glial cells, whereas grade II tumors have a texture that is only slightly different. Grade III tumors are malignant and have an aberrant look to the tissue, whereas grade IV tumors are the most severe stage of gliomas and have apparent tissue abnormalities [
11,
12]. Grade III tumors are malignant and have an abnormal appearance of tissue.
Meningiomas, on the other hand, form on the membranes inside the head that cover the brain and spinal cord. Meningiomas usually grow slowly, and most of them are harmless. Pituitary tumors come from the pituitary gland, which controls hormone output and the way the body works. These lumps can be harmless, harmless with bone growth, or cancerous. Pituitary tumors can cause problems that can lead to lasting hormone shortages and eye loss [
13].
Based on what has been said so far, it is important to find and classify brain tumors early in order to make an accurate diagnosis and choose the best treatment options to save patients’ lives. When a case is complicated, the grading stage can be hard and take a long time for doctors and experts. In these situations, experts usually need to look at the tumor and figure out where it is. They may also need to compare the tumor’s cells with those of nearby areas, add image filters if needed to make the images clearer for humans to understand, and finally figure out if it is a tumor and, if possible, its type and grade. This can take a long time, which shows how important it is to have computer-aided diagnosis (CAD) tools that can find brain tumors at an early stage, shortening the time it takes to diagnose and lowering the need for human intervention [
11,
12]. Recent advances in machine learning (ML), especially Deep Learning (DL), have changed how medical image patterns are found and put into groups. Machine learning has shown promise in a number of medical areas, such as predicting and diagnosing diseases, classifying images, and separating tissues. Convolutional Neural Network (CNNs) have become useful tools for processing images because they can make accurate diagnoses from a large number of incoming images. For representation learning, independent learning methods such as autoencoders have also been used. Several studies have used different methods and models to look into how to find brain tumors. However, some of these studies have problems, such as the fact that they do not compare their results to those of traditional machine learning methods or that they require complicated calculations. The knowledge of radiologists is needed to diagnose tumors from medical medical X-ray or CT Scanned images, and mistakes can happen. Computer-assisted interventions and computational intelligence methods can help doctors find and describe brain tumors more correctly, so they do not have to rely on their own opinions as much. DL and ML techniques, in particular, can be very helpful for studying, segmenting, and classifying cancer pictures, including those of brain tumors. By using these methods, tumors can be found accurately and reliably, making it possible to tell those apart from other diseases that look similar. Although various studies have been conducted in the field of DL-based brain tumor diagnosis, the literature review section demonstrates that the performance of existing models varies depending on the datasets, and there is room for improvement in the frameworks.
In this study, we present a functional system that combines the skills of an accurate and automatic classification DL model to classify whether a brain MRI tumor image has a tumor present or not, as well as identify three types of tumors such as glioma, meningioma, or pituitary classification. At present, DL models have achieved good success in image classification. In the case of feature extraction, these models can identify essential features of the object in the layer of these models, which makes it much easier for the classifier to identify the different features.
The major contributions of this paper are listed below:
- a.
The Resnet 50 and Inception V3 designs from CNN Architecture were selected to assess the effectiveness of DL models in the task of classifying tumors from brain MRI, as these designs have shown expertise in picture categorization.
- b.
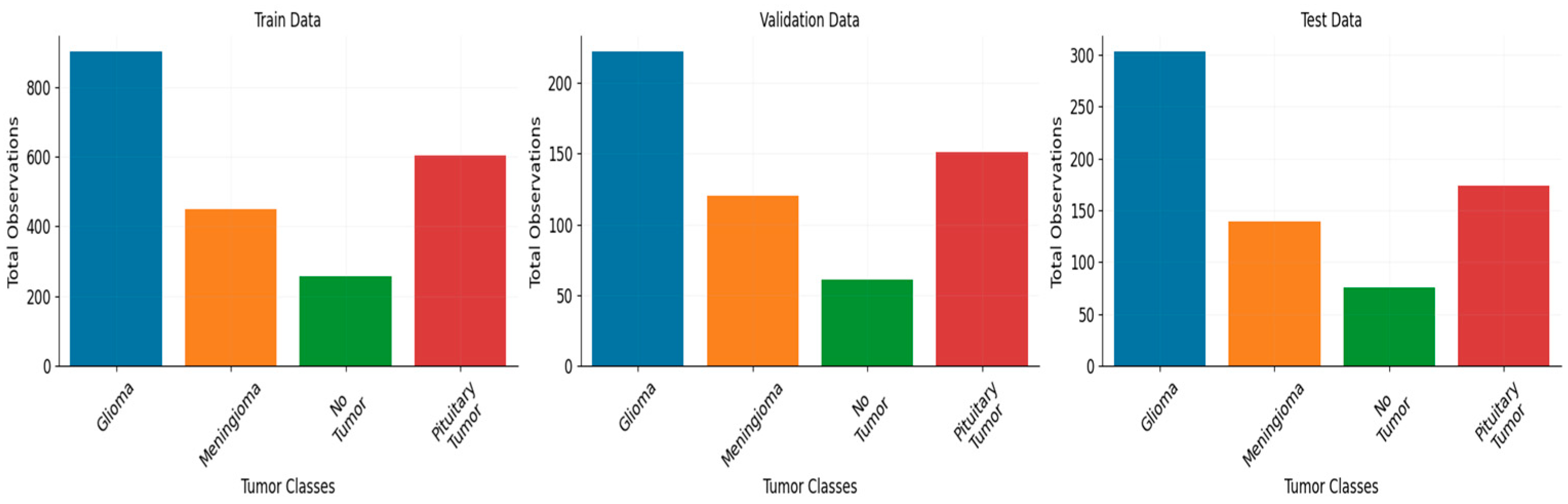
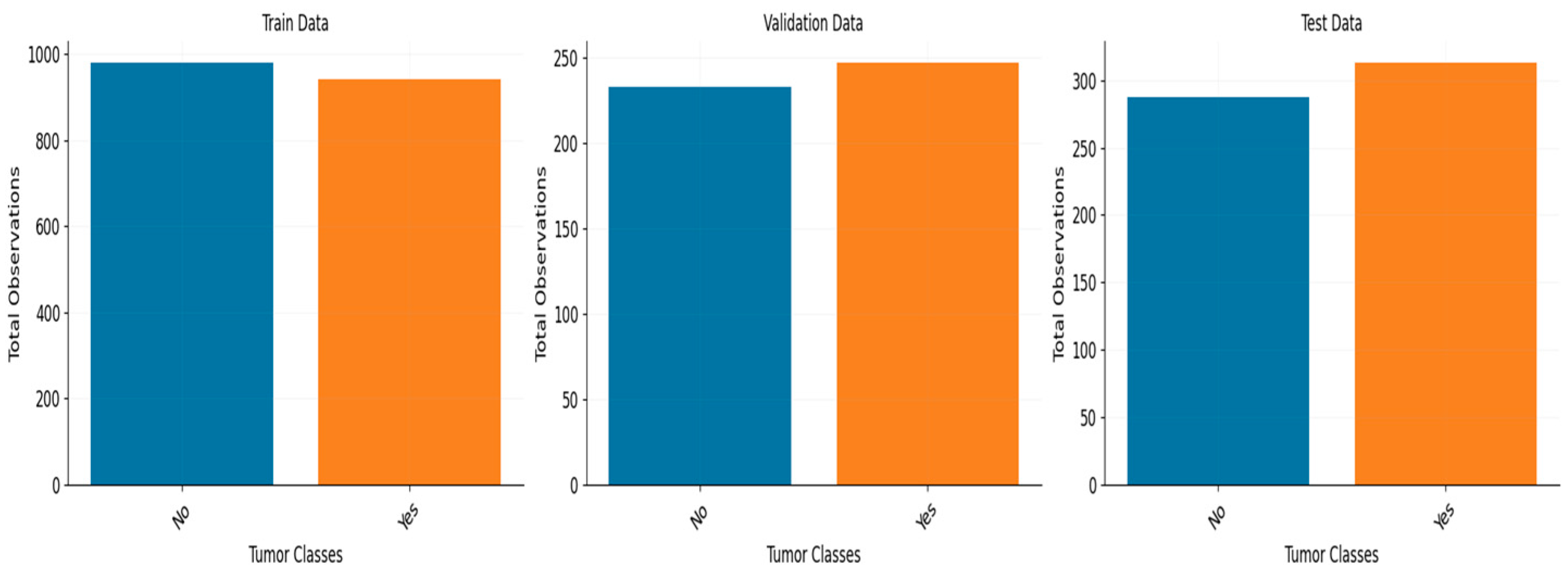
The performance of the model was assessed on two datasets. One dataset consists of 3459 MRI scans belonging to four distinct classes, while the second dataset has 3000 MRI images belonging to two distinct classes.
- c.
In order to enhance the efficiency of the models in this particular task, various techniques were employed, including Early Stopping and ReduceLROnPlateau. These techniques were utilized to optimize the model through hyperparameter tuning, which involved incorporating additional layers, experimenting with different loss functions and learning rates, and implementing dropout layers and regularization to mitigate overfitting. In both datasets, we observed exceptional performance that is at the forefront of current advancements.
The subsequent sections of the paper are structured in the following manner:
Section 2 of this study centers on the comprehensive analysis of the existing literature, while
Section 3 provides a concise overview of the datasets utilized in this research.
Section 5 provides a theoretical exposition of the models.
Section 6 provides an overview of the methodology employed in this study, as well as the proposed architecture.
Section 7 of the paper provides an in-depth analysis of the experimental findings, while
Section 8 serves as the concluding section, summarizing the key points and implications of the study.
2. The Literature Review
Ghosal et al. [
14] proposed a deep-neural-network-based approach using squeeze and excitation ResNet-101 for automatic brain tumor classification in MR images. The study focused on differentiating between the glioma, pituitary tumor, and meningioma. They employed zero-centering intensity normalization and data augmentation to improve performance. The experimental results showed significant improvements in precision, specificity, and sensitivity compared to other recent methods for brain tumor classification. Krishnapriya et al. [
15] conducted a study to explore the capability of pre-trained deep convolutional neural network (DCNN) models, specifically VGG-19, VGG-16, ResNet 50, and Inception V3, for categorizing brain MR images. The researchers employed data augmentation and transfer learning techniques to enhance the performance of these models in the classification task. Diaz-Pernas et al. [
16] introduced an algorithm for brain tumor segmentation and classification utilizing MRI scans of meningioma, glioma, and pituitary tumors that is fully automated. They employed CNN to operationalize the concept of a multi-scale strategy that is intrinsic to human cognitive processes. The researchers attained a 97% level of precision when analyzing a set of 3064-slice images obtained from a cohort of 233 patients. A CNN architecture was employed [
17] that consisted of 16 convolution layers, pooling and normalization layers, and a dropout layer that preceded the fully connected layer. The study revealed a 96% precision level when 68% of the images were utilized for training purposes, while the remaining pictures were allocated for validation and testing. The study conducted by Abd et al. [
18] involved the analysis of 25,000 MRI images of the brain using a differential (DCNN) for the purpose of identifying different types of brain tumors. They attained an exceptional overall performance, exhibiting a precision rate of 99.25% during the training phase. The study by Sajja and colleagues [
19] utilized Brat’s dataset, consisting of 577 T1-weighted brain tumors, to classify malignant and benign tumors through the implementation of the VGG16 network. The performers exhibited a level of inaccuracy of 96.70 during their performance. A CNN was proposed in [
20] to classify different types of brain cancers, including the glioma tumor, meningioma tumor, and pituitary tumor. The dataset consisted of 3064 T1-weighted contrast-enhanced MRI images. The CNN architecture underwent training to effectively employ multiple convolutional and pooling techniques. The researchers achieved a 94% accuracy rate through the implementation of a convolutional network that was resized based on convolutional filters/kernels of varying sizes. Abiwinanda et al. [
21] utilized 64 fully connected neurons and two convolution layers. They did not use the entire dataset but instead selected 700 images from each form of brain tumor in order to balance the data. For training, a subset of 500 tumor images from each class was used, and for testing, a subset of 200 images was used. They did not utilize data enhancement. Their classification accuracy was 84.1%. The classification CNN model proposed by Pashaei et al. [
22] consists of four convolution and normalization layers, three max-pooling layers, and a final completely connected layer. They utilized 70% of the dataset for training with no data augmentation and 30% for testing with 10-fold cross-validation. The accuracy of classification was 81.0%. Afshar et al. [
23] proposed the Capsule network (CapsNet) classification model for brain tumors. To improve accuracy, they modified the feature mappings in the convolutional layer of CapsNet. Using 64 feature maps and one convolutional layer of CapsNet, the maximum accuracy of 86.56 percent was achieved.
Table 1 summarizes the literature reviews. Ibrokhiov et al. [
24], in response to the rising incidence of pneumonia, particularly in the wake of the COVID-19 pandemic, introduced an advanced DL-based computer-aided diagnostic system, leveraging transfer learning and parallel computing techniques with VGG19 and ResNet 50 models, achieving an impressive average classification accuracy of 96.6% on the COVID-QU-Ex dataset. Edgar M et al. [
25] address the challenge of interpreting machine learning algorithms applied to medical image data, specifically in predicting brain tumor survival rates from MRI scans. By leveraging explainable AI techniques, such as Shapley overlays, in conjunction with CNN and the BraTS 2020 dataset, this research demonstrates the improved interpretability of key features, facilitating expert validation and enhancing the overall evaluation of predictive outcomes. Shokouhifar et al. [
26] present THENDEL, a three-stage DL model embedded in a camera scanning tool for noninvasive and rapid lymphedema arm volume measurement. THENDEL combines various feature extractors and regressors, with hyperparameters optimized using a swarm intelligence algorithm. The study successfully measured arm volumes in 730 arms from 365 women, showing strong reliability with a mean absolute error of 36.65 mL, a mean percent error of 1.69%, and a 0.992 correlation with actual volumes. Veeraiah et al. [
27] introduce MayGAN for Leukemia classification with 99.8% accuracy. Aryai et al. [
28] present MDML-RP for efficient health monitoring in WBANs, achieving substantial performance gains. Ibtisum et al. [
29] offer a comparative study on diverse Big Data tools, contributing to the evolving data analytics field.
6. Methodology and Proposed Architecture
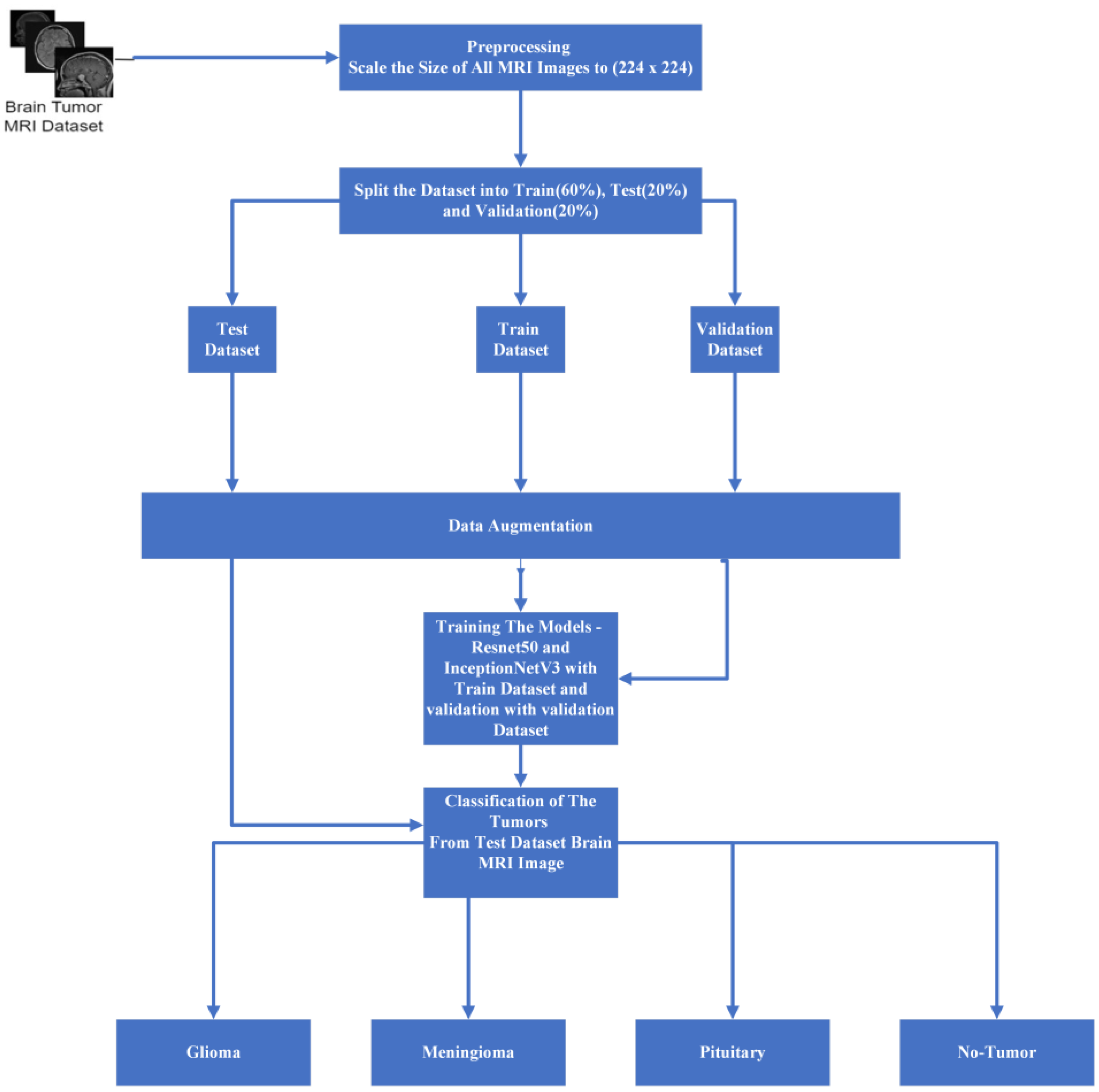
Figure 6 illustrates the comprehensive architecture of our proposed system, which encompasses multiple stages in our experimental process. Upon dataset extraction, we maintained a consistent size of 224 × 224 to ensure uniformity. To alleviate RAM constraints within the training, testing, and validation folders, we meticulously divided the data in accordance with the previously mentioned ratios. The data generation process, orchestrated within the data model, employs diverse data augmentation techniques aimed at mitigating model overfitting. After experimenting with various batch sizes during the loading phase, we settled on a batch size of 16 for optimal performance.
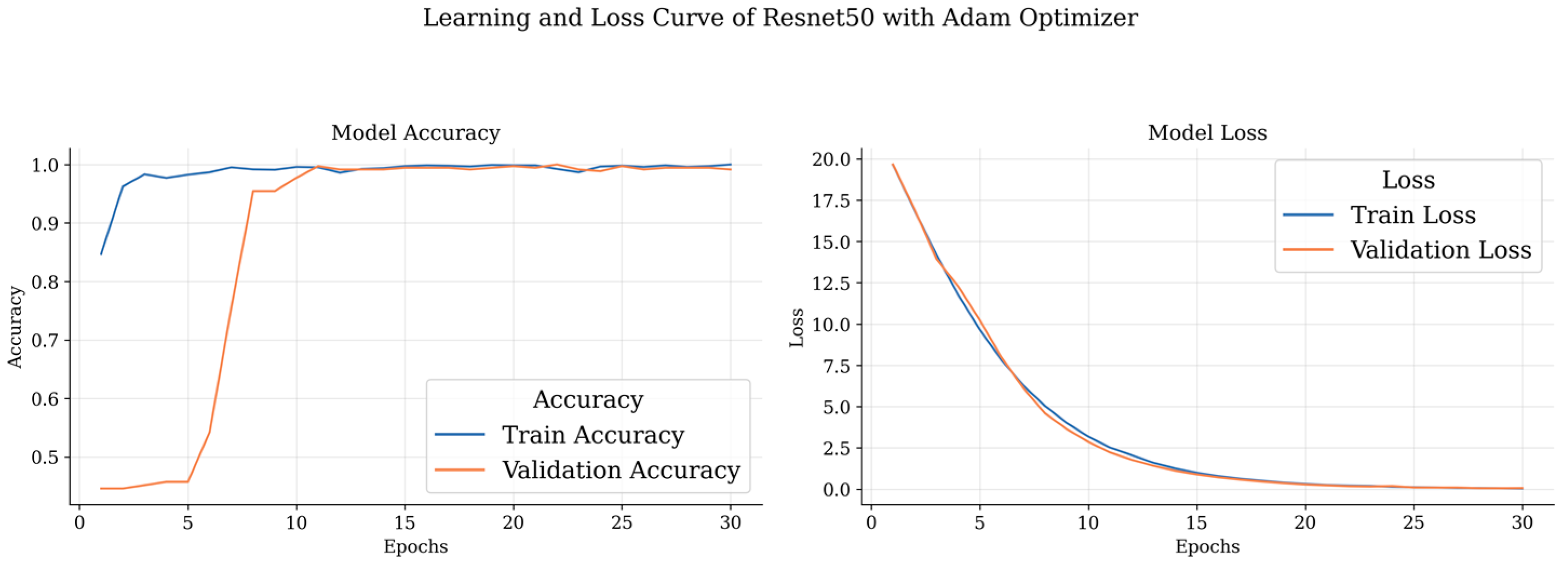
Our data has been trained with ResNet 50 and Inception V3 model optimization. Then, for the purpose of the experiment, we trained our data for up to thirty epochs. Several performance metrics were utilized to evaluate the system as a whole.
Our dataset underwent extensive training, with each model (ResNet 50 and Inception V3) being trained for up to 30 epochs. During this training period, we conducted experiments with three different optimizers: Adam, Nadam, and RMSprop. Additionally, categorical cross-entropy was employed as the loss function. It is noteworthy that our ResNet 50 model achieved its highest accuracy within the initial 12 epochs. To comprehensively assess the system’s performance, we employed a range of performance metrics, providing a holistic evaluation of its efficacy and capabilities.
During the training phase, the inner weights of the model were maintained as non-trainable, with the primary focus placed on training the introduced additional layers. This process involved a systematic introduction of key components, commencing with the incorporation of global average pooling, followed by the sequential implementation of dropout layers. Optimal convergence was determined via experimentation with dropout values ranging from 0.1 to 0.9, ultimately converging on a dropout value of 0.4.
In pursuit of further model optimization, a fully connected layer consisting of 1024 units was introduced, followed by the integration of another dropout layer. Subsequently, a connected layer comprising 512 units was introduced, followed by another dropout layer, culminating in the incorporation of a SoftMax classifier responsible for categorizing images into their respective classes. Vigilant monitoring of the model’s training progress on the validation dataset facilitated judicious adjustments to the fine-tuning strategies as necessitated.
In summary, for optimizing our ResNet 50 and Inception V3 models, we adopted a strategic approach to hyperparameter tuning, with a focus on the additional layers. We kept the base model weights static and experimented with dropout rates, settling on an effective rate of 0.4 after testing various options between 0.1 and 0.9. Our network architecture was enhanced with two fully connected layers, the first containing 1024 neurons and the second 512 neurons, each followed by dropout layers to combat overfitting. The models utilized Adam, Nadam, and RMSprop optimizers for their distinct advantages in convergence and efficiency, with categorical cross-entropy as our loss function. This configuration was refined through continuous validation to ensure optimal performance without overfitting.
7. Experimental Results
The experiments and the outcomes are described in this section. Using an Intel(R) Xeon(R) E5-2680 v4 processor clocked at 2.40 GHz and 32 GB of RAM, the experiments were run in JupyterLab. JupyterLab is the most recent web-based interactive development environment for notebooks, code, and data. It allows users to configure and organize workflows in data science, scientific computing, computational journalism, and machine learning through its adaptable user interface. Moreover, the experiment utilized the libraries Sklearn, numpy, Tensorflow, and Matplotlib.
Two types of transfer learning operations were employed in our study. First, fixed features were removed, and the model was trained using the data at the middle layer. Second, fine-tuning was performed by utilizing the net weights of ResNet 50 and Inception V3, where the fully connected layer at the end was replaced. Additionally, dense and regularization layers were incorporated.
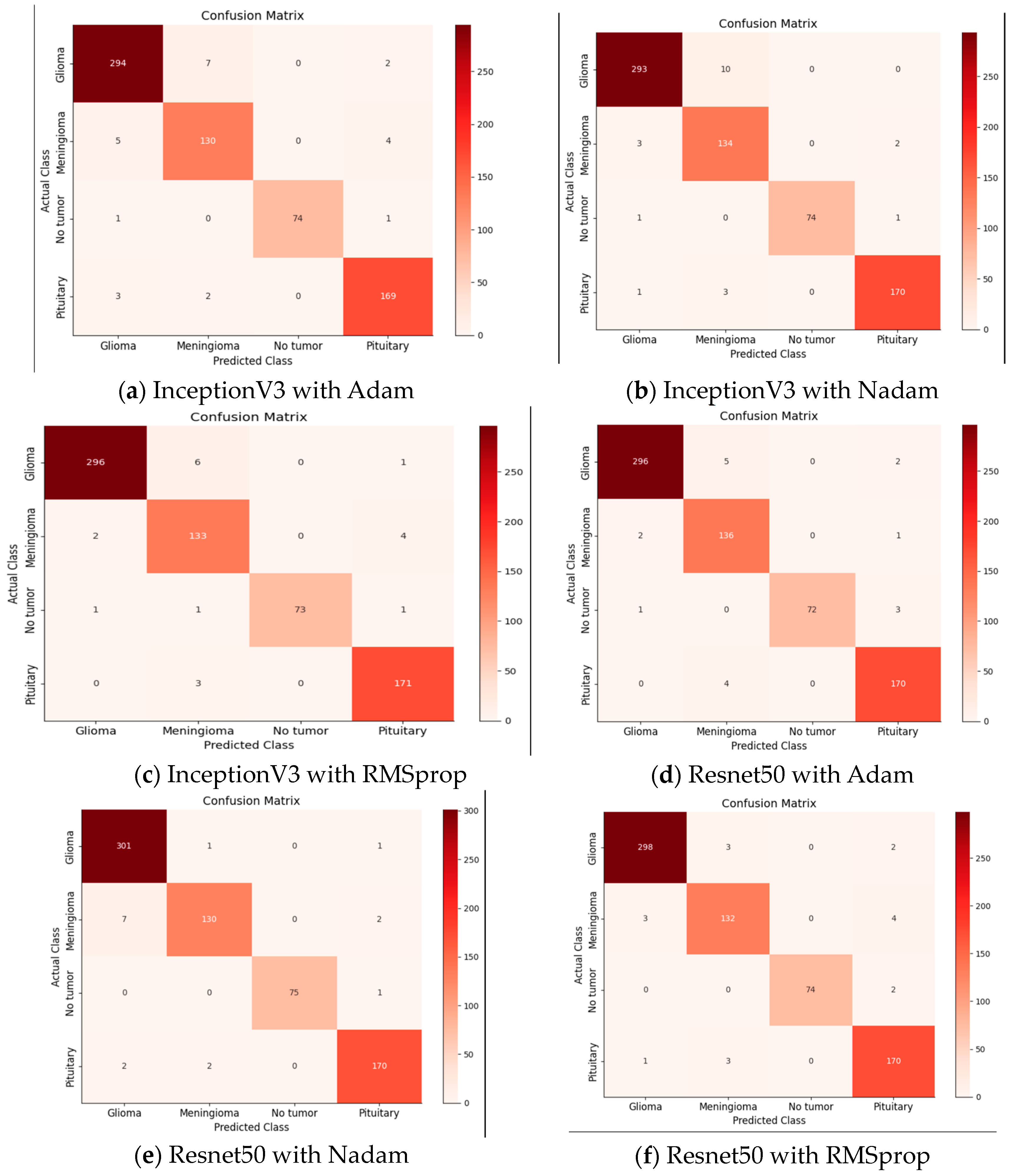
The primary objective of our model was to detect and analyze brain tumors. The classification of our data has involved differentiating between glioma tumors, meningioma tumors, no tumors, and pituitary tumors. To evaluate the performance of our model, we have constructed a 4 × 4 confusion matrix and calculated the true positives, false positives, true negatives, and false negatives. These metrics have provided insights into accurately identifying affected images and correctly detecting tumor presence.
During our data evaluation, our model demonstrated superior results. We have employed the Adam optimizer, which has facilitated quicker and more accurate result calculations. Despite high memory constraints, the Adam optimizer has exhibited efficient performance. In our experiments, we have compared the Adam and Nadam optimizers, both with an initial learning rate of 0.001. The training of the model has employed dynamic learning, wherein the learning rate was reduced after the improvement phase. We have utilized ReduceLROnPlateau with parameter values such as a factor of 0.3 and patience of 5 to monitor and adjust the learning rate based on the validation loss.
To prevent overfitting, an early stopping technique was applied, monitoring the validation loss with a tolerance of 10 s. If the validation loss has not decreased in the remaining 10 epochs, the training phase is concluded. The experiments were conducted for 50 epochs with a batch size of 20, resulting in a training time of approximately 35 min. The loss function employed in our multi-class classification was categorical cross-entropy.
The presented tables showcase the performance metrics of various DL models for brain tumor classification using different optimizers, namely, Adam, Nadam, and RMSprop.
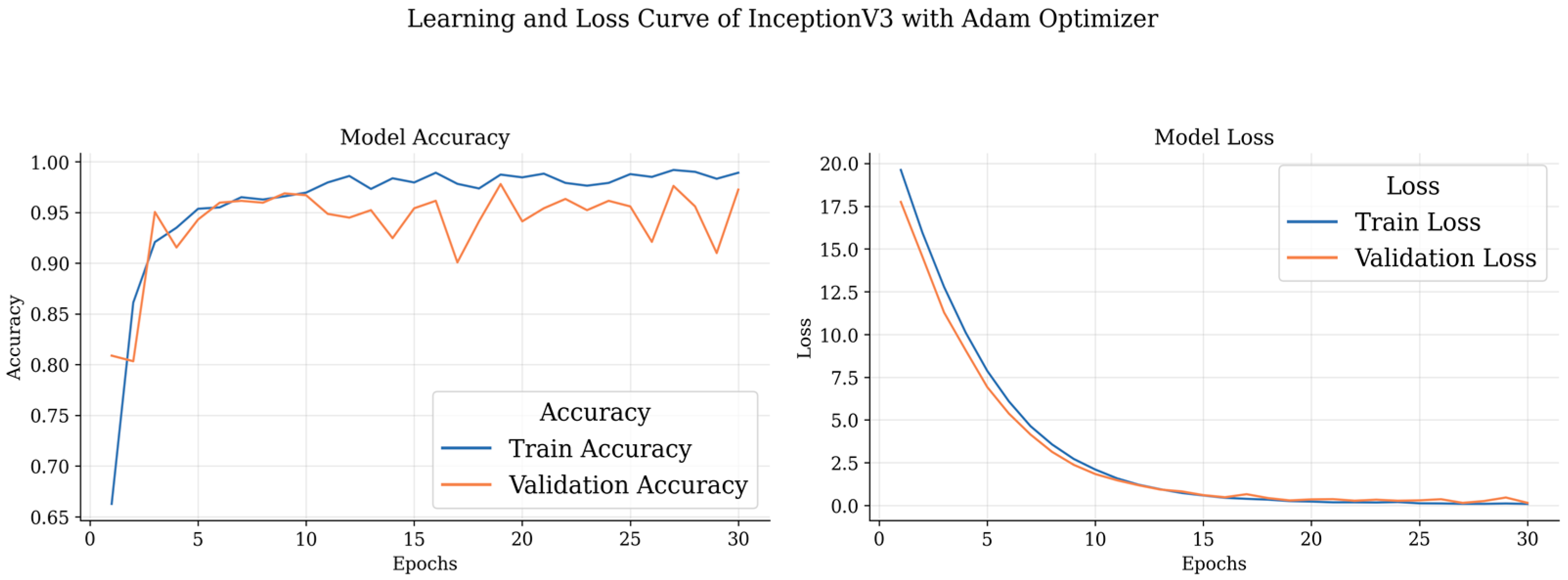
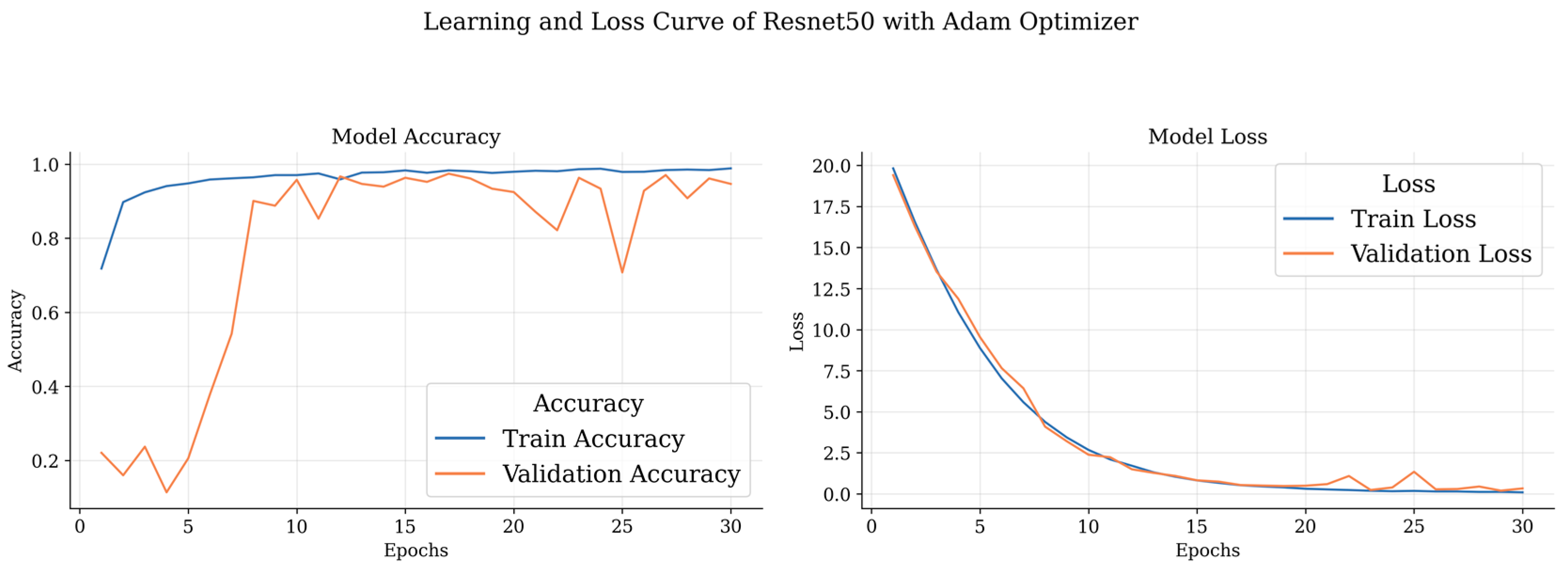
Table 4 outlines the results obtained by DL models with the Adam optimizer. Notably, the ResNet 50 model demonstrates outstanding precision and recall for different brain tumor classes, achieving an accuracy consistently above 97%. Remarkably, it achieves 98.97% precision, 97.69% recall, and an impressive F1 score of 98.33% for glioma classification. The meningioma class also exhibits excellent results, with precision, recall, and F1 scores exceeding 93%. Additionally, the non-tumor class achieves a perfect precision of 100%. The pituitary class shows commendable performance with an F1 score of 97.14% and an accuracy of 97.70%. On the other hand, Inception V3, while not surpassing ResNet 50, still demonstrates robust performance across classes, with accuracy ranging from 93.52% to 97.36%.
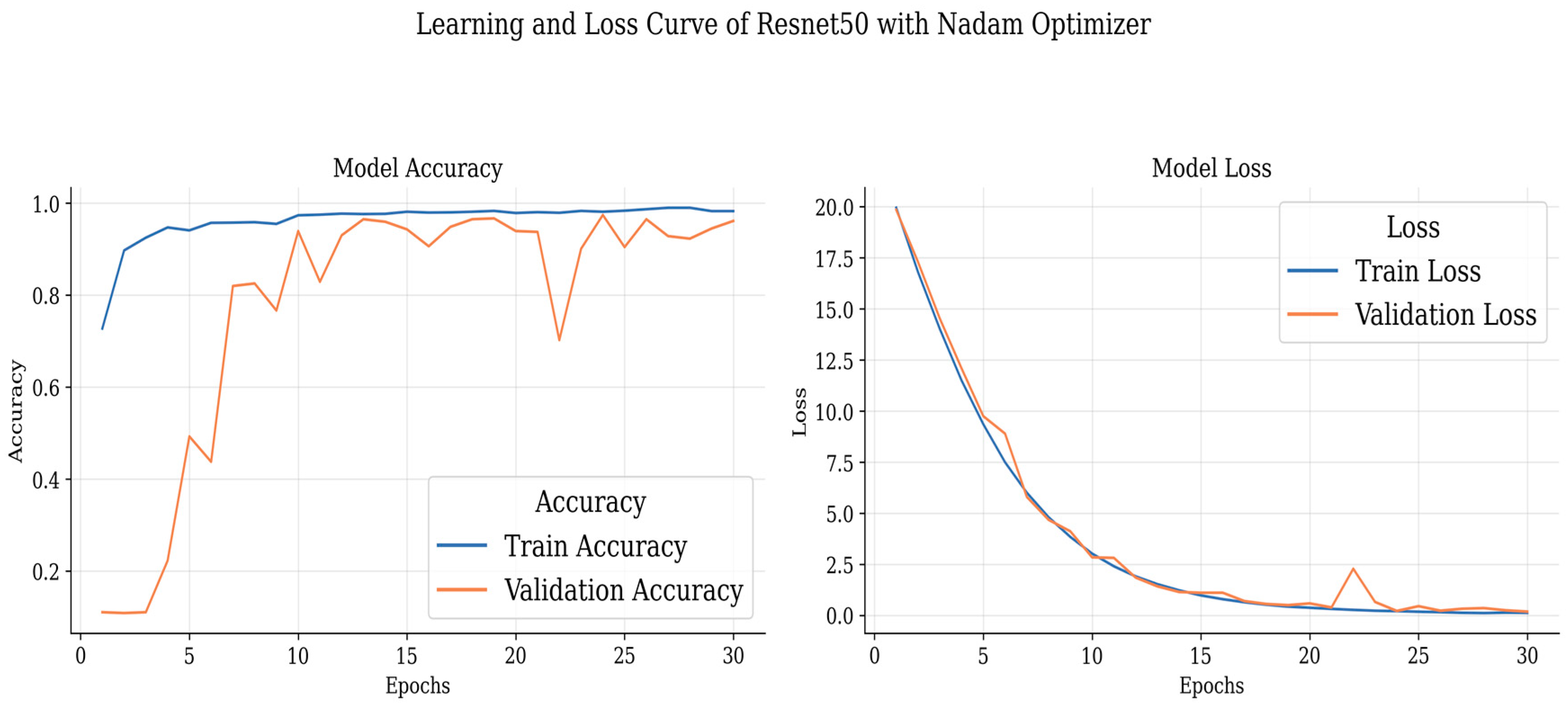
Table 5 highlights the performance metrics obtained using the Nadam optimizer. Once again, ResNet 50 stands out, achieving a precision of 97.09%, recall of 99.33%, and an impressive F1 score of 98.20% for Glioma classification, accompanied by a high accuracy of 99.34%. The meningioma class also performs well with precision, recall, and F1 score exceeding 93%. The non-tumor class maintains a perfect precision of 100%. Inception V3 demonstrates strong performance with an accuracy of 96.70% and an F1 score of 97.50% for glioma classification.
Table 6 presents the performance characteristics for the RMSprop optimizer. ResNet 50 continues to produce outstanding results for glioma classification, with precision, recall, and F1 score over 98%. The meningioma class attains excellent precision and recall levels of approximately 95%, but the non-tumor class maintains a flawless precision of 100%. With an F1 score of 98.33 percent for glioma categorization, Inception V3 fares highly. With high recall and F1 score values, the pituitary class displays impressive performance. From
Figure 7,
Figure 8 and
Figure 9, the accuracy and loss curves of the models trained on the four-class datasets can be observed.
These curves depict the model’s performance over epochs during the training process. The accuracy curve illustrates how well the model improves its predictions as it learns from the training data, while the loss curve shows the decrease in the model’s loss function over time. Analyzing these curves helps in understanding the model’s convergence and potential overfitting or underfitting issues, guiding further optimization and fine-tuning of the brain tumor detection model.
Figure 10 shows all the confusion matrices of the models with different optimizers, providing a comprehensive visual representation of their performance. The analysis of these matrices offers valuable insights into the strengths and weaknesses of each optimizer.
Table 7 presents the performance evaluation of several models and optimizers applied to a four-class dataset. ResNet 50 optimized with Nadam stands out for its remarkable 97.68 percent accuracy and 98.13 percent precision. Inception V3, when paired with RMSprop, exhibits solid performance, obtaining a 97.25 percent accuracy and a 97.02 percent F1 score. The results demonstrate that the choice of optimizer has a considerable impact on the overall performance of the model for this particular dataset, hence, offering helpful guidance for picking the optimal model–optimizer combination.
After achieving the highest performance with Resnet50, we intended to apply only this architecture with the same layers as the prior architecture on the two-class dataset.
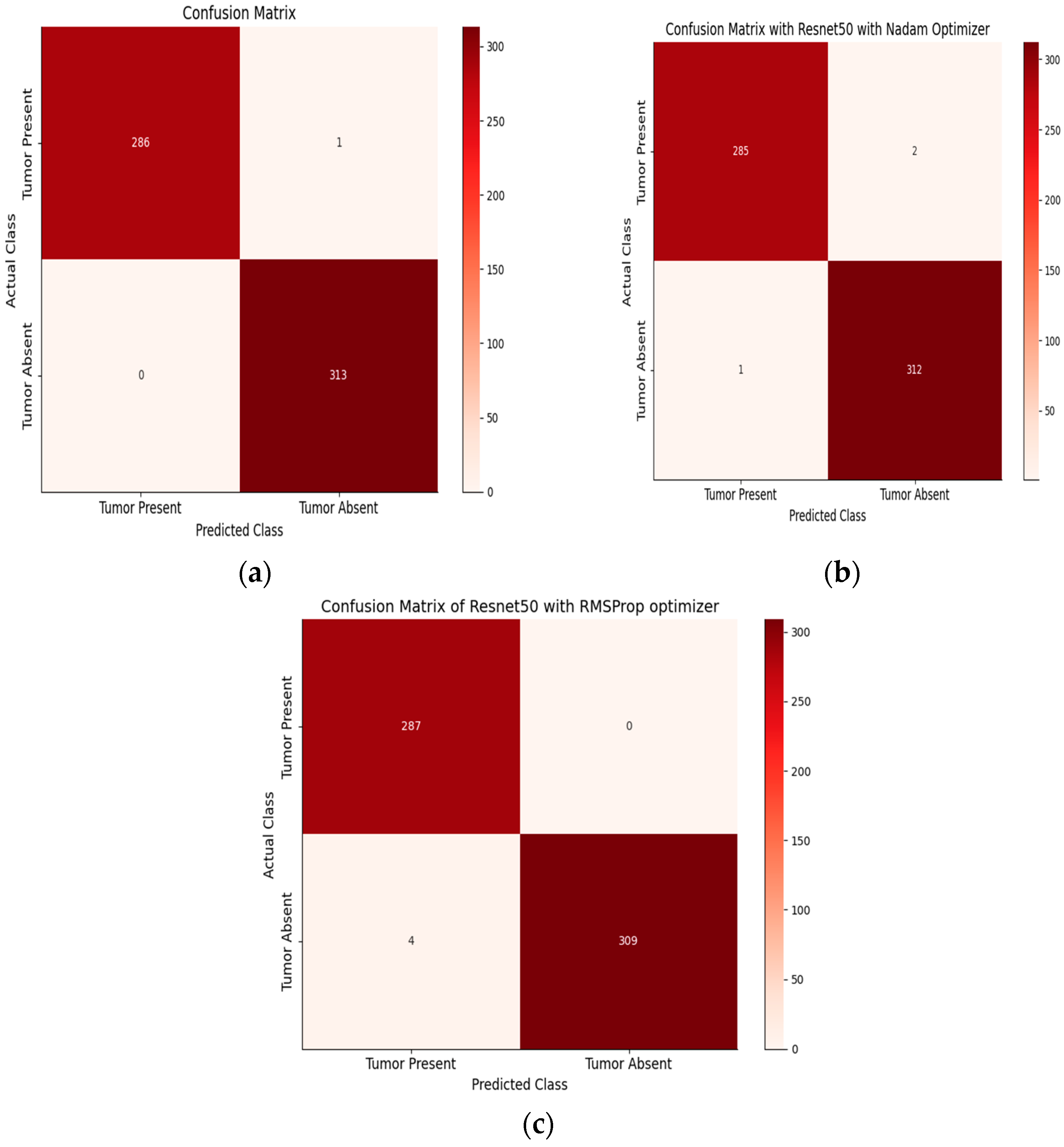
Table 8 shows the performance metrics for tumor classification on a two-class dataset using various models and optimizers. ResNet 50 with Adam achieves 100% precision and 99.65% recall for tumor positive, with an F1 score of 99.82% and an accuracy of 99.62%.
For tumor negative, it achieves 99.68% precision, 100% recall, and an F1 score of 99.81%, resulting in a perfect accuracy of 100%. With Nadam, ResNet 50 attains 99.65% precision, 99.30% recall, and a 99.47% F1 score for tumor positive, with an accuracy of 99.30%. For tumor negative, it achieves 99.36% precision, 99.68% recall, and a 99.52% F1 score, with an accuracy of 99.68%. With RMSprop, ResNet 50 demonstrates 98.96% precision, 99.65% recall, and a 99.30% F1 score for tumor positive, with an accuracy of 99.65%. For tumor negative, it achieves 99.67% precision, 99.04% recall, and a 99.35% F1 score, with an accuracy of 99.04%. Overall, ResNet 50 consistently performs well across all optimizer configurations, making it a suitable choice for accurate tumor classification in a two-class dataset. On the other hand,
Table 9 presents a detailed assessment of the performance of the Inception V3 model using various optimization algorithms for the task of tumor classification. The table focuses on two distinct classes: tumor positive and tumor negative, providing essential metrics in percentage form. Precision, which gauges the accuracy of positive predictions, exhibits consistently high values across all optimizer-model combinations, with values often exceeding 98%. This indicates that the model’s positive predictions are highly accurate. The recall metric, measuring the model’s ability to correctly identify positive instances, also shows strong performance, with values typically above 97%, indicating effective capture of positive cases. The F1 score, a harmonic balance of precision and recall, further highlights the model’s overall accuracy and balance in classification. Finally, the accuracy metric, which reflects the overall correctness of the model’s predictions, consistently demonstrates impressive results, frequently surpassing 98%. These results collectively underscore the robustness and effectiveness of the Inception V3 model in tumor classification, with varying optimizers demonstrating strong performance across both positive and negative classes.
Figure 11 and
Figure 12 showcase the performance evaluation of models trained on the two-class brain MRI tumor dataset.
Figure 10 illustrates accuracy and loss curves, offering insights into the model’s learning progress and convergence during training. On the other hand,
Figure 12 presents the confusion matrix, providing a comprehensive view of the model’s predictions for tumor present and tumor absent classes.
Table 10 provides a comprehensive performance comparison of two prominent DL models, ResNet 50 and Inception V3, utilizing different optimization algorithms on a two-class tumor dataset. For the ResNet 50 model, when optimized with Adam, it achieves a relatively low MAE of 1.6, showcasing the accuracy of its predictions. The model also demonstrates outstanding precision, recall, F1 score, and accuracy, all consistently above 99%. This indicates its exceptional ability to distinguish between the two tumor classes.
When the ResNet 50 model is optimized with Nadam and RMSprop, it continues to exhibit remarkable precision, recall, F1-score, and accuracy, although with slightly different loss values. This indicates that the choice of optimizer can affect training dynamics and model performance.
On the other hand, Inception V3, while still performing well, shows slightly lower precision, recall, F1 score, and accuracy compared to ResNet 50 across all optimizer settings. The MAE values for Inception V3 are also higher, suggesting that it might have slightly less accurate predictions. These results emphasize the importance of selecting the appropriate model and optimizer combination based on specific requirements and trade-offs in the context of tumor classification on this dataset.