Improvement in Disease Diagnosis in Computed Tomography Images by Correlating Organ Volumes with Disease Occurrences in Humans
Abstract
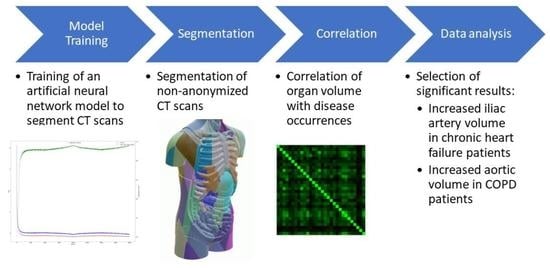
:1. Introduction
2. Materials and Methods
2.1. Model
2.2. Dataset for Model Training
2.3. Dataset Used for Correlation
2.4. Training
- Built-In: The built-in algorithm of the nnU-Net framework. The learning rate starts at the maximum learning rate and decreases polynomially using the formula
- Linear: The learning rate is varied cyclically over the course of training, rising linearly from zero to a maximal learning rate for the first part of a cycle, then decreasing linearly to zero during the second half of the cycle. The entire training consists of two cycles. When using this scheduler, the learning rate is cycled twice. The maximum learning rate is scheduled to be reached after half of the training. The practice of using multiple cycles was inspired by [7]. The formula for calculating the learning rate is
- Linear to Exponential: Modification of linear learning rate scheduling, which replaces the linear decrease in the second part with an exponential decrease, while only using one cycle. The maximum learning rate is scheduled to be reached after one quarter of the training. Here, the formula is
- Modified Linear to Exponential: To increase training efficiency in the early epochs and maintain greater training momentum during the second part of the training, we modified the learning rate schedule by taking the square root of the learning rate in the first part of training and decreasing the base from e to two for the exponential decay in the second half of the training. The modified formula is
3. Results
3.1. Network Training
3.2. Statistical Analysis
4. Discussion
4.1. Network Training
4.2. Statistical Analysis-Simple Correlation Coefficients
4.3. Statistical Analysis-Partial Correlation Coefficients
4.4. Conclusions
4.5. Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CT | Computed Tomography |
| NMDID | New Mexico Decedent Image Database |
Appendix A
| adrenal_gland_left | adrenal_gland_right | aorta |
| autochthon_left | autochthon_right | brain |
| clavicula_left | clavicula_right | colon |
| duodenum | esophagus | face |
| femur_left | femur_right | gallbladder |
| gluteus_maximus_left | gluteus_maximus_right | gluteus_medius_left |
| gluteus_medius_right | gluteus_minimus_left | gluteus_minimus_right |
| heart_atrium_left | heart_atrium_right | heart_myocardium |
| heart_ventricle_left | heart_ventricle_right | hip_left |
| hip_right | humerus_left | humerus_right |
| iliac_artery_left | iliac_artery_right | iliac_vena_left |
| iliac_vena_right | iliopsoas_left | iliopsoas_right |
| inferior_vena_cava | kidney_left | kidney_right |
| liver | lung_lower_lobe_left | lung_lower_lobe_right |
| lung_middle_lobe_right | lung_upper_lobe_left | lung_upper_lobe_right |
| pancreas | portal_vein_and_splenic_vein | pulmonary_artery |
| rib_left_1 | rib_left_10 | rib_left_11 |
| rib_left_12 | rib_left_2 | rib_left_3 |
| rib_left_4 | rib_left_5 | rib_left_6 |
| rib_left_7 | rib_left_8 | rib_left_9 |
| rib_right_1 | rib_right_10 | rib_right_11 |
| rib_right_12 | rib_right_2 | rib_right_3 |
| rib_right_4 | rib_right_5 | rib_right_6 |
| rib_right_7 | rib_right_8 | rib_right_9 |
| sacrum | scapula_left | scapula_right |
| small_bowel | spleen | stomach |
| trachea | urinary_bladder | vertebrae_C1 |
| vertebrae_C2 | vertebrae_C3 | vertebrae_C4 |
| vertebrae_C5 | vertebrae_C6 | vertebrae_C7 |
| vertebrae_L1 | vertebrae_L2 | vertebrae_L3 |
| vertebrae_L4 | vertebrae_L5 | vertebrae_T1 |
| vertebrae_T10 | vertebrae_T11 | vertebrae_T12 |
| vertebrae_T2 | vertebrae_T3 | vertebrae_T4 |
| vertebrae_T5 | vertebrae_T6 | vertebrae_T7 |
| vertebrae_T8 | vertebrae_T9 |
Appendix B

| Disease | Occurrences |
|---|---|
| Hypertension | 181 |
| Diabetes type II | 73 |
| Coronary artery disease | 56 |
| Hyperlipidemia | 43 |
| COPD | 36 |
| Asthma | 33 |
| Cirrhosis of the liver | 31 |
| Hepatitis C | 28 |
| Stroke | 26 |
| Diabetes type I | 22 |
| Arthritis | 19 |
| Chronic heart failure | 18 |
| Non-epileptic seizures | 14 |
| Autoimmune diseases | 12 |
| Myocardial infarction | 12 |
| Staphylococcus aureus | 7 |
| Osteoporosis | 3 |
| HIV/AIDS | 3 |

| Adrenal Gland | Aorta | Autochton | Clavicula | Colon | Duodenum | |
|---|---|---|---|---|---|---|
| Adrenal gland | 0.000 | −0.049 | 0.075 | −0.054 | −0.085 | −0.018 |
| Aorta | −0.049 | 0.000 | 0.053 | −0.054 | −0.034 | 0.059 |
| Autochton | 0.075 | 0.053 | 0.000 | −0.057 | 0.139 | −0.041 |
| Clavicula | −0.054 | −0.054 | −0.057 | 0.000 | 0.082 | −0.045 |
| Colon | −0.085 | −0.034 | 0.139 | 0.082 | 0.000 | 0.154 |
| Duodenum | −0.018 | 0.059 | −0.041 | −0.045 | 0.154 | 0.000 |
| Esophagus | −0.047 | −0.232 | 0.049 | −0.068 | 0.215 | −0.040 |
| Femur | 0.037 | 0.109 | −0.261 | −0.131 | 0.083 | −0.085 |
| Gluteal muscles | −0.010 | −0.080 | 0.358 | −0.031 | 0.120 | 0.038 |
| Heart | 0.130 | 0.029 | −0.170 | −0.043 | −0.066 | 0.015 |
| Hip | −0.014 | −0.022 | −0.142 | 0.018 | −0.037 | 0.088 |
| Humerus | 0.029 | −0.036 | 0.025 | 0.466 | −0.030 | 0.087 |
| Iliac artery | 0.053 | 0.332 | −0.016 | −0.053 | −0.030 | 0.106 |
| Iliac vena | −0.108 | 0.257 | −0.065 | 0.104 | −0.002 | 0.059 |
| Iliopsoas | −0.077 | −0.086 | 0.417 | −0.027 | −0.118 | −0.040 |
| Inferior vena cava | −0.024 | −0.050 | −0.054 | −0.046 | 0.118 | 0.052 |
| Kidney | 0.192 | 0.090 | 0.103 | 0.164 | −0.142 | −0.048 |
| Liver | 0.067 | −0.045 | 0.004 | −0.061 | 0.037 | 0.017 |
| Lung | −0.090 | 0.168 | 0.100 | −0.062 | −0.085 | 0.000 |
| Pancreas | 0.110 | 0.066 | −0.040 | 0.035 | −0.343 | 0.201 |
| Portal and splenic vein | 0.123 | 0.010 | 0.000 | −0.049 | 0.126 | 0.097 |
| Pulmonary artery | 0.065 | 0.117 | 0.108 | 0.076 | −0.014 | 0.017 |
| Ribs | −0.050 | −0.103 | 0.145 | 0.045 | −0.028 | 0.111 |
| Scapula | 0.093 | 0.089 | 0.116 | 0.428 | −0.074 | 0.020 |
| Small bowel | 0.088 | 0.042 | −0.004 | −0.021 | 0.351 | −0.090 |
| Spleen | 0.085 | −0.014 | 0.120 | 0.015 | 0.020 | 0.053 |
| Stomach | −0.057 | 0.084 | 0.097 | −0.030 | 0.079 | 0.050 |
| Trachea | −0.017 | 0.031 | −0.060 | 0.004 | 0.083 | −0.133 |
| Urinary bladder | −0.088 | −0.052 | 0.064 | −0.000 | −0.026 | 0.031 |
| Vertebrae | 0.017 | 0.105 | 0.103 | 0.044 | 0.092 | −0.116 |
| Diabetes type I | −0.024 | 0.003 | 0.003 | −0.020 | 0.062 | −0.034 |
| Diabetes type II | 0.121 | −0.029 | −0.039 | 0.103 | 0.019 | 0.020 |
| COPD | −0.074 | 0.139 | 0.028 | 0.048 | −0.116 | 0.016 |
| Non−epileptic seizures | −0.012 | 0.071 | 0.058 | −0.008 | −0.027 | −0.046 |
| Asthma | −0.048 | −0.023 | 0.021 | −0.088 | 0.081 | −0.027 |
| Hypertension | 0.097 | 0.066 | −0.047 | 0.090 | −0.024 | −0.025 |
| Arthritis | −0.012 | 0.021 | −0.017 | 0.099 | 0.082 | −0.085 |
| Chronic heart failure | −0.024 | −0.040 | 0.063 | −0.058 | 0.091 | −0.022 |
| Stroke | 0.039 | 0.040 | −0.023 | 0.027 | −0.014 | −0.086 |
| Myocardial infarction | 0.030 | −0.039 | 0.089 | 0.024 | 0.063 | −0.012 |
| Hyperlipidemia | −0.046 | −0.033 | −0.034 | 0.024 | −0.066 | 0.019 |
| HIV/AIDS | 0.024 | 0.001 | 0.059 | 0.004 | −0.028 | −0.016 |
| Hepatitis C | 0.013 | −0.012 | −0.001 | −0.012 | −0.045 | −0.056 |
| Osteoporosis | 0.006 | −0.032 | −0.035 | −0.017 | 0.013 | 0.005 |
| Cirrhosis of the liver | 0.069 | 0.052 | −0.004 | 0.085 | −0.054 | 0.024 |
| Coronary artery disease | −0.120 | −0.095 | −0.011 | 0.025 | −0.009 | 0.033 |
| Staphylococcus aureus | −0.006 | 0.092 | −0.012 | 0.048 | 0.074 | −0.185 |
| Autoimmune diseases | 0.064 | −0.031 | −0.007 | 0.065 | −0.044 | 0.059 |
| Esophagus | Femur | Gluteal Muscles | Heart | Hip | ||
| Adrenal gland | −0.047 | 0.037 | −0.010 | 0.130 | −0.014 | |
| Aorta | −0.232 | 0.109 | −0.080 | 0.029 | −0.022 | |
| Autochton | 0.049 | −0.261 | 0.358 | −0.170 | −0.142 | |
| Clavicula | −0.068 | −0.131 | −0.031 | −0.043 | 0.018 | |
| Colon | 0.215 | 0.083 | 0.120 | −0.066 | −0.037 | |
| Duodenum | −0.040 | −0.085 | 0.038 | 0.015 | 0.088 | |
| Esophagus | 0.000 | 0.019 | −0.003 | 0.074 | 0.038 | |
| Femur | 0.019 | 0.000 | 0.148 | 0.014 | 0.296 | |
| Gluteal muscles | −0.003 | 0.148 | 0.000 | 0.014 | 0.163 | |
| Heart | 0.074 | 0.014 | 0.014 | 0.000 | −0.086 | |
| Hip | 0.038 | 0.296 | 0.163 | −0.086 | 0.000 | |
| Humerus | 0.008 | 0.083 | −0.055 | −0.065 | −0.020 | |
| Iliac artery | −0.038 | 0.103 | −0.071 | 0.067 | 0.124 | |
| Iliac vena | 0.096 | 0.013 | 0.060 | 0.199 | 0.125 | |
| Iliopsoas | −0.024 | 0.047 | 0.480 | 0.064 | 0.080 | |
| Inferior vena cava | −0.210 | −0.180 | 0.010 | −0.031 | −0.061 | |
| Kidney | 0.050 | 0.001 | 0.079 | 0.065 | −0.017 | |
| Liver | −0.007 | −0.094 | 0.016 | 0.092 | 0.019 | |
| Lung | 0.206 | −0.053 | −0.202 | 0.075 | 0.063 | |
| Pancreas | 0.086 | 0.033 | 0.030 | −0.074 | −0.090 | |
| Portal and splenic vein | 0.107 | −0.149 | 0.096 | 0.088 | −0.047 | |
| Pulmonary artery | 0.188 | 0.026 | 0.100 | 0.388 | −0.055 | |
| Ribs | −0.089 | 0.167 | −0.068 | 0.003 | 0.004 | |
| Scapula | 0.108 | 0.083 | −0.000 | 0.119 | 0.058 | |
| Small bowel | 0.005 | 0.016 | 0.024 | −0.060 | −0.018 | |
| Spleen | 0.176 | −0.075 | 0.015 | 0.138 | −0.021 | |
| Stomach | 0.114 | −0.065 | 0.125 | 0.116 | −0.011 | |
| Trachea | 0.109 | −0.072 | −0.087 | 0.021 | −0.015 | |
| Urinary bladder | 0.066 | 0.040 | 0.043 | 0.100 | 0.017 | |
| Vertebrae | −0.025 | −0.076 | 0.013 | 0.008 | 0.536 | |
| Diabetes type I | 0.185 | −0.031 | −0.080 | −0.065 | 0.050 | |
| Diabetes type II | 0.077 | −0.040 | −0.009 | −0.016 | 0.070 | |
| COPD | 0.018 | 0.131 | 0.101 | −0.053 | −0.074 | |
| Non−epileptic seizures | 0.022 | 0.000 | −0.025 | 0.079 | 0.026 | |
| Asthma | −0.049 | 0.001 | 0.006 | −0.001 | 0.026 | |
| Hypertension | 0.040 | 0.043 | 0.094 | 0.075 | −0.117 | |
| Arthritis | 0.032 | 0.054 | 0.019 | 0.006 | 0.040 | |
| Chronic heart failure | −0.092 | −0.000 | −0.005 | 0.154 | −0.045 | |
| Stroke | 0.091 | −0.050 | −0.110 | −0.012 | 0.000 | |
| Myocardial infarction | 0.042 | 0.071 | 0.011 | 0.129 | −0.042 | |
| Hyperlipidemia | 0.041 | −0.023 | 0.038 | −0.044 | 0.003 | |
| HIV/AIDS | 0.012 | 0.121 | 0.018 | 0.114 | −0.028 | |
| Hepatitis C | −0.026 | 0.029 | 0.029 | 0.091 | −0.051 | |
| Osteoporosis | −0.014 | −0.152 | −0.041 | −0.053 | 0.051 | |
| Cirrhosis of the liver | 0.066 | −0.004 | −0.012 | 0.022 | 0.031 | |
| Coronary artery disease | −0.014 | 0.070 | 0.070 | 0.111 | −0.104 | |
| Staphylococcus aureus | 0.105 | −0.055 | −0.017 | −0.020 | 0.025 | |
| Autoimmune diseases | 0.055 | 0.102 | 0.000 | 0.028 | −0.055 | |
| Humerus | Iliac Artery | Iliac Vena | Iliopsoas | Inferior Vena Cava | ||
| Adrenal gland | 0.029 | 0.053 | −0.108 | −0.077 | −0.024 | |
| Aorta | −0.036 | 0.332 | 0.257 | −0.086 | −0.050 | |
| Autochton | 0.025 | −0.016 | −0.065 | 0.417 | −0.054 | |
| Clavicula | 0.466 | −0.053 | 0.104 | −0.027 | −0.046 | |
| Colon | −0.030 | −0.030 | −0.002 | −0.118 | 0.118 | |
| Duodenum | 0.087 | 0.106 | 0.059 | −0.040 | 0.052 | |
| Esophagus | 0.008 | −0.038 | 0.096 | −0.024 | −0.210 | |
| Femur | 0.083 | 0.103 | 0.013 | 0.047 | −0.180 | |
| Gluteal muscles | −0.055 | −0.071 | 0.060 | 0.480 | 0.010 | |
| Heart | −0.065 | 0.067 | 0.199 | 0.064 | −0.031 | |
| Hip | −0.020 | 0.124 | 0.125 | 0.080 | −0.061 | |
| Humerus | 0.000 | −0.036 | −0.022 | −0.019 | 0.056 | |
| Iliac artery | −0.036 | 0.000 | 0.173 | −0.013 | 0.119 | |
| Iliac vena | −0.022 | 0.173 | 0.000 | 0.108 | 0.131 | |
| Iliopsoas | −0.019 | −0.013 | 0.108 | 0.000 | −0.013 | |
| Inferior vena cava | 0.056 | 0.119 | 0.131 | −0.013 | 0.000 | |
| Kidney | −0.023 | −0.096 | 0.209 | −0.068 | 0.154 | |
| Liver | −0.053 | −0.135 | 0.127 | −0.100 | 0.034 | |
| Lung | −0.028 | −0.044 | 0.002 | 0.100 | −0.048 | |
| Pancreas | −0.004 | −0.018 | 0.127 | 0.104 | 0.079 | |
| Portal and splenic vein | 0.097 | 0.066 | 0.030 | −0.078 | −0.104 | |
| Pulmonary artery | 0.066 | 0.052 | −0.065 | −0.115 | 0.089 | |
| Ribs | −0.327 | −0.052 | −0.035 | −0.032 | 0.066 | |
| Scapula | 0.299 | 0.039 | −0.013 | 0.133 | −0.009 | |
| Small bowel | −0.051 | 0.178 | −0.186 | 0.002 | −0.016 | |
| Spleen | 0.016 | 0.037 | 0.072 | −0.079 | −0.008 | |
| Stomach | −0.017 | −0.100 | 0.100 | −0.071 | 0.084 | |
| Trachea | 0.034 | 0.040 | −0.018 | 0.066 | −0.063 | |
| Urinary bladder | −0.053 | 0.037 | −0.149 | −0.110 | −0.015 | |
| Vertebrae | −0.063 | 0.044 | −0.159 | −0.126 | 0.180 | |
| Diabetes type I | 0.023 | 0.021 | −0.041 | 0.036 | 0.050 | |
| Diabetes type II | −0.037 | 0.049 | −0.125 | −0.041 | 0.025 | |
| COPD | 0.052 | 0.065 | −0.038 | −0.107 | −0.025 | |
| Non−epileptic seizures | 0.053 | −0.032 | −0.075 | −0.002 | 0.075 | |
| Asthma | −0.026 | 0.044 | −0.016 | 0.003 | 0.002 | |
| Hypertension | −0.063 | 0.103 | −0.061 | −0.006 | 0.011 | |
| Arthritis | 0.039 | −0.042 | 0.001 | 0.012 | 0.094 | |
| Chronic heart failure | −0.066 | 0.137 | 0.007 | −0.099 | 0.032 | |
| Stroke | 0.104 | 0.018 | 0.082 | 0.055 | −0.003 | |
| Myocardial infarction | −0.038 | 0.122 | 0.003 | −0.062 | 0.040 | |
| Hyperlipidemia | −0.019 | −0.032 | 0.026 | −0.061 | −0.015 | |
| HIV/AIDS | −0.026 | −0.041 | −0.052 | −0.016 | 0.022 | |
| Hepatitis C | 0.059 | −0.028 | 0.039 | −0.003 | −0.003 | |
| Osteoporosis | −0.022 | 0.019 | 0.052 | 0.062 | −0.062 | |
| Cirrhosis of the liver | −0.006 | −0.045 | −0.028 | −0.071 | −0.064 | |
| Coronary artery disease | 0.004 | 0.170 | −0.038 | 0.020 | 0.088 | |
| Staphylococcus aureus | −0.003 | 0.115 | 0.017 | 0.031 | −0.027 | |
| Autoimmune diseases | 0.028 | 0.009 | 0.016 | 0.015 | −0.051 | |
| Adrenal gland | 0.192 | 0.067 | −0.090 | 0.110 | 0.123 | |
| Aorta | 0.090 | −0.045 | 0.168 | 0.066 | 0.010 | |
| Autochton | 0.103 | 0.004 | 0.100 | −0.040 | 0.000 | |
| Clavicula | 0.164 | −0.061 | −0.062 | 0.035 | −0.049 | |
| Kidney | Liver | Lung | Pancreas | Portal and Splenic Vein | ||
| Colon | −0.142 | 0.037 | −0.085 | −0.343 | 0.126 | |
| Duodenum | −0.048 | 0.017 | 0.000 | 0.201 | 0.097 | |
| Esophagus | 0.050 | −0.007 | 0.206 | 0.086 | 0.107 | |
| Femur | 0.001 | −0.094 | −0.053 | 0.033 | −0.149 | |
| Gluteal muscles | 0.079 | 0.016 | −0.202 | 0.030 | 0.096 | |
| Heart | 0.065 | 0.092 | 0.075 | −0.074 | 0.088 | |
| Hip | −0.017 | 0.019 | 0.063 | −0.090 | −0.047 | |
| Humerus | −0.023 | −0.053 | −0.028 | −0.004 | 0.097 | |
| Iliac artery | −0.096 | −0.135 | −0.044 | −0.018 | 0.066 | |
| Iliac vena | 0.209 | 0.127 | 0.002 | 0.127 | 0.030 | |
| Iliopsoas | −0.068 | −0.100 | 0.100 | 0.104 | −0.078 | |
| Inferior vena cava | 0.154 | 0.034 | −0.048 | 0.079 | −0.104 | |
| Kidney | 0.000 | 0.091 | −0.042 | −0.031 | 0.018 | |
| Liver | 0.091 | 0.000 | 0.109 | 0.168 | −0.125 | |
| Lung | −0.042 | 0.109 | 0.000 | −0.193 | 0.057 | |
| Pancreas | −0.031 | 0.168 | −0.193 | 0.000 | 0.239 | |
| Portal and splenic vein | 0.018 | −0.125 | 0.057 | 0.239 | 0.000 | |
| Pulmonary artery | −0.026 | −0.060 | −0.064 | 0.037 | −0.087 | |
| Ribs | 0.203 | −0.076 | 0.198 | 0.091 | 0.028 | |
| Scapula | −0.142 | 0.109 | 0.038 | −0.077 | −0.064 | |
| Small bowel | 0.209 | 0.247 | −0.082 | 0.150 | 0.029 | |
| Spleen | 0.121 | 0.173 | −0.094 | 0.113 | 0.067 | |
| Stomach | 0.084 | −0.096 | −0.030 | 0.147 | −0.057 | |
| Trachea | −0.044 | −0.106 | 0.078 | 0.042 | −0.048 | |
| Urinary bladder | 0.059 | 0.132 | 0.052 | 0.068 | 0.029 | |
| Vertebrae | −0.032 | −0.008 | 0.122 | 0.075 | 0.040 | |
| Diabetes type I | 0.112 | 0.051 | −0.013 | −0.060 | −0.063 | |
| Diabetes type II | 0.205 | 0.020 | −0.006 | 0.055 | −0.093 | |
| COPD | −0.017 | −0.041 | 0.174 | −0.029 | 0.018 | |
| Non−epileptic seizures | −0.014 | −0.023 | −0.115 | −0.028 | 0.060 | |
| Asthma | 0.003 | −0.070 | 0.053 | 0.065 | 0.044 | |
| Hypertension | −0.148 | −0.008 | −0.075 | −0.048 | 0.107 | |
| Arthritis | −0.049 | 0.044 | 0.003 | −0.033 | 0.071 | |
| Chronic heart failure | −0.117 | −0.035 | −0.060 | −0.046 | 0.015 | |
| Stroke | 0.037 | −0.003 | 0.020 | 0.120 | −0.058 | |
| Myocardial infarction | 0.001 | −0.014 | 0.049 | −0.003 | −0.007 | |
| Hyperlipidemia | 0.077 | −0.094 | 0.037 | 0.013 | −0.004 | |
| HIV/AIDS | −0.048 | 0.114 | 0.000 | 0.042 | −0.017 | |
| Hepatitis C | −0.097 | 0.047 | 0.057 | 0.098 | −0.014 | |
| Osteoporosis | 0.067 | 0.016 | 0.080 | −0.005 | −0.050 | |
| Cirrhosis of the liver | 0.258 | 0.097 | 0.031 | 0.096 | −0.148 | |
| Coronary artery disease | −0.011 | −0.059 | −0.027 | 0.010 | −0.003 | |
| Staphylococcus aureus | −0.092 | 0.110 | 0.044 | −0.070 | 0.031 | |
| Autoimmune diseases | −0.036 | 0.028 | −0.034 | −0.028 | −0.029 | |
| Pulmonary Artery | Ribs | Scapula | Small Bowel | Spleen | ||
| Adrenal gland | 0.065 | −0.050 | 0.093 | 0.088 | 0.085 | |
| Aorta | 0.117 | −0.103 | 0.089 | 0.042 | −0.014 | |
| Autochton | 0.108 | 0.145 | 0.116 | −0.004 | 0.120 | |
| Clavicula | 0.076 | 0.045 | 0.428 | −0.021 | 0.015 | |
| Colon | −0.014 | −0.028 | −0.074 | 0.351 | 0.020 | |
| Duodenum | 0.017 | 0.111 | 0.020 | −0.090 | 0.053 | |
| Esophagus | 0.188 | −0.089 | 0.108 | 0.005 | 0.176 | |
| Femur | 0.026 | 0.167 | 0.083 | 0.016 | −0.075 | |
| Gluteal muscles | 0.100 | −0.068 | −0.000 | 0.024 | 0.015 | |
| Heart | 0.388 | 0.003 | 0.119 | −0.060 | 0.138 | |
| Hip | −0.055 | 0.004 | 0.058 | −0.018 | −0.021 | |
| Humerus | 0.066 | −0.327 | 0.299 | −0.051 | 0.016 | |
| Iliac artery | 0.052 | −0.052 | 0.039 | 0.178 | 0.037 | |
| Iliac vena | −0.065 | −0.035 | −0.013 | −0.186 | 0.072 | |
| Iliopsoas | −0.115 | −0.032 | 0.133 | 0.002 | −0.079 | |
| Inferior vena cava | 0.089 | 0.066 | −0.009 | −0.016 | −0.008 | |
| Kidney | −0.026 | 0.203 | −0.142 | 0.209 | 0.121 | |
| Liver | −0.060 | −0.076 | 0.109 | 0.247 | 0.173 | |
| Lung | −0.064 | 0.198 | 0.038 | −0.082 | −0.094 | |
| Pancreas | 0.037 | 0.091 | −0.077 | 0.150 | 0.113 | |
| Portal and splenic vein | −0.087 | 0.028 | −0.064 | 0.029 | 0.067 | |
| Pulmonary artery | 0.000 | −0.051 | −0.131 | −0.010 | −0.013 | |
| Ribs | −0.051 | 0.000 | 0.408 | 0.059 | 0.007 | |
| Scapula | −0.131 | 0.408 | 0.000 | 0.113 | −0.038 | |
| Small bowel | −0.010 | 0.059 | 0.113 | 0.000 | −0.017 | |
| Spleen | −0.013 | 0.007 | −0.038 | −0.017 | 0.000 | |
| Stomach | 0.162 | 0.026 | 0.001 | 0.155 | −0.364 | |
| Trachea | 0.037 | −0.018 | 0.049 | −0.100 | −0.025 | |
| Urinary bladder | −0.042 | 0.042 | 0.011 | −0.114 | 0.026 | |
| Vertebrae | 0.145 | 0.212 | 0.183 | −0.044 | 0.079 | |
| Diabetes type I | −0.037 | −0.089 | −0.013 | 0.059 | −0.013 | |
| Diabetes type II | −0.055 | 0.011 | −0.081 | 0.076 | −0.060 | |
| COPD | 0.043 | −0.061 | −0.119 | 0.055 | 0.040 | |
| Non−epileptic seizures | −0.042 | 0.091 | −0.073 | 0.009 | 0.025 | |
| Asthma | 0.075 | −0.032 | 0.112 | −0.092 | 0.029 | |
| Hypertension | −0.031 | −0.002 | −0.035 | −0.011 | 0.040 | |
| Arthritis | 0.002 | 0.014 | −0.172 | 0.099 | −0.026 | |
| Chronic heart failure | −0.038 | 0.029 | 0.167 | −0.072 | 0.111 | |
| Stroke | −0.041 | 0.136 | −0.154 | 0.014 | −0.156 | |
| Myocardial infarction | −0.052 | −0.045 | 0.021 | 0.013 | −0.049 | |
| Hyperlipidemia | −0.045 | −0.012 | 0.010 | 0.065 | 0.004 | |
| HIV/AIDS | −0.007 | −0.042 | −0.063 | −0.023 | −0.053 | |
| Hepatitis C | −0.090 | 0.049 | −0.022 | 0.081 | −0.035 | |
| Osteoporosis | 0.050 | −0.042 | 0.035 | −0.010 | −0.038 | |
| Cirrhosis of the liver | −0.078 | −0.022 | −0.074 | 0.135 | −0.266 | |
| Coronary artery disease | −0.019 | 0.033 | −0.058 | −0.085 | 0.001 | |
| Staphylococcus aureus | −0.058 | 0.012 | −0.007 | 0.047 | 0.005 | |
| Autoimmune diseases | −0.096 | −0.052 | −0.136 | 0.004 | −0.043 | |
| Stomach | Trachea | Urinary Bladder | Vertebrae | |||
| Adrenal gland | −0.057 | −0.017 | −0.088 | 0.017 | ||
| Aorta | 0.084 | 0.031 | −0.052 | 0.105 | ||
| Autochton | 0.097 | −0.060 | 0.064 | 0.103 | ||
| Clavicula | −0.030 | 0.004 | −0.000 | 0.044 | ||
| Colon | 0.079 | 0.083 | −0.026 | 0.092 | ||
| Duodenum | 0.050 | −0.133 | 0.031 | −0.116 | ||
| Esophagus | 0.114 | 0.109 | 0.066 | −0.025 | ||
| Femur | −0.065 | −0.072 | 0.040 | −0.076 | ||
| Gluteal muscles | 0.125 | −0.087 | 0.043 | 0.013 | ||
| Heart | 0.116 | 0.021 | 0.100 | 0.008 | ||
| Hip | −0.011 | −0.015 | 0.017 | 0.536 | ||
| Humerus | −0.017 | 0.034 | −0.053 | −0.063 | ||
| Iliac artery | −0.100 | 0.040 | 0.037 | 0.044 | ||
| Iliac vena | 0.100 | −0.018 | −0.149 | −0.159 | ||
| Iliopsoas | −0.071 | 0.066 | −0.110 | −0.126 | ||
| Inferior vena cava | 0.084 | −0.063 | −0.015 | 0.180 | ||
| Kidney | 0.084 | −0.044 | 0.059 | −0.032 | ||
| Liver | −0.096 | −0.106 | 0.132 | −0.008 | ||
| Lung | −0.030 | 0.078 | 0.052 | 0.122 | ||
| Pancreas | 0.147 | 0.042 | 0.068 | 0.075 | ||
| Portal and splenic vein | −0.057 | −0.048 | 0.029 | 0.040 | ||
| Pulmonary artery | 0.162 | 0.037 | −0.042 | 0.145 | ||
| Ribs | 0.026 | −0.018 | 0.042 | 0.212 | ||
| Scapula | 0.001 | 0.049 | 0.011 | 0.183 | ||
| Small bowel | 0.155 | −0.100 | −0.114 | −0.044 | ||
| Spleen | −0.364 | −0.025 | 0.026 | 0.079 | ||
| Stomach | 0.000 | −0.074 | 0.023 | −0.063 | ||
| Trachea | −0.074 | 0.000 | 0.156 | 0.133 | ||
| Urinary bladder | 0.023 | 0.156 | 0.000 | −0.039 | ||
| Vertebrae | −0.063 | 0.133 | −0.039 | 0.000 | ||
| Diabetes type I | −0.034 | −0.029 | 0.056 | 0.017 | ||
| Diabetes type II | −0.054 | 0.024 | 0.099 | −0.047 | ||
| COPD | 0.004 | −0.111 | −0.124 | 0.102 | ||
| Non−epileptic seizures | 0.024 | 0.063 | 0.116 | −0.085 | ||
| Asthma | 0.021 | −0.077 | −0.037 | −0.135 | ||
| Hypertension | 0.034 | −0.086 | −0.022 | 0.121 | ||
| Arthritis | −0.087 | 0.036 | −0.001 | 0.009 | ||
| Chronic heart failure | 0.055 | 0.018 | 0.072 | −0.070 | ||
| Stroke | −0.105 | −0.068 | 0.057 | 0.053 | ||
| Myocardial infarction | −0.150 | −0.028 | −0.051 | −0.035 | ||
| Hyperlipidemia | 0.010 | −0.054 | −0.030 | −0.004 | ||
| HIV/AIDS | −0.047 | 0.094 | −0.014 | 0.064 | ||
| Hepatitis C | −0.071 | −0.034 | −0.025 | 0.020 | ||
| Osteoporosis | −0.040 | 0.050 | −0.013 | −0.027 | ||
| Cirrhosis of the liver | −0.128 | −0.000 | 0.108 | −0.048 | ||
| Coronary artery disease | −0.011 | −0.062 | −0.052 | 0.048 | ||
| Staphylococcus aureus | −0.002 | −0.135 | 0.025 | −0.033 | ||
| Autoimmune diseases | 0.112 | −0.090 | −0.001 | 0.160 |
References
- Takuma, Y.; Nouso, K.; Morimoto, Y.; Tomokuni, J.; Sahara, A.; Takabatake, H.; Matsueda, K.; Yamamoto, H. Portal Hypertension in Patients with Liver Cirrhosis: Diagnostic Accuracy of Spleen Stiffness. Radiology 2016, 279, 609–619. [Google Scholar] [CrossRef] [PubMed]
- Egger, J.; Gsaxner, C.; Pepe, A.; Pomykala, K.L.; Jonske, F.; Kurz, M.; Li, J.; Kleesiek, J. Medical deep learning—A systematic meta-review. Comput. Methods Programs Biomed. 2022, 221, 106874. [Google Scholar] [CrossRef] [PubMed]
- Wasserthal, J.; Meyer, M.; Breit, H.C.; Cyriac, J.; Yang, S.; Segeroth, M. TotalSegmentator: Robust segmentation of 104 anatomical structures in CT images. arXiv 2022, arXiv:2208.05868. [Google Scholar] [CrossRef]
- Edgar, H.; Daneshvari Berry, S.; Moes, E.; Adolphi, N.; Bridges, P.; Nolte, K. New Mexico Decedent Image Database; Office of the Medical Investigator, University of New Mexico: Albuquerque, NM, USA, 2020. [Google Scholar] [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image Segmentation. Nat. Methods 2020, 18, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention (MICCAI), Munich, Germany, 5–9 October 2015; LNCS; Springer: Berlin/Heidelberg, Germany, 2015; Volume 9351, pp. 234–241. [Google Scholar] [CrossRef] [Green Version]
- Smith, L.N. Cyclical Learning Rates for Training Neural Networks. In Proceedings of the 2017 IEEE Winter Conference on Applications of Computer Vision (WACV), Santa Rosa, CA, USA, 24–31 March 2017. [Google Scholar] [CrossRef]
- Egger, J.; Kapur, T.; Fedorov, A.; Pieper, S.; Miller, J.V.; Veeraraghavan, H.; Freisleben, B.; Golby, A.J.; Nimsky, C.; Kikinis, R. GBM volumetry using the 3D Slicer medical image computing platform. Sci. Rep. 2013, 3, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujikura, K.; Albini, A.; Barr, R.G.; Parikh, M.; Kern, J.; Hoffman, E.; Hiura, G.T.; Bluemke, D.A.; Carr, J.; Lima, J.A.; et al. Aortic enlargement in chronic obstructive pulmonary disease (COPD) and emphysema: The Multi-Ethnic Study of Atherosclerosis (MESA) COPD study. Int. J. Cardiol. 2021, 331, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Snider, G.L.; Kleinerman, J.; Thurlbeck, W.M.; Bengali, Z.H. The Definition of Emphysema. Am. Rev. Respir. Dis. 1985, 132, 182–185. [Google Scholar]
- Habib, S.L. Kidney atrophy vs hypertrophy in diabetes: Which cells are involved? Cell Cycle 2018, 17, 1683–1687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saad, M.; Ellis, C.L.; O’Neill, W.C. Renal Hypertrophy in Liver Failure. Kidney Int. Rep. 2018, 3, 1464–1467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Office of the Medical Investigator-UNM. Available online: https://hsc.unm.edu/omi/ (accessed on 18 May 2023).
- CT Scan Protocol. Available online: https://nmdid.unm.edu/resources/data-information (accessed on 18 May 2023).
- Torso Protocol. Available online: https://nmdid.unm.edu/docs/CT_OMI_protocol.pdf (accessed on 18 May 2023).


| Opposing Labels | Learning Rate Scheduling | Epochs | Aggregate Score |
|---|---|---|---|
| Separate | Built-in | 1000 | 0.9512 |
| Linear | 4000 | 0.9611 | |
| Linear to Exponential | 1000 | 0.9491 | |
| Quick Linear to Exponential | 1000 | 0.9592 | |
| Modified Linear to Exponential | 1000 | 0.8850 | |
| Combined | Linear to Exponential | 1000 | 0.9412 |
| TotalSegmentator model | 4000 | greater than 0.96 | |
| Opposing Labels | Learning Rate Scheduling | Epochs | Seconds per Epoch |
|---|---|---|---|
| Separate | Linear to Exponential | 1000 | 332.2 |
| Linear | 4000 | 712.1 | |
| Quick Linear to Exponential | 1000 | 668.7 | |
| Modified Linear to Exponential | 1000 | 649.4 | |
| Built-in | 1000 | 283.9 | |
| Combined | Modified | 1000 | 624.8 |
| Structure | Mean Volume | Standard Deviation | Variation Coefficient |
|---|---|---|---|
| Adrenal gland | 0.0054 | 0.0028 | 0.5179 |
| Aorta | 0.0844 | 0.0502 | 0.5946 |
| Autochton | 1.0526 | 0.3078 | 0.2924 |
| Clavicula | 0.0537 | 0.0170 | 0.3166 |
| Colon | 1.5131 | 0.7031 | 0.4647 |
| Duodenum | 0.0336 | 0.0169 | 0.5029 |
| Esophagus | 0.0403 | 0.0145 | 0.3604 |
| Femur | 0.4747 | 0.1120 | 0.2358 |
| Gluteal muscles | 1.6934 | 0.5710 | 0.3372 |
| Heart | 0.3239 | 0.1251 | 0.3861 |
| Hip | 0.7445 | 0.1629 | 0.2189 |
| Humerus | 0.0877 | 0.0531 | 0.6056 |
| Iliac artery | 0.0119 | 0.0084 | 0.7093 |
| Iliac vena | 0.0212 | 0.0146 | 0.6903 |
| Iliopsoas | 0.5318 | 0.2042 | 0.3839 |
| Inferior vena cava | 0.0110 | 0.0108 | 0.9834 |
| Kidney | 0.2686 | 0.1013 | 0.3769 |
| Liver | 0.8613 | 0.4221 | 0.4901 |
| Lung | 2.6232 | 0.7547 | 0.2877 |
| Pancreas | 0.0214 | 0.0195 | 0.9088 |
| Portal and splenic vein | 0.0047 | 0.0043 | 0.9149 |
| Pulmonary artery | 0.0291 | 0.0167 | 0.5718 |
| Ribs | 0.4137 | 0.1127 | 0.2723 |
| Scapula | 0.2155 | 0.0520 | 0.2412 |
| Small bowel | 0.9991 | 0.4253 | 0.4257 |
| Spleen | 0.7638 | 0.2866 | 0.3752 |
| Stomach | 0.4096 | 0.2961 | 0.7229 |
| Trachea | 0.0210 | 0.0129 | 0.6145 |
| Urinary bladder | 0.1902 | 0.1832 | 0.9630 |
| Vertebrae | 0.9109 | 0.1684 | 0.1848 |
| Structures | r | p-Value | |
|---|---|---|---|
| Hip | Vertebrae | 0.7353 | <0.05 |
| Autochthon | Gluteal muscles | 0.7116 | <0.05 |
| Iliopsoas | Gluteal muscles | 0.7102 | <0.05 |
| Autochthon | Iliopsoas | 0.6829 | <0.05 |
| Scapula | Ribs | 0.6481 | <0.05 |
| Scapula | Vertebrae | 0.6076 | <0.05 |
| Clavicula | Scapula | 0.5979 | <0.05 |
| Hip | Scapula | 0.5974 | <0.05 |
| Ribs | Vertebrae | 0.5954 | <0.05 |
| Hip | Ribs | 0.5917 | <0.05 |
| Scapula | Autochthon | 0.5044 | <0.05 |
| Clavicula | Humerus | 0.5003 | <0.05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
van Meegdenburg, T.; Kleesiek, J.; Egger, J.; Perrey, S. Improvement in Disease Diagnosis in Computed Tomography Images by Correlating Organ Volumes with Disease Occurrences in Humans. BioMedInformatics 2023, 3, 526-542. https://doi.org/10.3390/biomedinformatics3030036
van Meegdenburg T, Kleesiek J, Egger J, Perrey S. Improvement in Disease Diagnosis in Computed Tomography Images by Correlating Organ Volumes with Disease Occurrences in Humans. BioMedInformatics. 2023; 3(3):526-542. https://doi.org/10.3390/biomedinformatics3030036
Chicago/Turabian Stylevan Meegdenburg, Timo, Jens Kleesiek, Jan Egger, and Sören Perrey. 2023. "Improvement in Disease Diagnosis in Computed Tomography Images by Correlating Organ Volumes with Disease Occurrences in Humans" BioMedInformatics 3, no. 3: 526-542. https://doi.org/10.3390/biomedinformatics3030036
APA Stylevan Meegdenburg, T., Kleesiek, J., Egger, J., & Perrey, S. (2023). Improvement in Disease Diagnosis in Computed Tomography Images by Correlating Organ Volumes with Disease Occurrences in Humans. BioMedInformatics, 3(3), 526-542. https://doi.org/10.3390/biomedinformatics3030036