Deep Learning Architecture Optimization with Metaheuristic Algorithms for Predicting BRCA1/BRCA2 Pathogenicity NGS Analysis
Abstract
:1. Introduction
2. Materials and Methods
- Variant allele frequency ≥ 5%
- Variant reads ≥ 300 reads
- Amino acid change is different from synonymous (≠p.(=)). A synonymous variant will probably have a low influence on the gene because the amino acid does not change
- Polyphen (Polymorphism Phenotyping) score is in the range of [0.85, 1] (case of substitution variant)
- Grantham score is in a range of [5; 215] (in the case of substitution variant). [0, 50] = conservative, [51; 100] moderately conservative, [101; 150] moderately radical, over 150 radical
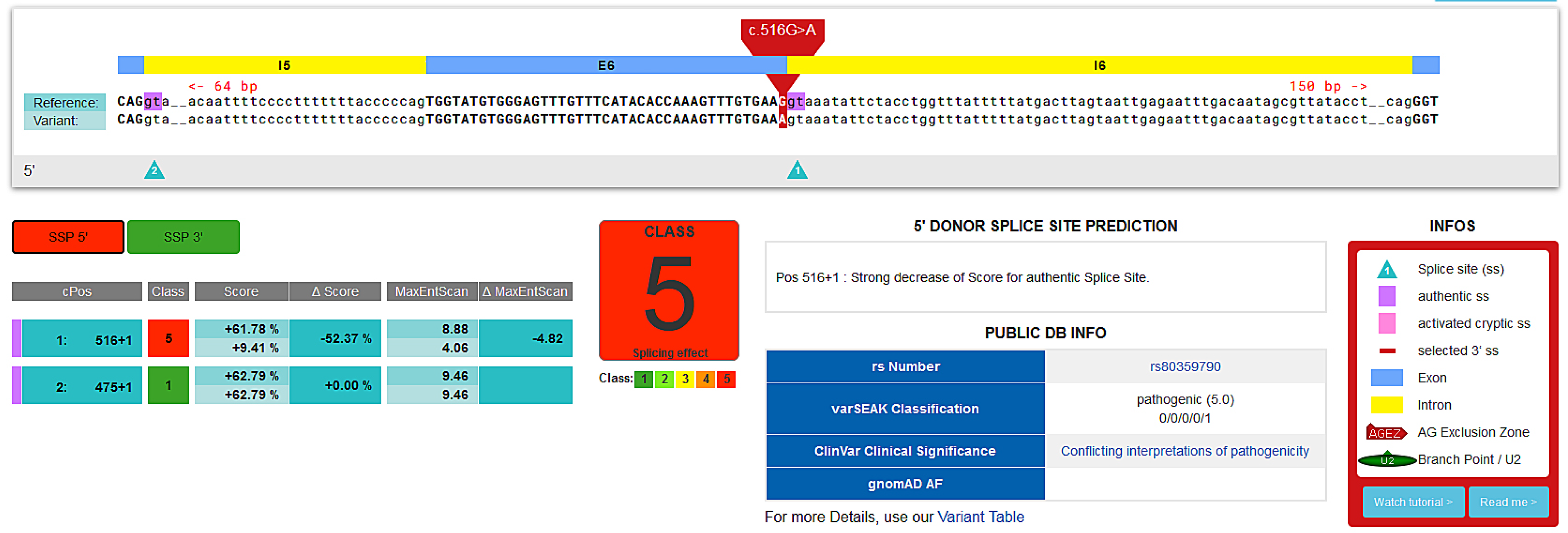
- Manual inspection on different databases and prediction tools: Arup database, VarSeak, Varsome, UMD Predictor, Cancer Genome Interpreter
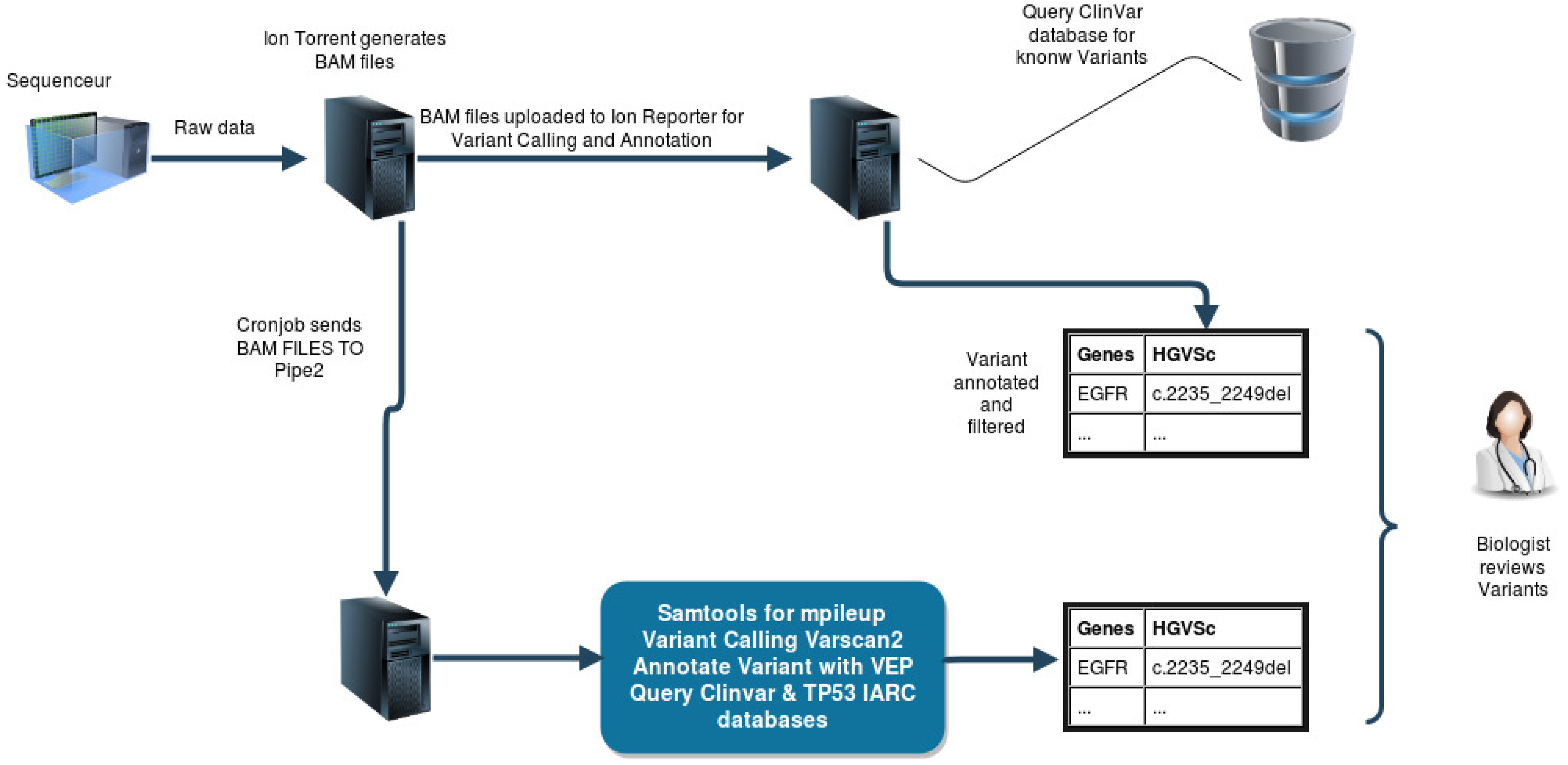
- We use the tool Integrative Genomics Viewer (IGV) to check if alignment sequences are clear and show no strand bias in the region where the variant is located. It allows us to eliminate false positives.
- We verify the presence of these pathogenic variants in our second-in-house pipeline to validate them
2.1. Programming Language
2.2. Code Availability
- install.packages(“particle.swarm.optimisation”)
- install_gitlab(“brunet.theo83140/genetics.algorithm”)
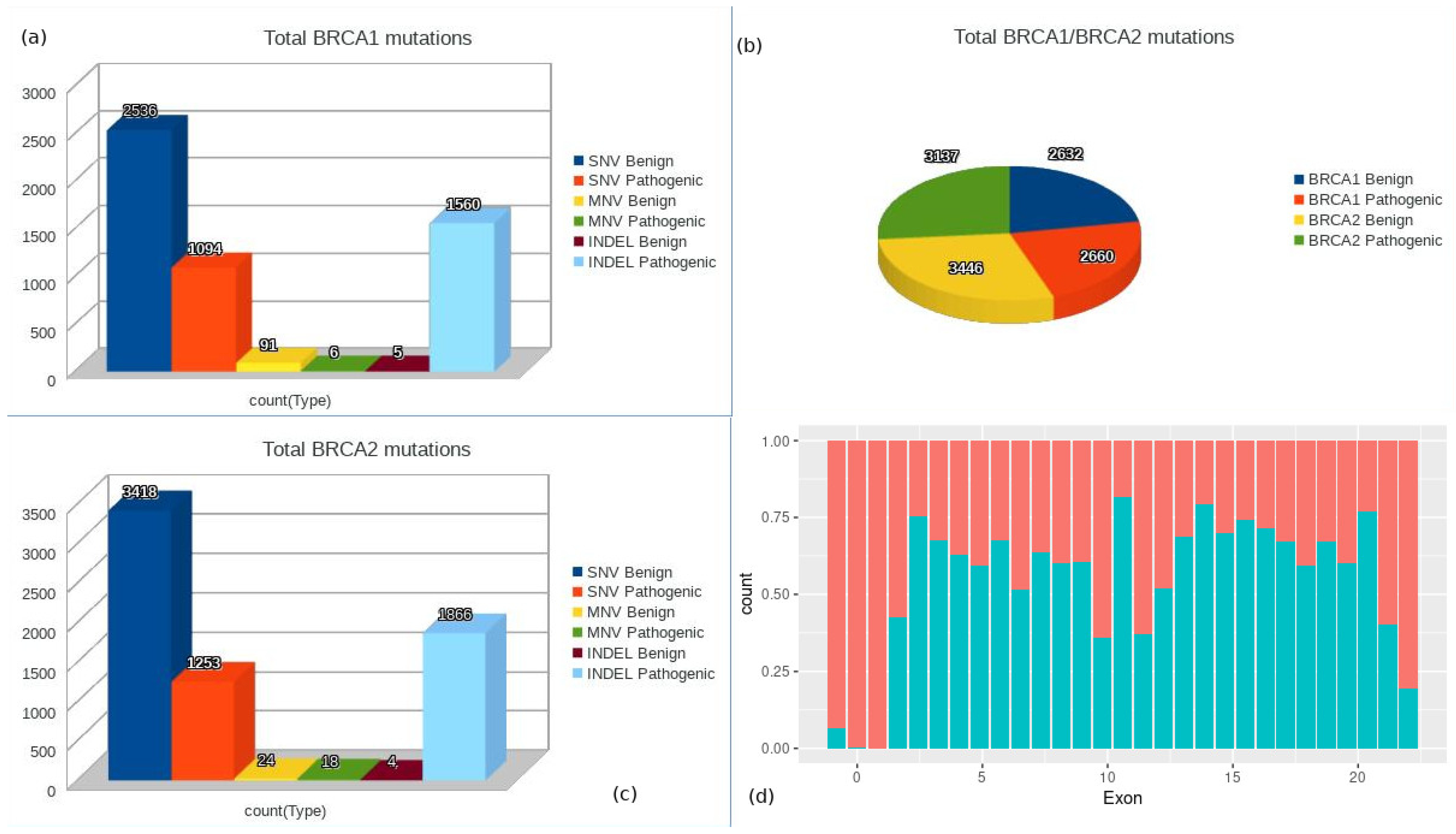
2.3. Data Selection
2.4. Problem Formulation
2.5. Data Encoding (Feature Construction)
- 0.0 to 0.15 → predicted to be benign
- 0.15 to 1.0 → possibly damaging
- 0.85 to 1.0 → more confidently predicted to be damaging
2.6. Data Normalization
2.7. Grid Search Method
- Monitor: validation loss (val_loss)
- Minimum change in the monitored quantity to qualify as an improvement (min_delta) = 0.0005
- Number of epochs with no improvement after which training will be stopped (patience) = 10
- Training will stop when the quantity monitored has stopped decreasing (mode) = ‘min’
2.8. Genetic Algorithm (GA)
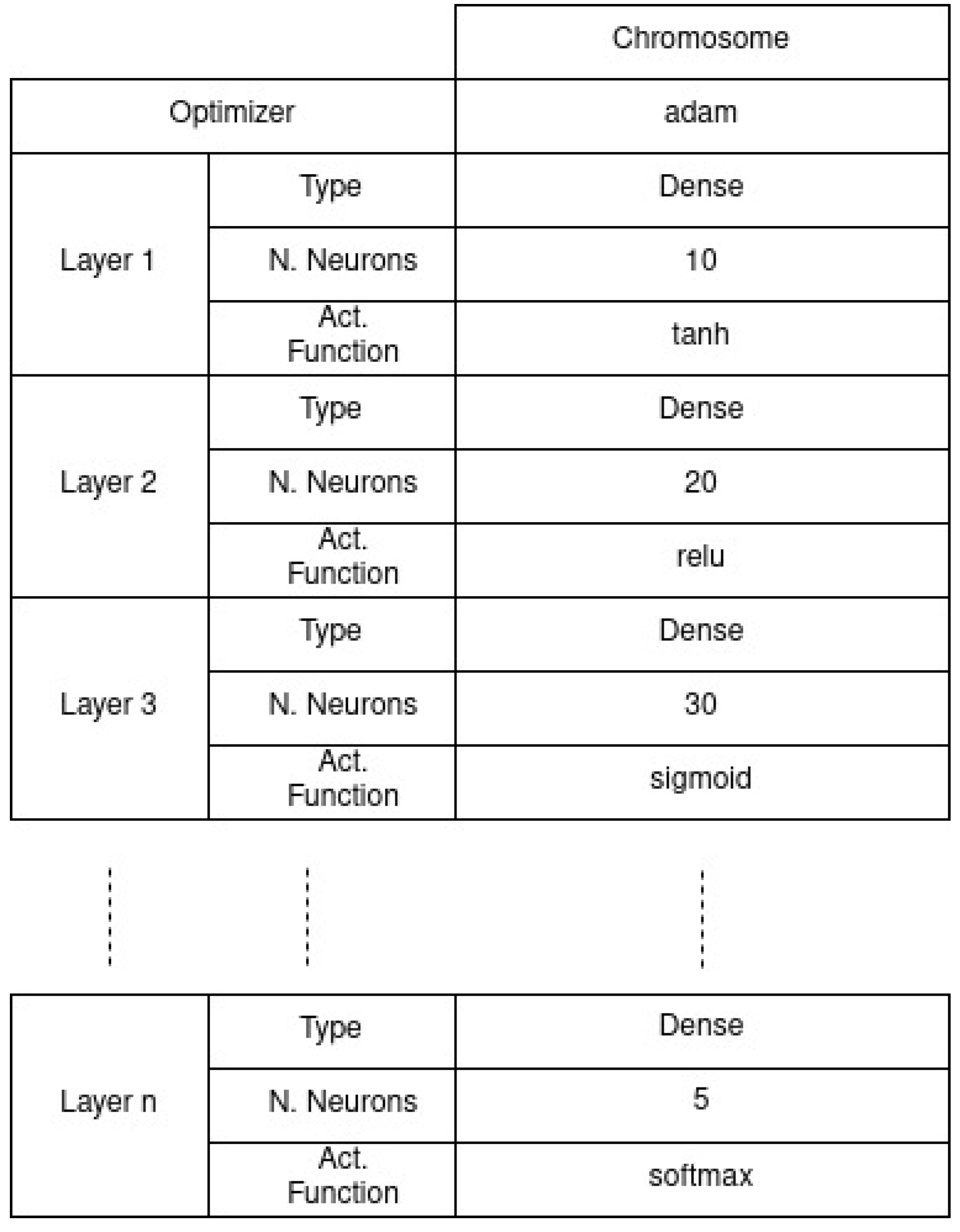
Chromosome Structure
- Optimizer functions: [adam, adamax, sgd]
- Hidden layer layout: [Number of neurons on a range of [1, 300], for each layer the activation function: [relu, elu, tanh, sigmoid] and dropout on a range of [0.1 to 0.9]
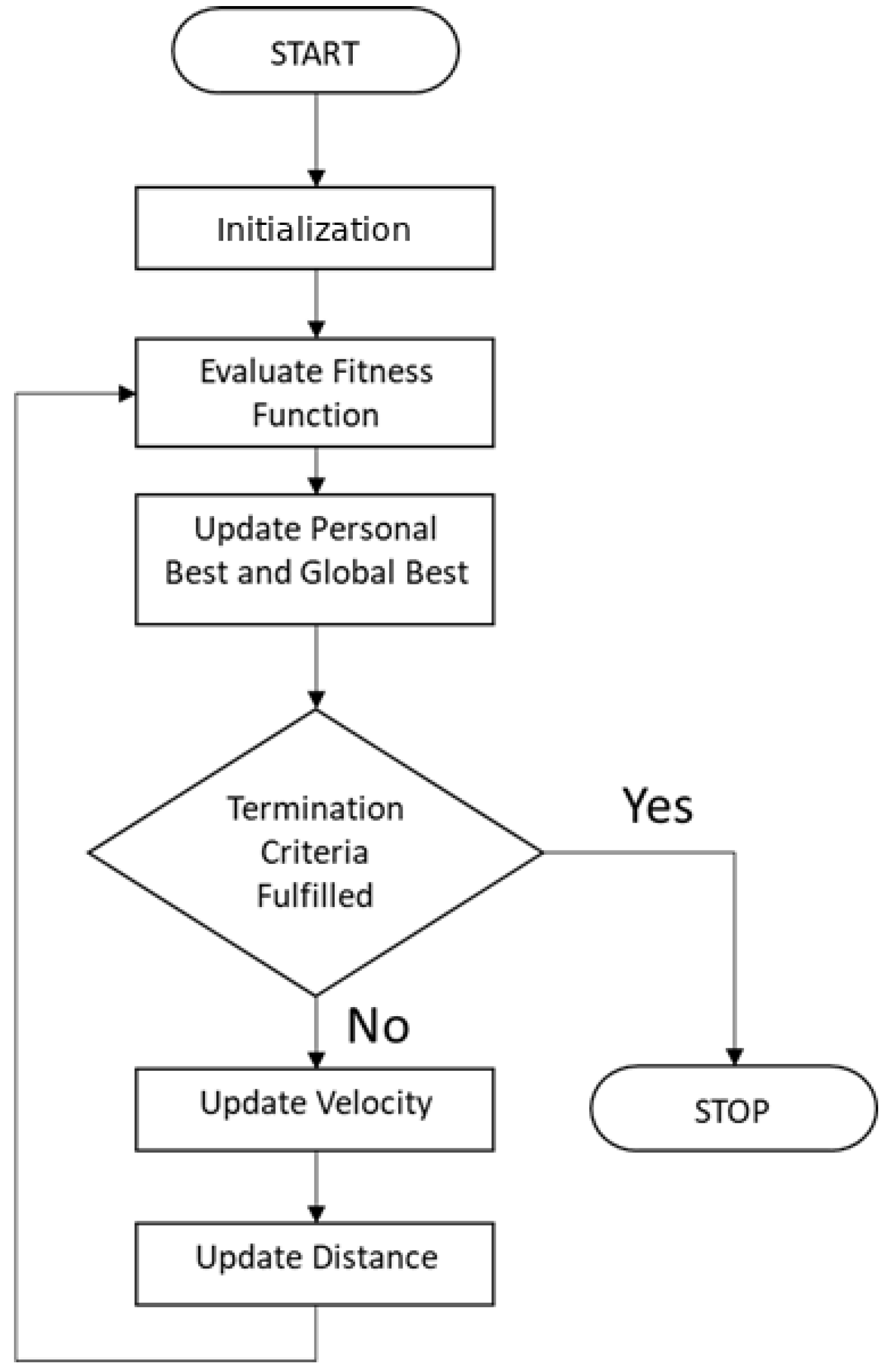
2.9. Particle Swarm Optimization (PSO)
- Its current speed
- His best solution
- The best solution obtained in its neighborhood
3. Results
3.1. Grid Search Results
3.2. GA Results
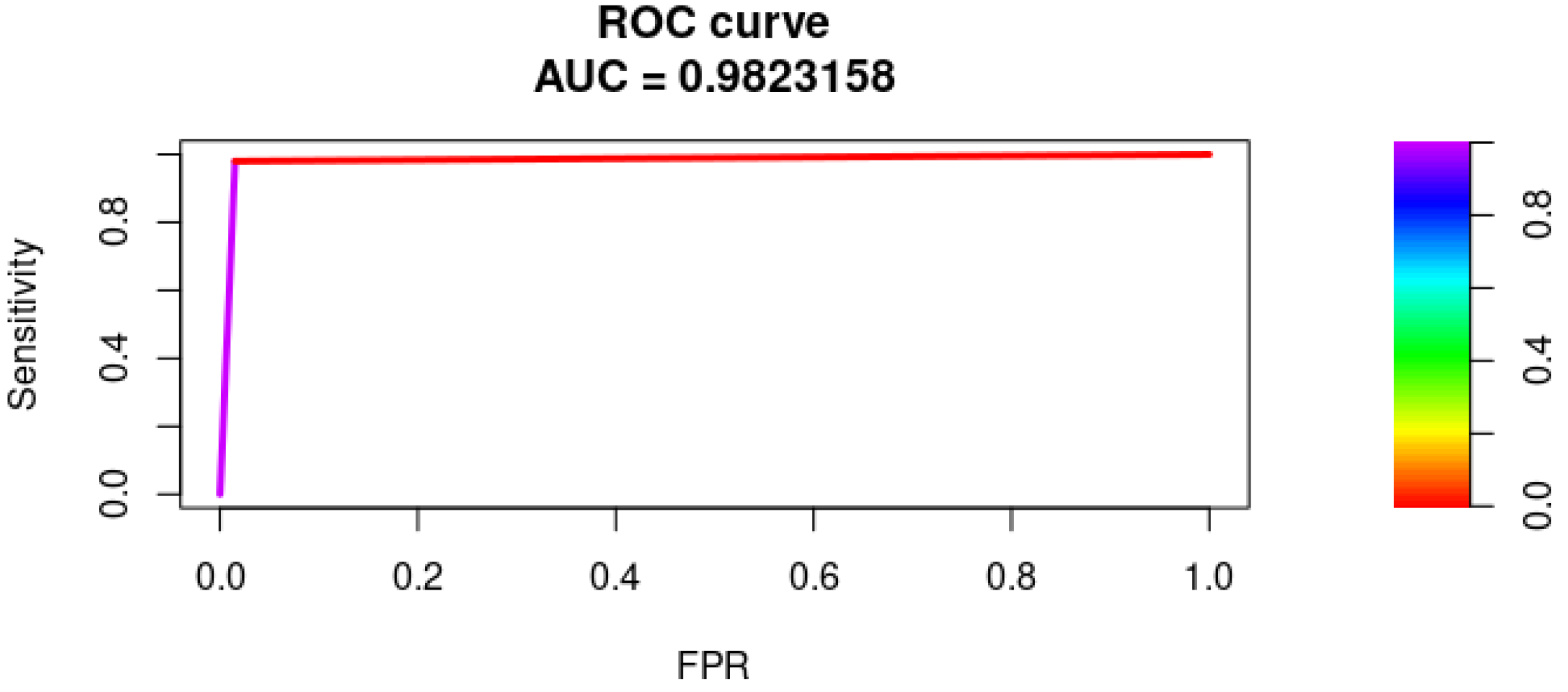
3.3. PSO Results
3.4. Feature Importance
3.5. Prediction on Repair Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hope, T.; Resheff, Y.S.; Lieder, I. Learning Tensorflow: A Guide to Building Deep Learning Systems; O’Reilly Media, Inc.: Newton, MA, USA, 2017. [Google Scholar]
- Carvalho, M.; Ludermir, T.B. Particle swarm optimization of neural network architectures andweights. In Proceedings of the 7th International Conference on Hybrid Intelligent Systems (HIS 2007), Kaiserslautern, Germany, 17–19 September 2007. [Google Scholar]
- Qolomany, B.; Maabreh, M.; Al-Fuqaha, A.; Gupta, A.; Benhaddou, D. Parameters optimization of deep learning models using particle swarm optimization. In Proceedings of the 2017 13th International Wireless Communications and Mobile Computing Conference (IWCMC), Valencia, Spain, 26–30 June 2017. [Google Scholar]
- Idrissi, M.A.J.; Ramchoun, H.; Ghanou, Y.; Ettaouil, M. Genetic algorithm for neural network architecture optimization. In Proceedings of the 2016 3rd International Conference on Logistics Operations Management, Fez, Morocco, 23–25 May 2016. [Google Scholar]
- Brownlee, J. Better Deep Learning: Train Faster, Reduce Overfitting, and Make Better Predictions; Machine Learning Mastery: Melbourne, Australia, 2018. [Google Scholar]
- Holland, J.H. Adaptation in Natural and Artificial Systems: An Introductory Analysis with Applications to Biology, Control, and Artificial Intelligence; MIT Press: Cambridge, MA, USA, 1992. [Google Scholar]
- Kennedy, J.; Eberhart, R. Particle swarm optimization. In Proceedings of the ICNN’95—International Conference on Neural Networks, Perth, Australia, 27 November–1 December 1995. [Google Scholar]
- Adyatama, A. RPubs-Introduction to Particle Swarm Optimization. Available online: https://rpubs.com/argaadya/intro-pso (accessed on 12 October 2021).
- Dagan, T.; Talmor, Y.; Graur, D. Ratios of Radical to Conservative Amino Acid Replacement are Affected by Mutational and Compositional Factors and May Not Be Indicative of Positive Darwinian Selection. Mol. Biol. Evol. 2002, 19, 1022–1025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grantham, R. Amino acid difference formula to help explain protein evolution. Science 1974, 185, 862–864. [Google Scholar] [CrossRef] [PubMed]
- Blum, A. Neural Networks in C++ an Object-Oriented Framework for Building Connectionist Systems; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 1992. [Google Scholar]
- Boger, Z.; Guterman, H. Knowledge extraction from artificial neural network models. In Proceedings of the 1997 IEEE International Conference on Systems, Man, and Cybernetics. Computational Cybernetics and Simulation, Orlando, FL, USA, 12–15 October 1997; Volume 4. [Google Scholar]
- Swingler, K. Applying Neural Networks: A Practical Guide; Morgan Kaufmann: Burlington, MA, USA, 1996. [Google Scholar]
- Linoff, G.S.; Berry, M.J. Data Mining Techniques: For Marketing, Sales, and Customer Relationship Management; John Wiley & Sons: Hoboken, NJ, USA, 2011. [Google Scholar]
- Heaton, J. Introduction to Neural Networks with Java, 2nd ed.; Heaton Research, Inc.: St. Louis, MO, USA, 2008. [Google Scholar]
- Beysolow, T., II. Introduction to Deep Learning Using R: A Step-by-Step Guide to Learning and Implementing Deep Learning Models Using R; Apress: New York, NY, USA, 2017. [Google Scholar]
- Mitchell, M. An Introduction to Genetic Algorithms; MIT Press: Cambridge, MA, USA, 1998. [Google Scholar]
- Gómez, N.; Mingo, L.F.; Bobadilla, J.; Serradilla, F.; Manzano, J.A.C. Particle Swarm Optimization models applied to Neural Networks using the R language. WSEAS Trans. Syst. 2010, 9, 192–202. [Google Scholar]
- Albaradei, S.; Thafar, M.; Alsaedi, A.; Van Neste, C.; Gojobori, T.; Essack, M.; Gao, X. Machine learning and deep learning methods that use omics data for metastasis prediction. Comput. Struct. Biotechnol. J. 2021, 19, 5008–5018. [Google Scholar] [CrossRef] [PubMed]
- Fakoor, R.; Ladhak, F.; Nazi, A.; Huber, M. Using deep learning to enhance cancer diagnosis and classification. In Proceedings of the International Conference on Machine Learning, Atlanta, GA, USA, 16–21 June 2013. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Pellegrino, E.; Jacques, C.; Beaufils, N.; Nanni, I.; Carlioz, A.; Metellus, P.; Ouafik, L.H. Machine learning random forest for predicting oncosomatic variant NGS analysis. Sci. Rep. 2021, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Boeringer, D.W.; Werner, D.H. A comparison of particle swarm optimization and genetic algorithms for a phased array synthesis problem. In Proceedings of the IEEE Antennas and Propagation Society International Symposium. Digest. Held in conjunction with: USNC/CNC/URSI North American Radio Sci. Meeting (Cat. No.03CH37450), Columbus, OH, USA, 22–27 June 2003. [Google Scholar]
- Khan, S.; Islam, N.; Jan, Z.; Din, I.U.; Rodrigues, J.J.P.C. A novel deep learning based framework for the detection and classification of breast cancer using transfer learning. Pattern Recognit. Lett. 2019, 125, 1–6. [Google Scholar] [CrossRef]
- Walsh, S.; de Jong, E.E.C.; van Timmeren, J.E.; Ibrahim, A.; Compter, I.; Peerlings, J.; Sanduleanu, S.; Refaee, T.; Keek, S.; Larue, R.T.H.M.; et al. Decision support systems in oncology. JCO Clin. Cancer Inform. 2019, 3, 1–9. [Google Scholar] [CrossRef] [PubMed]








| Row Names | Data Description | Data Coding | Data Type |
|---|---|---|---|
| Location | Position of the mutation on the gene (exon, intronic, splicing site) | Intronic = 0, utr_5’ = 1, utr_3’ = 1, splicesite_3’ = 2, splicesite_5’ = 2, exonic = 3 | Integer |
| Genes | BRCA1 or BRCA2 | BRCA1 = 0, BRCA2 = 1 | Integer |
| Transcript | Transcript ID | NM_007294.3 = 0, NM_007300.3 = 0, NM_000059.3 = 1 | Integer |
| Locus | Coordinates on the genome | No recoding | Big interger |
| Type | Type of mutation: single nucleoide variant (SNV), mutiple nucleotide variants (MNV), insertion/deletion (INDEL) | SNV = 0, MNV = 1, INDEL = 2 | Integer |
| Exon | Exon number. 1 for the first exon in gene, 2 for the second one, …, n | when it is intronic. | Integer |
| Freq | Variant allele frequency (frequency of the mutation) | No recoding of the values. | Float |
| MAF | Minor Allele Frequency | MAF value if it exists, −1 when there is no MAF | Float |
| Coverage | Sequencing coverage (0 if < 300 reads, 1 if > 300 reads) | No recoding | Integer |
| protdesc | Mutation effect on the protein (amino acid change) | Intronic or splice site (p.? = 0), same acid amino (p.(=) = 1), acid amino change = 2 | Integer |
| Polyphen | Polyphen score (from −1 to 1) | Polyphen score when it exists, −1 when it is not applicable | Float |
| Grantham | Grantham score of the mutation | Grantham value from 5 to 215. −1 when it is not applicable | |
| Variant.effect | Effect of the mutation on the reading frame (frameshift, missense …) | Frameshitf = 3, missense = 1, nonsense = 2, synonymous = 0, unknown = −1 | |
| aaref | Amino acid (before the mutation) | Arg = 1, His = 2, Lys = 3, Asp = 4, Glu = 5, Ser = 6, Thr = 7, Asn = 8, Gln = 9, Trp = 10, Sec = 11, Gly = 12, Pro = 13, Ala = 14, Val = 15, Ile = 16, Leu = 17, Met = 18, Phe = 19, Tyr = 20, Cys = 21 | Integer (more detail on Table 2) |
| aamut | Amino acid (after the mutation) | Arg = 1, His = 2, Lys = 3, Asp = 4,Glu = 5, Ser = 6, Thr = 7, Asn = 8, Gln = 9, Trp = 10, Sec = 11, Gly = 12, Pro = 13, Ala = 14, Val = 15, Ile = 16, Leu = 17, Met = 18, Phe = 19, Tyr = 20, Cys = 21, fs = 22, Ter = 22, del = 22 | Interger (more detail on Table 2) |
| isMut | Potential pathogenic variant. Decision of the biologist on the mutation variant | Benign/uncertain significance = 0, pathogenic = 1 | Boolean |
| Chemical Properties | Amino Acids |
|---|---|
| Acidic | Aspartic (Asp), Glutamic (Glu) |
| Aliphatic | Alanine (Ala), Glycine (Glycine), Isoleucine (Ile), Leucine (Leu), Valine (Val) |
| Amide | Asparagine (Asn), Glutamine (Gln) |
| Aromatic | Phenylalanine (Phe), Tryptophan (Trp), Tyrosine (Tyr) |
| Basic | Arginine (Arg), Histidine (His), Lysine (Lys) |
| Hydroxyl | Serine (Ser), Threonine (Thr) |
| Imino | Proline (Pro) |
| Sulfur | Cysteine (Cys), Methionine (Met) |
| Parameters | Range | Number of Possibilities |
|---|---|---|
| Number of neurons per hidden layers | [1; 50] | 50 |
| Number of hidden layers | [1: 5] | 5 |
| Activation functions | [relu, elu, tanh, sigmoid] | 4 |
| Training functions | Sgd, adam, adamax | 3 |
| Dropout | [0.1; 0.9] | 9 |
| Total of combination | 27,000 |
| Parameters | First Run (GA1) | Second Run (GA2) |
|---|---|---|
| Elitism | 0.3 | 0.3 |
| Random selection | 0.1 | 0.1 |
| Population size | 30 | 50 |
| Mutation rate | 0.25 | 0.25 |
| Number of generations | 20 | 40 |
| Parameters | Number of Hidden Nodes per Hidden Layers | Activation Functions | Number of Hidden Layers | Optimizer |
|---|---|---|---|---|
| Values | 1 to 300 | Relu, elu, sigmoid, tanh | 1 to 20 | Adam, adamax, sgd |
| Parameters | Number of Hidden Nodes per Hidden Layers | Activation Functions | Number of Hidden Layers | Optimizer |
|---|---|---|---|---|
| Values | 1 to 300 | Relu, elu, sigmoid, tanh | 1 to 6 | Adam, adamax, sgd |
| Parameters | PSO (Run 1) | PSO (Run 2) |
|---|---|---|
| Inertia | 0.5 | 0.5 |
| c1 | [0:4] | [0:4] |
| c1 | [0:4] | [0:4] |
| Number of particles | 30 | 50 |
| Number of iterations | 20 | 50 |
| Models | Number of hidden nodes per hidden layers | Number of hidden layers | Activation function | Optimizer |
|---|---|---|---|---|
| GA (run 1) | 297 | 6 | relu | adam |
| PSO (run 1) | 275 | 6 | relu | adam |
| Models | H1 | F1 | H2 | F2 | H3 | F3 | H4 | F4 | H4 | F5 | H6 | F6 | Optimizer |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GA run 2 | 212 | Elu | 134 | Relu | 246 | Sigmoid | 285 | Relu | 61 | Relu | 196 | Relu | Adam |
| PSO run 2 | 45 | Relu | 156 | Relu | 132 | Relu | 284 | Elu | 105 | Tanh | 169 | Tanh | Adam |
| Models | Precision | Sensitivity | Specificity | Accuracy | AUC |
|---|---|---|---|---|---|
| GA (run 1) | 0.9897 | 0.9822 | 0.9899 | 0.9861 | 0.9848 |
| GA (run 2) | 0.9896 | 0.9879 | 0.9901 | 0.9892 | 0.9869 |
| PSO (run 1) | 0.9965 | 0.9863 | 0.9966 | 0.9852 | 0.9858 |
| PSO (run 2) | 0.9898 | 0.9821 | 0.9883 | 0.9915 | 0.9865 |
| Genes | Coding | Protdesc | isMut | PSO1 | Pb Benign | Pb Pathogenic | Comment |
|---|---|---|---|---|---|---|---|
| ARID1A | c.1351-22A>C | p.? | 0 | 0 | 1 | 0 | Benign |
| ARID1A | c.2420-18G>C | p.? | 0 | 0 | 1 | 0 | Benign |
| ARID1A | c.3999_4001del | p.Gln1334del | 0 | 1 | 0.04 | 0.96 | Benign |
| ATM | c.72+36TAA>T | p.? | 0 | 0 | 1 | 0 | Benign |
| ATM | c.902-18T>A | p.? | 0 | 0 | 0.73 | 0.27 | Benign |
| ATM | c.1607+47GAC>G | p.? | 0 | 0 | 1 | 0 | Benign |
| ATM | c.1809_1810dup | p.Pro604fs | 1 | 1 | 0 | 1 | Pathogenic |
| ATM | c.1810C>T | p.Pro604Ser | 0 | 0 | 1 | 0 | Benign |
| ATM | c.2119T>C | p.Ser707Pro | 0 | 0 | 1 | 0 | Benign |
| ATM | c.3078-77C>T | p.? | 0 | 0 | 1 | 0 | Benign |
| TP53 | c.645T>G | p.Ser215Arg | 1 | 0 | 0.72 | 0.28 | Pathogenic |
| ATM | c.79G>A | p.Val27Ile | 0 | 0 | 1 | 0 | Benign |
| PALB2 | c.3114-51T>A | p.? | 0 | 0 | 1 | 0 | Benign |
| PALB2 | c.1684+42TGAA>A | p.? | 0 | 0 | 1 | 0 | Benign |
| BRIP1 | c.3411T>C | p.(=) | 0 | 0 | 1 | 0 | Benign |
| BRIP1 | c.206-2TA>A | p.? | 0 | 1 | 0.06 | 0.94 | Pathogenic |
| NBN | c.1124+18C>T | p.? | 0 | 0 | 1 | 0 | Benign |
| NBN | c.553G>C | p.Glu185Gln | 0 | 0 | 1 | 0 | Benign |
| NBN | c.381T>C | p.(=) | 0 | 0 | 1 | 0 | Benign |
| ARID1A | c.854del | p.Gly285fs | 1 | 1 | 0 | 1 | Pathogenic |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pellegrino, E.; Brunet, T.; Pissier, C.; Camilla, C.; Abbou, N.; Beaufils, N.; Nanni-Metellus, I.; Métellus, P.; Ouafik, L. Deep Learning Architecture Optimization with Metaheuristic Algorithms for Predicting BRCA1/BRCA2 Pathogenicity NGS Analysis. BioMedInformatics 2022, 2, 244-267. https://doi.org/10.3390/biomedinformatics2020016
Pellegrino E, Brunet T, Pissier C, Camilla C, Abbou N, Beaufils N, Nanni-Metellus I, Métellus P, Ouafik L. Deep Learning Architecture Optimization with Metaheuristic Algorithms for Predicting BRCA1/BRCA2 Pathogenicity NGS Analysis. BioMedInformatics. 2022; 2(2):244-267. https://doi.org/10.3390/biomedinformatics2020016
Chicago/Turabian StylePellegrino, Eric, Theo Brunet, Christel Pissier, Clara Camilla, Norman Abbou, Nathalie Beaufils, Isabelle Nanni-Metellus, Philippe Métellus, and L’Houcine Ouafik. 2022. "Deep Learning Architecture Optimization with Metaheuristic Algorithms for Predicting BRCA1/BRCA2 Pathogenicity NGS Analysis" BioMedInformatics 2, no. 2: 244-267. https://doi.org/10.3390/biomedinformatics2020016
APA StylePellegrino, E., Brunet, T., Pissier, C., Camilla, C., Abbou, N., Beaufils, N., Nanni-Metellus, I., Métellus, P., & Ouafik, L. (2022). Deep Learning Architecture Optimization with Metaheuristic Algorithms for Predicting BRCA1/BRCA2 Pathogenicity NGS Analysis. BioMedInformatics, 2(2), 244-267. https://doi.org/10.3390/biomedinformatics2020016






