Developing High-Efficiency PCR Mini-Barcoding to Enforce Conservation Efforts Against Illegal Trade and Habitat Loss of Endangered Taxus L. in the Himalayas
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling of Intact and Environmental Materials
2.2. Primer Selection
2.3. DNA Extraction Protocol
2.4. PCR Optimization and DNA Sequencing
2.5. BLAST Search Criteria
3. Results
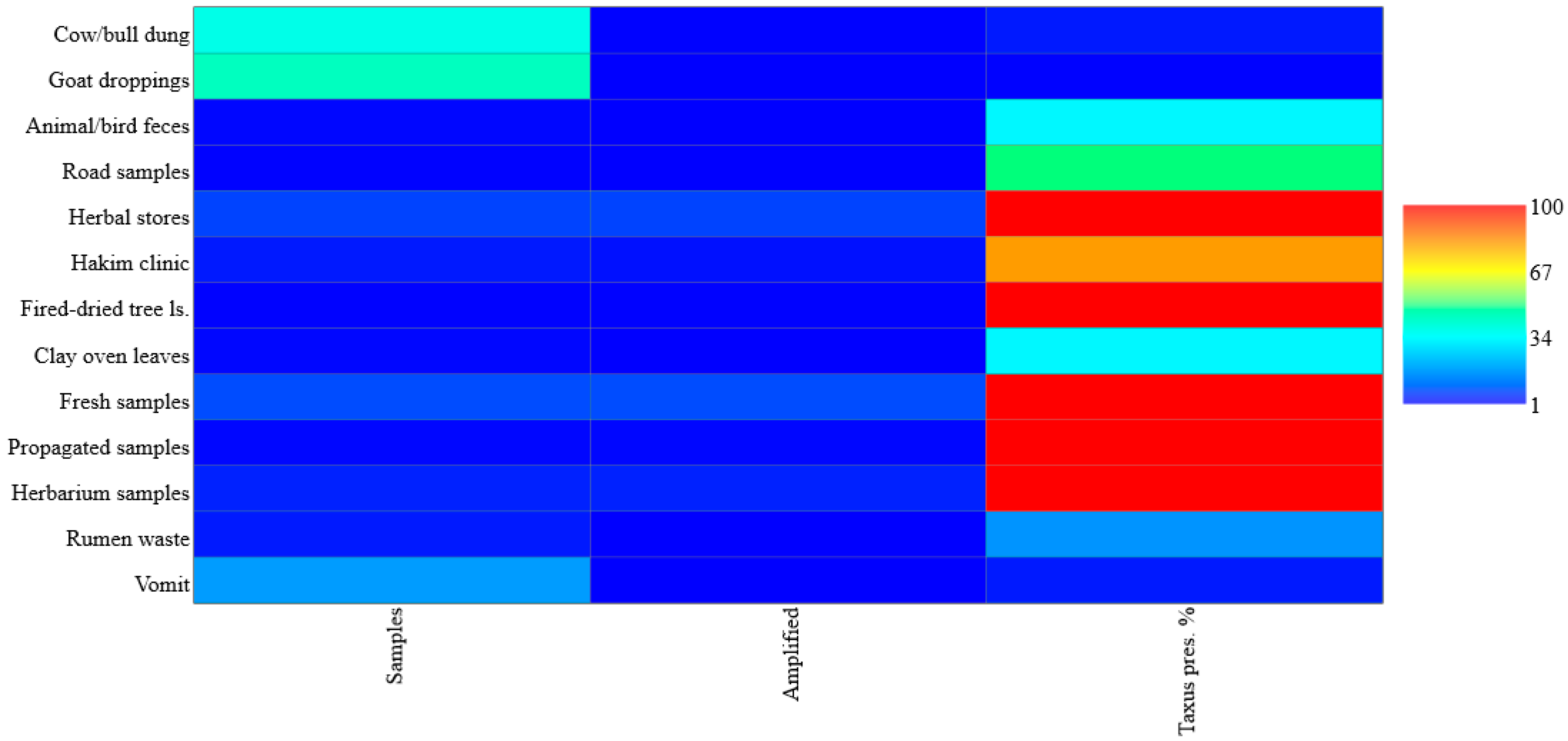
3.1. Sampling Distribution and Statistical Analysis
3.2. PCR Amplification of Taxus DNA Samples
3.2.1. Initial Amplification of Custom-Designed Primers for Taxus Samples
3.2.2. Diverse Sample Amplification
3.3. BLAST Analysis of Amplified Sequences
3.4. Recognized Species Diversity
3.5. Mini-Barcoding-Based DNA Amplification Success and Detection of Crimes
4. Discussion
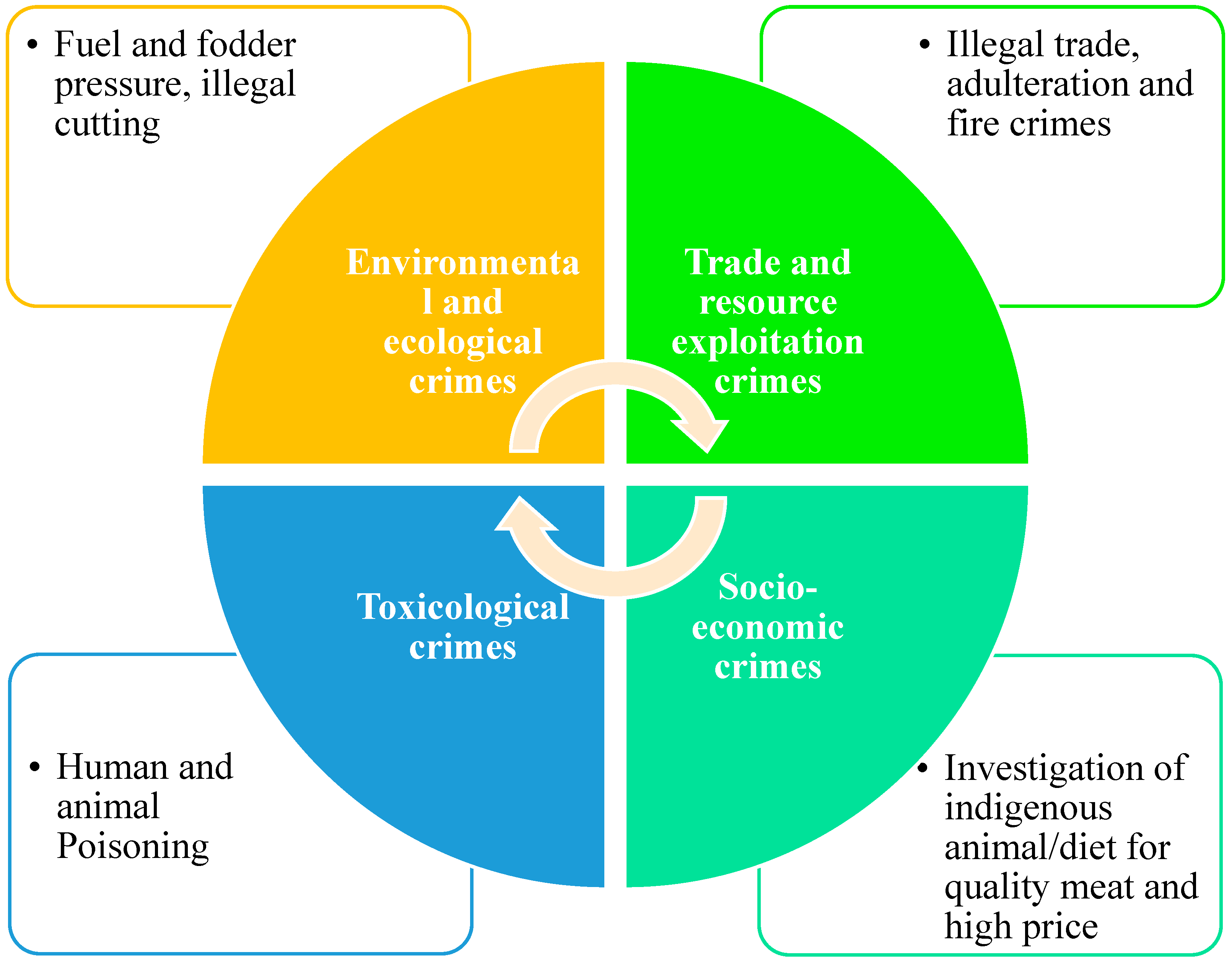
4.1. Forensic Utility of Taxus Identification in Pakistan
4.1.1. Application to Poisoning and Illegal Trade
4.1.2. Meeting Regional Forensic Demands
4.2. Effectiveness of Mini-Barcoding and Meta Barcoding for Taxus Identification
4.2.1. Management of Degraded Samples
4.2.2. Comparison with International Methods
4.3. Contribution to Global Databases
4.4. Policy Recommendations and Future Research
4.5. Botanical Forensics and Wildlife Protection: Legal and Conservation Challenges in Pakistan
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huffman, J.E.; Wallace, J.R. Wildlife Forensics: Methods and Applications; John Wiley & Sons: Hoboken, NJ, USA, 2011. [Google Scholar] [CrossRef]
- Galton, F. Finger Prints; MacMillan and Co.: London, UK; New York, NY, USA, 1892. [Google Scholar]
- Ashbaugh, D.R. Quantitative-Qualitative Friction Ridge Analysis: An Introduction to Basic and Advanced Ridgeology; CRC Press: Boca Raton, FL, USA, 1999. [Google Scholar]
- Caplan, R.M. How fingerprints came into use for personal identification. J. Am. Acad. Dermatol. 1990, 23, 109–114. [Google Scholar] [CrossRef]
- Ulery, B.T.; Hicklin, R.A.; Buscaglia, J.; Roberts, M.A. Accuracy and reliability of forensic latent fingerprint decisions. Proc. Natl. Acad. Sci. USA 2011, 108, 7733–7738. [Google Scholar] [CrossRef]
- Ulery, B.T.; Hicklin, R.A.; Buscaglia, J.; Roberts, M.A. Repeatability and reproducibility of decisions by latent fingerprint examiners. PLoS ONE 2012, 7, e32800. [Google Scholar] [CrossRef]
- Bell, S.; Sah, S.; Albright, T.D.; Gates, S.J., Jr.; Denton, M.B.; Casadevall, A. A call for more science in forensic science. Proc. Natl. Acad. Sci. USA 2018, 115, 4541–4544. [Google Scholar] [CrossRef] [PubMed]
- Lacey Act (1900 et seq) 16 U.S.C; U.S. GPO: Washington, DC, USA, 1990; pp. 3371–3378.
- Migratory Bird Treaty Act (1918 et seq) 16 U.S.C; U.S. GPO: Washington, DC, USA, 1918; pp. 703–712.
- Bald and Golden Eagle Protection Act (1940 et seq) 16 U.S.C; 668a–d; U.S. GPO: Washington, DC, USA, 1940.
- Marine Mammal Protection Act (1972) 16 U.S.C; §§1361–1383b, 1401–1406; U.S. GPO: Washington, DC, USA, 1972; pp. 1411h–1421h.
- CITES. Convention on International Trade in Endangered Species of Wild Fauna and Flora. 993 U.N.T.S. 243, 3 March 1973. [Google Scholar]
- Endangered Species Act of 1973 (1973 et seq); U.S. GPO: Washington, DC, USA, 1973. [CrossRef]
- Anderson, R.S. The Lacey Act: America’s premier weapon in the fight against unlawful wildlife trafficking. Pub. Land L. Rev. 1995, 16, 27. [Google Scholar]
- Burnham-Curtis, M.K.; Trail, P.W.; Kagan, R.; Moore, M.K. Wildlife forensics: An overview and update for the prosecutor. US Att’ys Bull. 2015, 63, 53. [Google Scholar]
- Breyer, S. Science in the courtroom. Issues Sci. Technol. 2000, 16, 52–56. [Google Scholar]
- Spjut, R.W. Taxonomy and nomenclature of Taxus (Taxaceae). J. Bot. Res. Inst. Tex. 2007, 1, 203–289. [Google Scholar]
- Cope, E.A. Taxaceae: The genera and cultivated species. Bot. Rev. 1998, 64, 291–322. [Google Scholar] [CrossRef]
- Shaheen, H.; Attique, A.; Riaz, M.T.; Manzoor, M.; Khan, R.W.A.; Riaz, M.T. From biodiversity hotspot to conservation hotspot: Assessing distribution, population structure, associated flora and habitat geography of threatened Himalayan Yew in temperate forest ecosystems of Kashmir. Biodivers. Conserv. 2024, 33, 553–577. [Google Scholar] [CrossRef]
- Bangelesa, F.; Abel, D.; Pollinger, F.; Rai, P.; Ziegler, K.; Ebengo, D.; Tshimanga, R.M.; Ali, M.M.; Knight, J.; Paeth, H. Projected changes in rainfall amount and distribution in the Democratic Republic of Congo–Evidence from an ensemble of high-resolution climate simulations. Weather Clim. Extrem. 2023, 42, 100620. [Google Scholar] [CrossRef]
- Tiwari, O.P.; Sharma, C.M. Anthropogenic disturbance impact on forest composition and dominance-diversity: A case of an ecosensitive region of Garhwal Himalaya, India. Acta Ecol. Sin. 2023, 43, 662–673. [Google Scholar] [CrossRef]
- Poudel, R.C.; Moeller, M.; Gao, L.M.; Ahrends, A.; Baral, S.R.; Liu, J.; Thomas, P.; Li, D.Z. Using morphological, molecular and climatic data to delimitate yews along the Hindu Kush-Himalaya and adjacent regions. PLoS ONE 2012, 7, e46873. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, V. Taxus wallichiana Zucc. (Himalayan yew): A medicinal plant exhibiting antibacterial properties. Adv. Microbiol. Infect. Dis. Public Health 2023, 17, 145–153. [Google Scholar] [CrossRef]
- Gajurel, J.P.; Werth, S.; Shrestha, K.K.; Scheidegger, C. Species distribution modeling of Taxus wallichiana (Himalayan yew) in Nepal Himalaya. Asian J. Conserv. Biol. 2014, 3, 127–134. [Google Scholar]
- Mehta, P.; Sekar, K.C.; Bhatt, D.; Tewari, A.; Bisht, K.; Upadhyay, S.; Negi, V.S.; Soragi, B. Conservation and prioritization of threatened plants in Indian Himalayan Region. Biodivers. Conserv. 2020, 29, 1723. [Google Scholar] [CrossRef]
- Din, S.; Ali, H.; Panagopoulos, T.; Alam, J.; Malik, S.; Sher, H. AI-Driven Conservation of the Endangered Twisted Yew (Taxus contorta Griff.) in the Western Himalaya. Sustainability 2025, 17, 8541. [Google Scholar] [CrossRef]
- WWF-Pakistan. Annual Report 2020; Nawaz, R., Shahzad, S.M., Eds.; WWF-Pakistan: Lahore, Pakistan, 2020. [Google Scholar]
- Government of Khyber Pakhtunkhwa. Billion Tree Tsunami Afforestation Project; Climate Change, Forestry, Environment and Wildlife Department: Peshawar, Pakistan, 2014.
- Government of Pakistan. Ten Billion Trees Tsunami Programme—Phase-I: Up-Scaling of Green Pakistan Programme (Revised); Ministry of Climate Change and Environmental Coordination: Islamabad, Pakistan, 2019.
- Ali, H.A.I.D.A.R.; Ahmad, H.; Marwat, K.B.; Yousaf, M.; Gul, B.; Khan, I. Trade potential and conservation issues of medicinal plants in District Swat, Pakistan. Pak. J. Bot. 2012, 44, 1905–1912. [Google Scholar]
- Iqbal, J.; Meilan, R.; Khan, B. Assessment of risk, extinction, and threats to Himalayan yew in Pakistan. Saudi J. Biol. Sci. 2020, 27, 762–767. [Google Scholar] [CrossRef]
- Gillani, S.W.; Ahmad, M.; Zafar, M.; Haq, S.M.; Waheed, M.; Manzoor, M.; Shaheen, H.; Sultana, S.; Rehman, F.U.; Makhkamov, T. An insight into indigenous ethnobotanical knowledge of medicinal and aromatic plants from Kashmir Himalayan region. Ethnobot. Res. Appl. 2024, 28, 1–21. [Google Scholar] [CrossRef]
- Dhyani, S. Are Himalayan ecosystems facing hidden collapse? Assessing the drivers and impacts of change to aid conservation, restoration and conflict resolution challenges. Biodivers. Conserv. 2023, 32, 3731–3764. [Google Scholar] [CrossRef]
- Kumar, B.; Singh, K.; Sharma, J.; Gairola, S. A comprehensive review of fuelwood resources and their use pattern in rural villages of Western Himalaya, India. Plant Arch. 2020, 20, 1949–1958. [Google Scholar] [CrossRef]
- Ratsch, C. Encyclopedia of Psychoactive Plants: Botany, Ethnopharmacology, and Application AT Verlag; Park Street Press: Paris, France, 1998. [Google Scholar]
- Wilson, C.R.; Sauer, J.M.; Hooser, S.B. Taxines: A review of the mechanism and toxicity of yew (Taxus spp.) alkaloids. Toxicon 2001, 39, 175–185. [Google Scholar] [CrossRef]
- Frohne, D.; Pribilla, O. Fatal poisoning with Taxus baccata. Archiv für Toxikologie 1965, 21, 150–162. [Google Scholar] [CrossRef]
- Beike, J.; Karger, B.; Meiners, T.; Brinkmann, B.; Köhler, H. LC-MS determination of Taxus alkaloids in biological specimens. Int. J. Leg. Med. 2003, 117, 335–339. [Google Scholar] [CrossRef]
- Mroczek, T.; Glowniak, K. Solid-phase extraction and simplified high-performance liquid chromatographic determination of 10-deacetylbaccatin III and related taxoids in yew species. J. Pharm. Biomed. Anal. 2001, 26, 89–102. [Google Scholar] [CrossRef]
- Mußhoff, F.; Jacob, B.; Fowinkel, C.; Daldrup, T. Suicidal yew leave ingestion—Phloroglucindimethylether (3,5-dimethoxyphenol) as a marker for poisoning from Taxus baccata. Int. J. Leg. Med. 1993, 106, 45–50. [Google Scholar] [CrossRef]
- Li, R. Forensic Biology: Identification and DNA Analysis of Biological Evidence; CRC Press: Boca Raton, FL, USA, 2008. [Google Scholar] [CrossRef]
- Holland, M.M.; Parsons, T.J. Mitochondrial DNA Sequence Analysis-Validation and Use for Forensic Casework. Forensic Sci. Rev. 1999, 11, 21–50. [Google Scholar] [PubMed]
- Levy-Booth, D.J.; Campbell, R.G.; Gulden, R.H.; Hart, M.M.; Powell, J.R.; Klironomos, J.N.; Pauls, K.P.; Swanton, C.J.; Trevors, J.T.; Dunfield, K.E. Cycling of extracellular DNA in the soil environment. Soil Biol. Biochem. 2007, 39, 2977–2991. [Google Scholar] [CrossRef]
- Minamoto, T.; Yamanaka, H.; Takahara, T.; Honjo, M.N.; Kawabata, Z.I. Surveillance of fish species composition using environmental DNA. Limnology 2012, 13, 193–197. [Google Scholar] [CrossRef]
- Miya, M.; Sato, Y.; Fukunaga, T.; Sado, T.; Poulsen, J.Y.; Sato, K.; Minamoto, T.; Yamamoto, S.; Yamanaka, H.; Araki, H.; et al. MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: Detection of more than 230 subtropical marine species. R. Soc. Open Sci. 2015, 2, 150088. [Google Scholar] [CrossRef]
- Deiner, K.; Bik, H.M.; Mächler, E.; Seymour, M.; Lacoursière-Roussel, A.; Altermatt, F.; Creer, S.; Bista, I.; Lodge, D.M.; De Vere, N.; et al. Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol. Ecol. 2017, 26, 5872–5895. [Google Scholar] [CrossRef]
- Yamamoto, S.; Masuda, R.; Sato, Y.; Sado, T.; Araki, H.; Kondoh, M.; Minamoto, T.; Miya, M. Environmental DNA metabarcoding reveals local fish communities in a species-rich coastal sea. Sci. Rep. 2017, 7, 40368. [Google Scholar] [CrossRef]
- Cristescu, M.E.; Hebert, P.D. Uses and misuses of environmental DNA in biodiversity science and conservation. Annu. Rev. Ecol. Evol. Syst. 2018, 49, 209–230. [Google Scholar] [CrossRef]
- Taberlet, P.; Bonin, A.; Zinger, L.; Coissac, E. Environmental DNA: For Biodiversity Research and Monitoring; Oxford University Press: Oxford, UK, 2018. [Google Scholar]
- Ruppert, K.M.; Kline, R.J.; Rahman, M.S. Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: A systematic review in methods, monitoring, and applications of global eDNA. Glob. Ecol. Conserv. 2019, 17, e00547. [Google Scholar] [CrossRef]
- Calderón-Sanou, I.; Münkemüller, T.; Boyer, F.; Zinger, L.; Thuiller, W. From environmental DNA sequences to ecological conclusions: How strong is the influence of methodological choices? J. Biogeogr. 2020, 47, 193–206. [Google Scholar] [CrossRef]
- Banerjee, P.; Dey, G.; Antognazza, C.M.; Sharma, R.K.; Maity, J.P.; Chan, M.W.; Huang, Y.H.; Lin, P.Y.; Chao, H.C.; Lu, C.M.; et al. Reinforcement of environmental DNA based methods (sensu stricto) in biodiversity monitoring and conservation: A review. Biology 2021, 10, 1223. [Google Scholar] [CrossRef] [PubMed]
- Pääbo, S.; Poinar, H.; Serre, D.; Jaenicke-Després, V.; Hebler, J.; Rohland, N.; Kuch, M.; Krause, J.; Vigilant, L.; Hofreiter, M. Genetic analyses from ancient DNA. Annu. Rev. Genet. 2004, 38, 645–679. [Google Scholar] [CrossRef]
- Hansen, A.J.; Mitchell, D.L.; Wiuf, C.; Paniker, L.; Brand, T.B.; Binladen, J.; Gilichinsky, D.A.; Rønn, R.; Willerslev, E. Crosslinks rather than strand breaks determine access to ancient DNA sequences from frozen sediments. Genetics 2006, 173, 1175–1179. [Google Scholar] [CrossRef]
- Kress, W.J. Plant DNA barcodes: Applications today and in the future. J. Syst. Evol. 2017, 55, 291–307. [Google Scholar] [CrossRef]
- Alaeddini, R.; Walsh, S.J.; Abbas, A. Forensic implications of genetic analyses from degraded DNA—A review. Forensic Sci. Int. Genet. 2010, 4, 148–157. [Google Scholar] [CrossRef]
- Latham, K.E.; Miller, J.J. DNA recovery and analysis from skeletal material in modern forensic contexts. Forensic Sci. Res. 2019, 4, 51–59. [Google Scholar] [CrossRef]
- Bhoyar, L.; Mehar, P.; Chavali, K. Assessing the forensic implications of DNA degradation for PMI estimation using comet assay: A systematic review. J. Forensic Leg. Med. 2025, 109, 102801. [Google Scholar] [CrossRef] [PubMed]
- Parveen, I.; Gafner, S.; Techen, N.; Murch, S.J.; Khan, I.A. DNA barcoding for the identification of botanicals in herbal medicine and dietary supplements: Strengths and limitations. Planta Medica 2016, 82, 1225–1235. [Google Scholar] [CrossRef] [PubMed]
- Meusnier, I.; Singer, G.A.; Landry, J.F.; Hickey, D.A.; Hebert, P.D.; Hajibabaei, M. A universal DNA mini-barcode for biodiversity analysis. BMC Genom. 2008, 9, 214. [Google Scholar] [CrossRef] [PubMed]
- Srirama, R.; Gurumurthy, B.R.; Senthilkumar, U.; Ravikanth, G.; Shaanker, R.U.; Shivanna, M.B. Are mini DNA-barcodes sufficiently informative to resolve species identities? An in silico analysis using Phyllanthus. J. Genet. 2014, 93, 823–829. [Google Scholar] [CrossRef]
- Møller, P.; Azqueta, A.; Boutet-Robinet, E.; Koppen, G.; Bonassi, S.; Milić, M.; Gajski, G.; Costa, S.; Teixeira, J.P.; Costa Pereira, C.; et al. Minimum Information for Reporting on the Comet Assay (MIRCA): Recommendations for describing comet assay procedures and results. Nat. Protoc. 2020, 15, 3817–3826. [Google Scholar] [CrossRef]
- Coyle, H.M.; Ladd, C.; Palmbach, T.; Lee, H.C. The green revolution: Botanical contributions to forensics and drug enforcement. Croat. Med. J. 2001, 42, 340–345. [Google Scholar]
- Chandra, R.; Sharma, V. Forensic botany: An emerging discipline of plant sciences. Indian Bot. Blog-O-J. 2014, 1–8. [Google Scholar]
- Yoon, C.K. Botanical witness for the prosecution. Science 1993, 260, 894–895. [Google Scholar] [CrossRef]
- Pillay, V.V.; Sasidharan, A. Oleander and datura poisoning: An update. Indian J. Crit. Care Med. 2019, 23 (Suppl. S4), S250. [Google Scholar] [CrossRef]
- Khajja, B.S.; Sharma, M.; Singh, R.; Mathur, G.K. Forensic study of Indian toxicological plants as botanical weapon (BW): A review. J. Environ. Anal. Toxicol. 2011, 1, 112. [Google Scholar]
- Mahmoudian, M.; Salehian, P.; Jalilpour, H. Toxicity of Peganum harmala: Review and a case report. Iran. J. Pharmacol. Ther. (IJPT) 2002, 1, 1–4. [Google Scholar]
- Dubey, N.K.; Dwivedy, A.K.; Chaudhari, A.K.; Das, S. Common toxic plants and their forensic significance. In Natural Products and Drug Discovery; Elsevier: Amsterdam, The Netherlands, 2018; pp. 349–374. [Google Scholar]
- Friess, D.A.; Webb, E.L. Bad data equals bad policy: How to trust estimates of ecosystem loss when there is so much uncertainty? Environ. Conserv. 2011, 38, 1–5. [Google Scholar] [CrossRef]
- Flores-Palacios, A.; Valencia-Díaz, S. Local illegal trade reveals unknown diversity and involves a high species richness of wild vascular epiphytes. Biol. Conserv. 2007, 136, 372–387. [Google Scholar] [CrossRef]
- Challender, D.W.; Hywood, L. African pangolins under increased pressure from poaching and intercontinental trade. Traffic Bull. 2012, 24, 53–55. [Google Scholar]
- Caillabet, O.S. The Trade in Tokay Geckos Gekko Gecko in South-East Asia: With a Case Study on Novel Medicinal Claims in Peninsular Malaysia; TRAFFIC: Petaling Jaya, Malaysia, 2013. [Google Scholar]
- Mateen, R.M.; Tariq, A.; Rasool, N. Forensic science in Pakistan; present and future. Egypt J. Forensic. Sci. 2018, 8, 45. [Google Scholar] [CrossRef]
- Julian, R.; Kelty, S. Forensic Science and Justice: From Crime Scene to Court and Beyond. Curr. Issues Crim. Justice 2012, 24, 1–6. [Google Scholar] [CrossRef]
- Garland, N.M.; Stuckey, G.B. Criminal Evidence for the Law Enforcement Officer; Glencoe/McGraw-Hill Post Secondary: Columbus, OH, USA, 2000. [Google Scholar]
- Giannelli, P.C. The abuse of scientific evidence in criminal cases: The need for independent crime laboratories. Va. J. Soc. Pol’y L. 1996, 4, 439. [Google Scholar]
- Lindquist, C.A. Criminalistics education and the role of the criminalistics educator. Forensic Sci. Rev. 1995, 7, 61–75. [Google Scholar]
- Margot, P. Forensic science on trial-What is the law of the land? Aust. J. Forensic Sci. 2011, 43, 89–103. [Google Scholar] [CrossRef]
- Berghaus, G. DNA-Technology and Its Forensic Application; Springer: New York, NY, USA, 1991. [Google Scholar]
- DeForest, P.; Petraco, N.; Kobilinsky, L. Chapter 5 in Chemistry and Crime—From Sherlock Holmes to Today’s Courtroom. In Chemistry and the Challenge of Crime; Gerber, S.M., Ed.; ACS Symposium Series; American Chemical Society: Washington, DC, USA, 1983; pp. 45–63. [Google Scholar]
- Berg, B.; Horgan, J. Criminal Investigation. In Forensic Economics in Competition Law Enforcement; Centre for Law and Economics: Amsterdam, The Netherlands, 1998. [Google Scholar]
- Weston, P.; Lushbaugh, C. Criminal Investigation, Basic Perspectives, 9th ed.; Prentice Hall: Upper Saddle River, NJ, USA, 2003. [Google Scholar]
- Steadman, G.W. Survey of DNA Crime Laboratories, 1998; US Department of Justice, Office of Justice Programs, Bureau of Justice Statistics: Washington, DC, USA, 2000.
- Lappas, N.T. Forensic Science Laboratories in the United States: A Survey. J. Forensic Sci. Soc. 1978, 18, 171–180. [Google Scholar] [CrossRef]
- Fradella, H.F.; Owen, S.S.; Burke, T.W. Building bridges between criminal justice and the forensic sciences to create forensic studies programs. J. Crim. Justice Educ. 2007, 18, 261–282. [Google Scholar] [CrossRef]
- Tilstone, W.J. Education, training and assessment in forensic science. J. Forensic Sci. Soc. 1991, 31, 95–100. [Google Scholar] [CrossRef]
- Grover, N.; Tyagi, I. Development of Forensic Science and Criminal Prosecution-India. Nature 1910, 23, 76. [Google Scholar]
- Riaz, Z. Forensic DNA Testing and Its Conduct Problems in Sindh Police: Evaluations and Suggestions. Master’s Thesis, Department of Criminology, University of Sindh, Jamshoro, Pakistan, 2008. [Google Scholar]
- Connor, J.M. Forensic economics: An introduction with special emphasis on price fixing. Joclec 2008, 4, 31–59. [Google Scholar] [CrossRef]
- Platt, R. Crime Scene: The Ultimate Guide to Forensic Science; DK Publishing: London, UK, 2003. [Google Scholar]
- Taberlet, P.; Prud’Homme, S.M.; Campione, E.; Roy, J.; Miquel, C.; Shehzad, W.; Gielly, L.; Rioux, D.; Choler, P.; Clément, J.C.; et al. Soil sampling and isolation of extracellular DNA from large amount of starting material suitable for metabarcoding studies. Mol. Ecol. 2012, 21, 1816–1820. [Google Scholar] [CrossRef]
- Taberlet, P.; Coissac, E.; Pompanon, F.; Gielly, L.; Miquel, C.; Valentini, A.; Vermat, T.; Corthier, G.; Brochmann, C.; Willerslev, E. Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 2007, 35, e14. [Google Scholar] [CrossRef]
- Dohm, J.C.; Lottaz, C.; Borodina, T.; Himmelbauer, H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008, 36, e105. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Oshima, T.; Morimoto, T.; Ikeda, S.; Yoshikawa, H.; Shiwa, Y.; Ishikawa, S.; Linak, M.C.; Hirai, A.; Takahashi, H.; et al. Sequence-specific error profile of Illumina sequencers. Nucleic Acids Res. 2011, 39, e90. [Google Scholar] [CrossRef]
- Coghlan, M.L.; Maker, G.; Crighton, E.; Haile, J.; Murray, D.C.; White, N.E.; Byard, R.W.; Bellgard, M.I.; Mullaney, I.; Trengove, R.; et al. Combined DNA, toxicological and heavy metal analyses provides an auditing toolkit to improve pharmacovigilance of traditional Chinese medicine (TCM). Sci. Rep. 2015, 5, 17475. [Google Scholar] [CrossRef]
- Srivastava, D.; Manjunath, K. DNA barcoding of endemic and endangered orchids of India: A molecular method of species identification. Pharmacogn. Mag. 2020, 16, 290–299. [Google Scholar] [CrossRef]
- Jiao, L.; He, T.; Dormontt, E.E.; Zhang, Y.; Lowe, A.J.; Yin, Y. Applicability of chloroplast DNA barcodes for wood identification between Santalum album and its adulterants. Holzforschung 2019, 73, 209–218. [Google Scholar] [CrossRef]
- Shah, A.; Li, D.Z.; Möller, M.; Gao, L.M.; Hollingsworth, M.L.; Gibby, M. Delimitation of Taxus fuana Nan Li & RR Mill (Taxaceae) based on morphological and molecular data. Taxon 2008, 57, 211–222. [Google Scholar] [CrossRef]
- Liu, L.; Shu, X.; Ren, L.; Zhou, H.; Li, Y.; Liu, W.; Zhu, C.; Liu, L. Determination of the early time of death by computerized image analysis of DNA degradation: Which is the best quantitative indicator of DNA degradation? J. Huazhong Univ. Sci. Technol. 2007, 27, 362–366. [Google Scholar] [CrossRef] [PubMed]
- Shukla, R.K. Forensic application of comet assay: An emerging technique. Forensic Sci. Res. 2017, 2, 180–184. [Google Scholar] [CrossRef] [PubMed]
- Gaudio, D.; Fernandes, D.M.; Schmidt, R.; Cheronet, O.; Mazzarelli, D.; Mattia, M.; O’Keeffe, T.; Feeney, R.N.; Cattaneo, C.; Pinhasi, R. Genome-wide DNA from degraded petrous bones and the assessment of sex and probable geographic origins of forensic cases. Sci. Rep. 2019, 9, 8226. [Google Scholar] [CrossRef]
- de Lapuente, J.; Lourenço, J.; Mendo, S.A.; Borràs, M.; Martins, M.G.; Costa, P.M.; Pacheco, M. The Comet Assay and its applications in the field of ecotoxicology: A mature tool that continues to expand its perspectives. Front. Genet. 2015, 6, 180. [Google Scholar] [CrossRef]
- Massey, S.E. Comparative microbial genomics and forensics. Microbiol. Spectr. 2016, 4, 237–276. [Google Scholar] [CrossRef] [PubMed]
- Meyers, R.A. (Ed.) Molecular Biology and Biotechnology: A Comprehensive Desk Reference; John Wiley & Sons: Hoboken, NJ, USA, 2021. [Google Scholar]
- Bannick, K. Mechanisms to Combat DNA Degradation. Honors Project No. 566. 2021. Available online: https://scholarworks.bgsu.edu/honorsprojects/566 (accessed on 17 July 2025).
- Bonfigli, A.; Cesare, P.; Volpe, A.R.; Colafarina, S.; Forgione, A.; Aloisi, M.; Zarivi, O.; Poma, A.M. Estimation of DNA degradation in archaeological human remains. Genes 2023, 14, 1238. [Google Scholar] [CrossRef]
- Mohamed, M.A.; El Sherbeny, M.; Farag, D.A. A comparative study of two gel-based techniques to detect the relationship between post-mortem interval and nuclear DNA degradation in different tissues. Egypt. Dent. J. 2020, 66, 175–186. [Google Scholar] [CrossRef]
- Chandrasekaran, A.R.; Halvorsen, K. Nuclease degradation analysis of DNA nanostructures using gel electrophoresis. Curr. Protoc. Nucleic Acid Chem. 2020, 82, e115. [Google Scholar] [CrossRef]
- Figueroa-González, G.; Pérez-Plasencia, C. Strategies for the evaluation of DNA damage and repair mechanisms in cancer. Oncol. Lett. 2017, 13, 3982–3988. [Google Scholar] [CrossRef] [PubMed]
- Tozzo, P.; Scrivano, S.; Sanavio, M.; Caenazzo, L. The role of DNA degradation in the estimation of post-mortem interval: A systematic review of the current literature. Int. J. Mol. Sci. 2020, 21, 3540. [Google Scholar] [CrossRef]
- Langie, S.A.; Azqueta, A.; Collins, A.R. The comet assay: Past, present, and future. Front. Genet. 2015, 6, 266. [Google Scholar] [CrossRef]
- Williams, T.; Soni, S.; White, J.; Can, G.; Javan, G.T. Evaluation of DNA degradation using flow cytometry: Promising tool for postmortem interval determination. Am. J. Forensic Med. Pathol. 2015, 36, 104–110. [Google Scholar] [CrossRef]
- Cina, S.J. Flow cytometric evaluation of DNA degradation: A predictor of postmortem interval? Am. J. Forensic Med. Pathol. 1994, 15, 300–302. [Google Scholar] [CrossRef]
- Henderson, D.S. DNA Repair Protocols Mammalian Systems, 2nd ed.; Humana Press: Totowa, NJ, USA, 2006. [Google Scholar]
- Huerta, S.; Goulet, E.J.; Huerta-Yepez, S.; Livingston, E.H. Screening and detection of apoptosis. J. Surg. Res. 2007, 139, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Vermes, I.; Haanen, C.; Steffens-Nakken, H.; Reutellingsperger, C. A novel assay for apoptosis flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled annexin V. J. Immunol. Methods 1995, 184, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Pietkiewicz, S.; Schmidt, J.H.; Lavrik, I.N. Quantification of apoptosis and necroptosis at the single cell level by a combination of Imaging Flow Cytometry with classical Annexin V/propidium iodide staining. J. Immunol. Methods 2015, 423, 99–103. [Google Scholar] [CrossRef]
- Basiji, D.; O’Gorman, M.R. Imaging flow cytometry. J. Immunol. Methods 2015, 423, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Gavrieli, Y.; Sherman, Y.; Ben-Sasson, S.A. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J. Cell Biol. 1992, 119, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Walker, P.R.; Carson, C.; Leblanc, J.; Sikorska, M. Labeling DNA damage with terminal transferase: Applicability, specificity, and limitations. In In Situ Detection of DNA Damage: Methods and Protocols; Humana Press: Totowa, NJ, USA, 2002; pp. 3–19. [Google Scholar] [CrossRef]
- Loo, D.T. TUNEL assay: An overview of techniques. In In Situ Detection of DNA Damage: Methods Protocols; Humana Press: Totowa, NJ, USA, 2002; pp. 21–30. [Google Scholar] [CrossRef]
- Kumari, S.; Rastogi, R.P.; Singh, K.L.; Singh, S.P.; Sinha, R.P. DNA damage: Detection strategies. EXCLI J 2008, 7, 44–62. [Google Scholar] [CrossRef]
- Hwang, J.S.; Kobayashi, C.; Agata, K.; Ikeo, K.; Gojobori, T. Detection of apoptosis during planarian regeneration by the expression of apoptosis-related genes and TUNEL assay. Gene 2004, 333, 15–25. [Google Scholar] [CrossRef]
- Tripathi, A.K.; Pareek, A.; Singla-Pareek, S.L. TUNEL assay to assess extent of DNA fragmentation and programmed cell death in root cells under various stress conditions. Bio-Protocol 2017, 7, e2502. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Wittwer, C.T. The MIQE guidelines: Minimum information for publication of Quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Lindahl, T. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef]
- Schulze Johann, K.; Bauer, H.; Wiegand, P.; Pfeiffer, H.; Vennemann, M. Detecting DNA damage in stored blood samples. Forensic Sci. Med. Pathol. 2023, 19, 50–59. [Google Scholar] [CrossRef]
- McCord, B.; Opel, K.; Funes, M.; Zoppis, S.; Meadows Jantz, L. An investigation of the effect of DNA degradation and inhibition on PCR amplification of single source and mixed forensic samples. In National Archive of Criminal Justice Data; US Department of Justice: Washington, DC, USA, 2011; pp. 1–65. [Google Scholar]
- Gall, J.G.; Pardue, M.L. Formation and detection of RNA-DNA hybrid molecules in cytological preparations. Proc. Natl. Acad. Sci. USA 1969, 63, 378–383. [Google Scholar] [CrossRef]
- Levsky, J.M.; Singer, R.H. Fluorescence in situ hybridization: Past, present and future. J. Cell Sci. 2003, 116, 2833–2838. [Google Scholar] [CrossRef] [PubMed]
- Halling, K.C.; Kipp, B.R. Fluorescence in situ hybridization in diagnostic cytology. Hum. Pathol. 2007, 38, 1137–1144. [Google Scholar] [CrossRef]
- Mardis, E.R. Next-generation DNA sequencing methods. Annu. Rev. Genom. Hum. Genet. 2008, 9, 387–402. [Google Scholar] [CrossRef]
- Briggs, A.W.; Good, J.M.; Green, R.E.; Krause, J.; Maricic, T.; Stenzel, U.; Lalueza-Fox, C.; Rudan, P.; Brajković, D.; Kućan, Ž.; et al. Targeted retrieval and analysis of five Neandertal mtDNA genomes. Science 2009, 325, 318–321. [Google Scholar] [CrossRef]
- Overballe-Petersen, S.; Orlando, L.; Willerslev, E. Next-generation sequencing offers new insights into DNA degradation. Trends Biotechnol. 2012, 30, 364–368. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314. [Google Scholar] [PubMed]
- Karger, B.L.; Snyder, L.R.; Horvath, C. An Introduction to Separation Science; Wiley: New York, NY, USA, 1973. [Google Scholar]
- Hughes-Stamm, S.R.; Ashton, K.J.; van Daal, A. Assessment of DNA degradation and the genotyping success of highly degraded samples. Int. J. Leg. Med. 2011, 125, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Rohland, N.; Hofreiter, M. Ancient DNA extraction from bones and teeth. Nat. Protoc. 2007, 2, 1756–1762. [Google Scholar] [CrossRef]
- Santella, R.M. Immunological methods for detection of carcinogen-DNA damage in humans. Cancer Epidemiol. Biomark. Prev. 1999, 8, 733–739. [Google Scholar]
- Yatabe, Y. ALK FISH and IHC: You cannot have one without the other. J. Thorac. Oncol. 2015, 10, 548–550. [Google Scholar] [CrossRef]
- Kriste, A.G.; Martincigh, B.S.; Salter, L.F. A sensitive immunoassay technique for thymine dimer quantitation in UV-irradiated DNA. J. Photochem. Photobiol. A Chem. 1996, 93, 185–192. [Google Scholar] [CrossRef]
- Rindgen, D.; Turesky, R.J.; Vouros, P. Determination of in vitro formed DNA adducts of 2-amino-1-methyl-6-phenylimidazo [4,5-b] pyridine using capillary liquid chromatography/electrospray ionization/tandem mass spectrometry. Chem. Res. Toxicol. 1995, 8, 1005–1013. [Google Scholar] [CrossRef]
- Mullins, E.A.; Rubinson, E.H.; Pereira, K.N.; Calcutt, M.W.; Christov, P.P.; Eichman, B.F. An HPLC–tandem mass spectrometry method for simultaneous detection of alkylated base excision repair products. Methods 2013, 64, 59–66. [Google Scholar] [CrossRef]
- Toyokuni, S. Oxidative stress as an iceberg in carcinogenesis and cancer biology. Arch. Biochem. Biophys. 2016, 595, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Dizdaroglu, M.İ.R.A.L.; Coskun, E.; Jaruga, P. Measurement of oxidatively induced DNA damage and its repair, by mass spectrometric techniques. Free Radic. Res. 2015, 49, 525–548. [Google Scholar] [CrossRef]
- Sato, K.; Greenberg, M.M. Selective detection of 2-deoxyribonolactone in DNA. J. Am. Chem. Soc. 2005, 127, 2806–2807. [Google Scholar] [CrossRef]
- Reddy, P.T.; Jaruga, P.; Nelson, B.C.; Lowenthal, M.; Dizdaroglu, M. Stable isotope-labeling of DNA repair proteins, and their purification and characterization. Protein Expr. Purif. 2011, 78, 94–101. [Google Scholar] [CrossRef]
- Fojta, M.; Daňhel, A.; Havran, L.; Vyskočil, V. Recent progress in electrochemical sensors and assays for DNA damage and repair. TrAC Trends Anal. Chem. 2016, 79, 160–167. [Google Scholar] [CrossRef]
- Boon, E.M.; Ceres, D.M.; Drummond, T.G.; Hill, M.G.; Barton, J.K. Mutation detection by electrocatalysis at DNA-modified electrodes. Nat. Biotechnol. 2000, 18, 1096–1100. [Google Scholar] [CrossRef] [PubMed]
- Boal, A.K.; Barton, J.K. Electrochemical detection of lesions in DNA. Bioconjugate Chem. 2005, 16, 312–321. [Google Scholar] [CrossRef]
- Haines, A.M.; Webb, S.L.; Wallace, J.R. Conservation forensics: The intersection of wildlife crime, forensics, and conservation. In Wildlife Biodiversity Conservation: Multidisciplinary and Forensic Approaches; Springer International Publishing: Cham, Switzerland, 2021; pp. 125–146. [Google Scholar] [CrossRef]
- Díaz-Delgado, R.; Mücher, S. Editorial of special issue “Drones for biodiversity conservation and ecological monitoring”. Drones 2019, 3, 47. [Google Scholar] [CrossRef]
- Augusteyn, J.; Pople, A.; Rich, M. Evaluating the use of thermal imaging cameras to monitor the endangered greater bilby at Astrebla Downs National Park. Aust. Mammal. 2020, 42, 329–340. [Google Scholar] [CrossRef]
- Darras, K.F.; Balle, M.; Xu, W.; Yan, Y.; Zakka, V.G.; Toledo-Hernández, M.; Sheng, D.; Lin, W.; Zhang, B.; Lan, Z.; et al. Eyes on nature: Embedded vision cameras for terrestrial biodiversity monitoring. Methods Ecol. Evol. 2024, 15, 2262–2275. [Google Scholar] [CrossRef]
- Dominy, N.J.; Duncan, B. GPS and GIS methods in an African rain forest: Applications to tropical ecology and conservation. Conserv. Ecol. 2002, 5, 6. [Google Scholar] [CrossRef]
- Hill, A.P.; Prince, P.; Piña Covarrubias, E.; Doncaster, C.P.; Snaddon, J.L.; Rogers, A. AudioMoth: Evaluation of a smart open acoustic device for monitoring biodiversity and the environment. Methods Ecol. Evol. 2018, 9, 1199–1211. [Google Scholar] [CrossRef]
- Hanisch, E.; Johnston, R.; Longnecker, N. Cameras for conservation: Wildlife photography and emotional engagement with biodiversity and nature. Hum. Dimens. Wildl. 2019, 24, 267–284. [Google Scholar] [CrossRef]
- Buxton, R.T.; McKenna, M.F.; Clapp, M.; Meyer, E.; Stabenau, E.; Angeloni, L.M.; Crooks, K.; Wittemyer, G. Efficacy of extracting indices from large-scale acoustic recordings to monitor biodiversity. Conserv. Biol. 2018, 32, 1174–1184. [Google Scholar] [CrossRef]
- Ross, R.; Anderson, B.; Bienvenu, B.; Scicluna, E.L.; Robert, K.A. WildTrack: An IoT system for tracking passive-RFID microchipped wildlife for ecology research. Automation 2022, 3, 426–438. [Google Scholar] [CrossRef]
- Foley, C.J.; Sillero-Zubiri, C. Open-source, low-cost modular GPS collars for monitoring and tracking wildlife. Methods Ecol. Evol. 2020, 11, 553–558. [Google Scholar] [CrossRef]
- Brown, D.D.; LaPoint, S.; Kays, R.; Heidrich, W.; Kümmeth, F.; Wikelski, M. Accelerometer-informed GPS telemetry: Reducing the trade-off between resolution and longevity. Wildl. Soc. Bull. 2012, 36, 139–146. [Google Scholar] [CrossRef]
- Turner, W. Sensing biodiversity. Science 2014, 346, 301–302. [Google Scholar] [CrossRef]
- Urbano, F.; Viterbi, R.; Pedrotti, L.; Vettorazzo, E.; Movalli, C.; Corlatti, L. Enhancing biodiversity conservation and monitoring in protected areas through efficient data management. Environ. Monit. Assess. 2024, 196, 12. [Google Scholar] [CrossRef]
- Dijk, N. Advanced Wildlife Camera Trapping Using Embedded AI Machine Vision. Bachelor’s Thesis, University of Twente, Enschede, The Netherlands, 2023. [Google Scholar]
- Pettorelli, N.; Williams, J.; Schulte to Bühne, H.; Crowson, M. Deep learning and satellite remote sensing for biodiversity monitoring and conservation. Remote Sens. Ecol. Conserv. 2025, 11, 123–132. [Google Scholar] [CrossRef]
- Rodríguez, J.P.; Brotons, L.; Bustamante, J.; Seoane, J. The application of predictive modelling of species distribution to biodiversity conservation. Divers. Distrib. 2007, 13, 243–251. [Google Scholar] [CrossRef]
- Sarkar, S.; Pressey, R.L.; Faith, D.P.; Margules, C.R.; Fuller, T.; Stoms, D.M.; Moffett, A.; Wilson, K.A.; Williams, K.J.; Williams, P.H.; et al. Biodiversity conservation planning tools: Present status and challenges for the future. Annu. Rev. Environ. Resour. 2006, 31, 123–159. [Google Scholar] [CrossRef]
- Raghav, Y.Y.; Vyas, V. Harnessing Cloud Computing and IoT for Wildlife Conservation. In Integration of Cloud Computing and IoT: Trends, Case Studies and Applications; CRC Press: Boca Raton, FL, USA, 2024; Volume 142. [Google Scholar]
- Ullah, F.; Saqib, S.; Xiong, Y.C. Integrating artificial intelligence in biodiversity conservation: Bridging classical and modern approaches. Biodivers. Conserv. 2025, 34, 45–65. [Google Scholar] [CrossRef]
- Hebert, P.D.; Cywinska, A.; Ball, S.L.; DeWaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. London. Ser. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [PubMed]
- Fahner, N.A.; Shokralla, S.; Baird, D.J.; Hajibabaei, M. Large-scale monitoring of plants through environmental DNA metabarcoding of soil: Recovery, resolution, and annotation of four DNA markers. PLoS ONE 2016, 11, e0157505. [Google Scholar] [CrossRef]
- Hunter, M.E.; Oyler-McCance, S.J.; Dorazio, R.M.; Fike, J.A.; Smith, B.J.; Hunter, C.T.; Reed, R.N.; Hart, K.M. Environmental DNA (eDNA) sampling improves occurrence and detection estimates of invasive Burmese pythons. PLoS ONE 2015, 10, e0121655. [Google Scholar] [CrossRef]
- TRAFFIC. Combating Wildlife Crime Linked to the Internet: Global Trends and China’s Experiences; TRAFFIC: Cambridge, UK, 2019. [Google Scholar]
- WWF. Over 100 Wildlife Rangers Died on Duty in Past Year. 2018. Available online: https://phys.org/news/2018-07-wildlife-rangers-died-duty-year.html (accessed on 17 July 2025).
- Ogden, R.; Dawnay, N.; McEwing, R. Wildlife DNA forensics—Bridging the gap between conservation genetics and law enforcement. Endanger. Species Res. 2009, 9, 179–195. [Google Scholar] [CrossRef]
- Harris, L.; Shiraishi, H. Understanding the Global Caviar Market. Results of a Rapid Assessment of Trade in Sturgeon Caviar. TRAFFIC and WWF Joint Report. 2018. Available online: https://www.traffic.org/site/assets/files/9805/global_caviar_market-1.pdf (accessed on 17 July 2025).
- Williamson, D.F. Caviar and Conservation: Status, Management and Trade of North American Sturgeon and Paddlefish; TRAFFIC North America; World Wildlife Fund: Washington, DC, USA, 2003. [Google Scholar]
- Gore, M.L. Global risks, conservation, and criminology. In Conservation Criminology; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 2017; pp. 1–23. [Google Scholar] [CrossRef]
- Kahler, J.S.; Gore, M.L. Conservation crime science. In Conservation Criminology; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 2017; pp. 25–43. [Google Scholar] [CrossRef]

| Taxon | Gene | Primer | Secquence | Length |
|---|---|---|---|---|
| T. canadensis | matK | F | GCAGAAAGCTCTGGTTCCAC | 20 |
| R | CGATCGGGAAAAGATCCATA | |||
| Rbcl | F | TTTGGATTCAAAGCCCTACG | ||
| R | AGTCCACCACGGAGACATTC | |||
| T. wallichiana | ITS | F | GCGGTAGGATCATTGTCGTT | |
| R | GGCACTCGCCCTTGTAATAA | |||
| T. contorta | trnL-F | F | ATTTTGAATGGGCAATCCTG | |
| R | GGCAAGCCTAGAACAACTCAA | 21 | ||
| TS | F | GCAGATGAGCTGGTTGTGAA | 20 | |
| R | ATTCGATACCCCATGATCCA | |||
| trnS-trnQ | F | CTTGCCAAGCAAAGGCTAAG | ||
| R | CGGAGAATCAAAATGCCAGT |
| Criteria | S. S. (5000) E. (0.05) | S.S. (5000) E. (100) | S. (100) E. (0.05) | Query Length | |
|---|---|---|---|---|---|
| Hits | Taxon | Eukaryota/Taxus L. | Hits | ||
| matK | 270 | Taxaceae | 1396/382 | Taxus L. (100%) | 117 |
| rbcL | 2454 | Gymnosperm/fern | 7637/557 | 200 | |
| ITS | 159 | Taxus L. | 1209/1030 | 175 | |
| Group | Taxon | Hits | Taxa | A/hits |
|---|---|---|---|---|
| Species | T. baccata (101), T. brevifolia (27), T. calcicola (11), T. canadensis (45), T. celebica (1), T. chinensis (171), T. contorta (26), T. cuspidata (70), T. floridana (33), T. florinii (13), T. fuana (43), T. globosa (46), T. mairei (548), T. phytonii (369), T. sumatrana (15), T. wallichiana (68) | 1547 | 16 | 97 |
| Variety | T. cuspidata var. cuspidata (6), T. cuspidata var. latifolia (6), T. cuspidata var. nana (10), T. wallichiana var. wallichiana (90), T. wallichiana var. yunnanensis (12) | 124 | 5 | 25 |
| Hybrid | T. x hunnewelliana (4), T. x media (9) | 13 | 2 | 6.5 |
| Variant | T. sp. ‘Emei type’ (9), T. sp. ‘Huangshan type’ (17), T. sp. ‘Mandiya type’ (2), T. sp. ‘Qinling type’ (148), T. sp. ‘Yunman type’ (2), T. sp. SW-2025a (62), Emb. envir. sample (5) | 245 | 7 | 35 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Din, S.; Ali, H.; Panagopoulos, T.; Alam, J.; Malik, S.; Sher, H. Developing High-Efficiency PCR Mini-Barcoding to Enforce Conservation Efforts Against Illegal Trade and Habitat Loss of Endangered Taxus L. in the Himalayas. Conservation 2025, 5, 62. https://doi.org/10.3390/conservation5040062
Din S, Ali H, Panagopoulos T, Alam J, Malik S, Sher H. Developing High-Efficiency PCR Mini-Barcoding to Enforce Conservation Efforts Against Illegal Trade and Habitat Loss of Endangered Taxus L. in the Himalayas. Conservation. 2025; 5(4):62. https://doi.org/10.3390/conservation5040062
Chicago/Turabian StyleDin, Salahud, Haidar Ali, Thomas Panagopoulos, Jan Alam, Saira Malik, and Hassan Sher. 2025. "Developing High-Efficiency PCR Mini-Barcoding to Enforce Conservation Efforts Against Illegal Trade and Habitat Loss of Endangered Taxus L. in the Himalayas" Conservation 5, no. 4: 62. https://doi.org/10.3390/conservation5040062
APA StyleDin, S., Ali, H., Panagopoulos, T., Alam, J., Malik, S., & Sher, H. (2025). Developing High-Efficiency PCR Mini-Barcoding to Enforce Conservation Efforts Against Illegal Trade and Habitat Loss of Endangered Taxus L. in the Himalayas. Conservation, 5(4), 62. https://doi.org/10.3390/conservation5040062







